Abstract
Lactate dehydrogenase (LDH) is present in the amitochondriate parasitic protist Trichomonas vaginalis and some but not all other trichomonad species. The derived amino acid sequence of T. vaginalis LDH (TvLDH) was found to be more closely related to the cytosolic malate dehydrogenase (MDH) of the same species than to any other LDH. A key difference between the two T. vaginalis sequences was that Arg91 of MDH, known to be important in coordinating the C-4 carboxyl of oxalacetate/malate, was replaced by Leu91 in LDH. The change Leu91Arg by site-directed mutagenesis converted TvLDH into an MDH. The reverse single amino acid change Arg91Leu in TvMDH, however, gave a product with no measurable LDH activity. Phylogenetic reconstructions indicate that TvLDH arose from an MDH relatively recently.
The 2-ketoacid:NAD(P) oxidoreductases, malate dehydrogenase (MDH) and lactate dehydrogenase (LDH) comprise a broad enzyme family present in all three domains of life (1). In eukaryotic cells, LDH is localized in the cytosol whereas MDHs are present in the cytosol, mitochondria, chloroplasts, and peroxisomes (2). The tertiary structure of the enzymes is highly conserved within the MDH/LDH family (1, 3–5). The substrates of these enzymes are organic acids with one (pyruvate/lactate) or two (oxalacetate/malate) carboxyl groups. Most enzymes of this family have high substrate specificity for either lactate or malate, although some less-specific variants are also known; for example, in Mycoplasma genitalium, the same protein is believed to have both MDH and LDH activity (6).
Extensive kinetic sequencing and site-directed mutagenesis studies led to a solid understanding of the catalytic mechanism (3, 7). The residues involved in the enzyme reactions that are responsible for the respective specificities of MDH and LDH have been determined (3, 8, 9). In the substrate-binding cleft, MDHs have a positively charged extra arginine residue that forms hydrogen bonds with the second carboxyl group of the dicarboxylic acid substrate (3, 8). In most LDHs, the same position is occupied by an uncharged glutamine residue (3). Site-directed mutagenesis studies have shown that enzymes with LDH specificity can be easily converted to MDH by mutating this glutamine to arginine (1, 3, 10). A reverse conversion of MDH into LDH has been achieved by replacing the arginine with glutamine in the MDH of certain organisms (11) but not of others (12).
Behind this structural and functional uniformity lies an extensive diversity in the primary structure of members of the MDH/LDH family, with amino acid identities often <20% in pairwise comparisons (3). Phylogenetic reconstructions show that members of the MDH and LDH families form separate monophyletic lineages that probably arose from an early gene duplication, predating the divergence of the three domains of life (1, 5, 13). The MDH lineage separates into two rather divergent major subfamilies (3). The first contains mitochondrial and peroxisomal enzymes together with most eubacterial homologs (mMDH) and also includes an NAD-specific chloroplast enzyme (14). The second clade comprises eukaryotic NAD-specific cytosolic and NADP-specific chloroplast enzymes with a few eubacterial members [cytosolic MDH (cMDH) and NADP-specific chloroplast MDH (chlMDH)] (1, 3, 5, 15, 16). Archaebacterial and some eubacterial MDHs occupy intermediate phylogenetic positions and are more closely related to the LDH subfamily than to other MDHs (3, 17, 18).
Although the structures and catalytic mechanisms of MDH and LDH are highly similar, even permitting their mutual conversion by experimental means in some cases, so far no LDH has been detected in the two MDH subfamilies (1), suggesting that “… there must be enough of a selective advantage due to supplementary substitutions to prevent a duplicate copy of MDH from replacing a native LDH gene (or vice versa)” (ref. 1, p. 360).
We report here the sequencing, expression, protein structure modeling, and evolutionary analysis of LDH genes from Trichomonas vaginalis, a genitourinary parasite of humans. T. vaginalis belongs to the Parabasala (19), a group of unicellular eukaryotes without typical mitochondria and with intensive glycolytic fermentation by the Embden-Meyerhof-Parnas pathway (20, 21). Lactate, formed by LDH, is a major end product of this process (22–25). This cytosolic enzyme (26) has been characterized (27, 28). The T. vaginalis LDH (TvLDH) sequences established in the present study showed an unexpectedly high similarity to TvMDH (29) and a significant divergence from other LDHs, the functional analogs of TvLDH. Phylogenetic analysis suggested that TvLDH originated from a relatively recent duplication and subsequent modification of a cMDH gene. This protein is an example of recent convergent enzyme evolution and shows that the MDH-LDH barrier had been breached in this direction at least once in the evolution of unicellular eukaryotes.
MATERIALS AND METHODS
Organism.
T. vaginalis NIH-C1 strain (American Type Culture Collection 30001) has been used throughout this study. The cells were cultivated, collected, and washed as described (26). Genomic DNA (gDNA) was extracted by standard methods. The gDNA library in λ-ZapII was obtained from P. J. Johnson (University of California at Los Angeles).
Numbering of Amino Acid Residues.
Numbering of amino acid residues is based on the derived sequence of TvLDH (29). Most authors use the numbering proposed in ref. 30; thus, for comparison, the corresponding numbers also are given, in square brackets.
Cloning and Site-Directed Mutagenesis of LDH and MDH Genes.
A preparation highly enriched in LDH but also with MDH activity was obtained from T. vaginalis homogenates (29). SDS/PAGE revealed two closely spaced protein bands, ≈37 kDa, with the upper band assumed to be TvLDH and the lower one TvMDH. The material was transferred electrophoretically to a nitrocellulose membrane. The bands were excised and subjected to in situ fragmentation. Several fragments were purified and sequenced by automatic Edman degradation.
Based on the peptide sequences obtained from the putative TvLDH band, two PCR primers [sense: codons 58–64, LDHP8 (5′-Y GGN GCN TTY CAR CAY Y-3′]; antisense: codons 295–298, LDHM7 (5′-ACR TGD ATR TGN CCY TC-3′)] were designed. With T. vaginalis gDNA as template, these primers consistently gave a 0.8-kbp PCR product. Several clones were isolated by screening the gDNA library with the use of this PCR product as a hybridization probe. A gene sequence determined earlier (29) was used to isolate a complete copy of the TvMDH gene.
ORFs corresponding to TvLDH and TvMDH were inserted into the expression vector pQE31 (Qiagen, Valencia, CA). This construct codes for six histidine residues adjacent to the amino terminus of the ORF. The constructs were transfected into Escherichia coli (XL1-Blue). The plasmids were isolated, were characterized by restriction fragment analysis and sequencing, and were retransfected into E. coli M15(pREP4).
The Quikchange Site-Directed mutagenesis system (Stratagene) was used to mutate selected codons in the TvLDH and TvMDH genes. The primers were used (i) to change L90 to R in TvLDH1: 5′-GCT TCA ATG CCA CGC AAG CCA GGT CAA GTT CGC-3′ and 5′-GCG AAC TTG ACC TGG CTT GCG TGG CAT TGA AGC-3′; and (ii) to change R90 to L in TvMDH: 5′-C GTT GGC TCA TTC CCA CTT AAG GAT GGC ATG GAC CG-3′ and 5′- CG GTC CAT GCC ATC CTT AAG TGG GAA TGA GCC AAC G-3′ (bold face italics denote the changed nucleotides). The same approach has been used to obtain further single or multiple mutations. The mutant plasmids were transfected into the same host as the wild-type genes, and their sequences were verified.
Molecular Modeling.
Three-dimensional (3D) models of TvLDH1 and TvMDH were built by comparative protein structure modeling with the program modeller-5 (31, 32). The program is freely available to academic researchers at http://guitar.rockefeller.edu. The input consisted of the template structures and the alignment of the target sequence with these structures. The output, obtained without any user intervention, was a 3D model of the target with all nonhydrogen atoms. This model was derived by minimizing violations of many distance and dihedral angle restraints extracted from the template structures. The models calculated in this study passed the tests in the prosaii (33) and procheck (34) programs.
The closest template structure found with the sequence_search command of modeller was Sus scrofa cMDH [Protein Data Bank (PDB), 4mdh], which shared 51 and 43% amino acid identity to the target TvMDH and TvLDH1 sequences, respectively. Unfortunately, the loop in 4mdh (Asp92–Leu100) that forms a considerable part of the active site cleft was not well defined by the crystallographic analysis (4), consistent with a poor prosaii energy profile for the region. Also, conformation of the loop in 4mdh is different from that in other MDH; in particular, one of the two crucial Arg side chains, which presumably interacts with the substrate (7), has an unusual orientation and sticks out of the active site pocket. Thus, the loop was built by using MDH from E. coli (35) (PDB 1emd; residues 76–98), and the remaining parts of the TvLDH and TvMDH sequences were built by using 4mdh (Fig. 1).
Figure 1.
Alignment of the derived amino acid sequences of TvLDH and TvMDH (29) with selected cytosolic-type and chloroplast MDHs, as well as with two mMDH and two LDH sequences. Numbering of residues is based on the TvLDH sequence. Amino terminal extensions were deleted from the sequences of Flaveria trinervia and Zea mays chlMDH and S. scrofa LDH (68, 27, and 14 residues, respectively). Red columns highlight positions with identical residues over all sequences. Green columns highlight positions with identical and conserved residues. A residue position was defined as conserved if at least 7 of the following 10 physicochemical properties were preserved, as defined by amas (39): hydrophobic, polar, small, proline, positive, negative, charged, tiny, aliphatic, and aromatic. The blanks correspond to gaps in the alignment. The arrows underneath the alignment indicate residues that have at least one atom within 5 Å of any substrate atom. The modeled secondary structure of the TvLDH sequence is shown below the alignment: α-helices are marked with cylinders, and β-strands are marked with arrows. The structurally variable active site loop, modeled based on 1emd, is underlined. The plot was created by the program alscript (40). Sequence (GenBank) and protein data bank accession numbers are as follows: (i) cMDH: Arabidopsis thaliana, AC000104; Chlamydia trachomatis, AE001311; Echinococcus granulosus, L08894 (Q04820); G. lamblia, AF076964; Medicago sativa, AF020272; Mesembryanthemum crystallinum, X96539; Mus musculus, P14152; Mycobacterium leprae, U15180; Mycobacterium tuberculosis, AL021006; S. scrofa, A32472 (PDB, 4mdhA); Thermus aquaticus, P10584 (PDB, 1bmdA); T. vaginalis, U38692; Z. mays, AF007581; (ii) chlMDH (NADP): F. trinervia, U22533; Z. mays, P15719; (iii) mMDH: E. coli, U04742 (PDB, 1emd); S. scrofa, 999617 (PDB, 1mld); (iv) LDH: B. stearothermophilus, M14788; S. scrofa, (PDB, 5ldh and 9ldbA). Structures evaluated but not included in the alignment are (i) mMDH: S. scrofa (PDB, 1mdlA); (ii) LDH: Squalus acanthias 1 (PDB, 1ldm), Squalus acanthias 2 (PDB, 3ldh); M. musculus (PDB, 2ldx).
The citrate substrate analog in the 1emd structure was replaced with malate and lactate to obtain the TvMDH and TvLDH models in complex with their substrates, respectively. The models also included the NAD cofactor, inherited from the 4mdh template. The substrates and the cofactor were docked as rigid bodies mimicking the corresponding conformations and positions in the template structures as much as possible. Several restraints, in addition to those automatically derived by modeller, were imposed on the relative orientation of the substrates and the enzymes to incorporate previous knowledge about the substrate–enzyme interactions (4, 7–9). They were upper bound distance restraints of 3.5 Å on the following pairs of atoms: His186:NE1 and Malate:O2, Arg161:NH1 and Malate:O1A, Arg161:NH2 and Malate:O1B. The distance between the catalytic Asp158:OD2 and His186:NE2 also was not allowed to be >3.5 Å.
Phylogenetic Analysis.
The amino acid sequences initially were aligned manually with all known cMDH and selected chlMDH, mMDH, and LDH sequences with the ed program of the must package (36). Database accession numbers are listed in the legend to Fig. 1. The alignment was refined by considering known 3D structures of 2-ketoacid:NAD(P) oxidoreductases; specifically, gaps were placed outside secondary structure segments. Phylogenetic reconstruction of TvLDH, TvMDH, cMDH, and chlMDH sequences was done with a maximum likelihood method (protml) (37). The Jones-Taylor-Thorston model of amino acid substitutions was used (38). Local bootstrap proportions were determined with the rell method (37).
Expression of LDHs and MDHs.
E. coli M15 cells harboring the pQE31 plasmid were inoculated into 6 ml of LB broth containing 100 μg⋅ml−1 ampicillin and 25 μg⋅ml−1 kanamycin. Overnight cultures were transferred into 300 ml of the same medium and were grown at 37°C until an OD value of 0.9 at 600 nm was reached. Isopropyl-β-d-thiogalactopyranoside was added to a final concentration of 200 μM, and the cultures were further grown at 23–25°C for 15 hours. The expressed enzymes were isolated and purified by Ni–nitrilotriacetic acid resin chromatography according to the supplier’s instructions (Qiagen).
Enzyme Assays.
LDH and MDH activities were determined in the direction of keto acid reduction by monitoring the oxidation of NADH at 340 nm at 30°C. The standard reaction mixture contained 150 μM NADH and 250 μM substrate in 50 mM triethanolamine buffer (pH 7.6). In determining the substrate affinities, the keto-acid concentration was varied within the range of 5 μM–9 mM, with NADH kept constant at 150 μM, or the NADH concentration was varied in the range of 30 μM–0.9 mM, with the keto-acid substrate kept constant at the optimal concentration determined for each enzyme (TvLDH, 2 mM pyruvate or 3 mM oxalacetate; TvMDH, 200 μM oxalacetate; TvLDH-L91R, 2 mM pyruvate or 5 mM oxalacetate; TvMDH-R91L, 200 μM oxalacetate). pH optima were determined at optimum NADH and substrate concentrations. Km and Vmax values of the enzymes were estimated from Lineweaver-Burk plots.
RESULTS
Derived Amino Acid Sequences.
Both TvLDH1 and TvLDH2 clones contained ORFs of 999 bp, corresponding to putative translation products of 333 amino acid residues (Fig. 1). The two ORFs shared an amino acid identity of 93.5%. All three peptides obtained from the upper protein band were present in these sequences, showing that the cloned genes were expressed. Southern blots revealed at least four additional copies of TvLDH, which were not sequenced (data not shown). The sequences of peptides from the lower protein band were present in the published sequence of T. vaginalis cMDH (29), confirming the identity of this band (results not shown).
The derived TvLDH1 and TvLDH2 amino acid sequences were, respectively, 49.2 and 49.5% identical (323 shared amino acid positions) to the TvMDH sequence. A crucial difference was noted in the mobile loop (residues 90–105), where Leu occupied the position of the critical Arg91 of TvMDH (Fig. 1). This residue also was seen in one of the peptides, showing that it was not a sequencing artifact.
Alignment and Phylogenetic Reconstruction.
Alignment of TvLDH with NAD-specific cMDH sequences and NADP-specific chlMDH sequences required the insertion of only a few gaps (Fig. 1). Only 29% of all residues were unique to TvLDH1 and TvLDH2. The three T. vaginalis dehydrogenases shared single residue gaps after positions 31[47] and 212[218] and had a proline inserted at position 272[275A]. A four-residue insertion was shared by all eukaryotic cMDH sequences at positions 201–204[209A-210B] whereas at positions 276[278A] and 292[294], the T. vaginalis sequences shared a one-residue insertion with some of the other sequences. The overall sequence identity of TvLDH to the 14 evaluated cMDH and chlMDH sequences was 33.5–44.5%. No other LDH was present in this enzyme subfamily. Alignment of TvLDH with mMDH sequences required the introduction of longer gaps in some regions. Typical LDH sequences from other organisms proved even more distant.
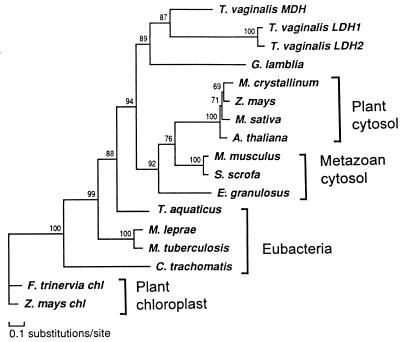
The alignment of the sequences in the cMDH and chlMDH group was analyzed with a maximum likelihood method. The reconstruction suggested that TvLDH and TvMDH shared a most recent common ancestor (Fig. 2). The high bootstrap proportion of the corresponding node suggests the robustness of this relationship. The only other available protist cMDH, that of Giardia lamblia (15), was the closest outgroup to the T. vaginalis sequences. The remaining sequences were distributed in several other groups that corresponded to defined biological entities: eubacteria, plant cytosolic enzymes, vertebrate cytosolic enzymes, and chloroplast enzymes. The only unexpected, but poorly supported, position was shown by cMDH of the cestode, Echinococcus granulosus, which was an outlier to the common ancestor of plant and vertebrate enzymes. The relationships among the latter were poorly defined.
Figure 2.
Phylogenetic reconstruction of cytosolic-type malate dehydrogenases with the maximum likelyhood method protml (37), showing the nested position of T. vaginalis lactate dehydrogenase within this enzyme subfamily. The tree was arbitrarily rooted with two chloroplast NADP-MDH sequences. Local bootstrap proportions are shown at the individual nodes.
Molecular Modeling.
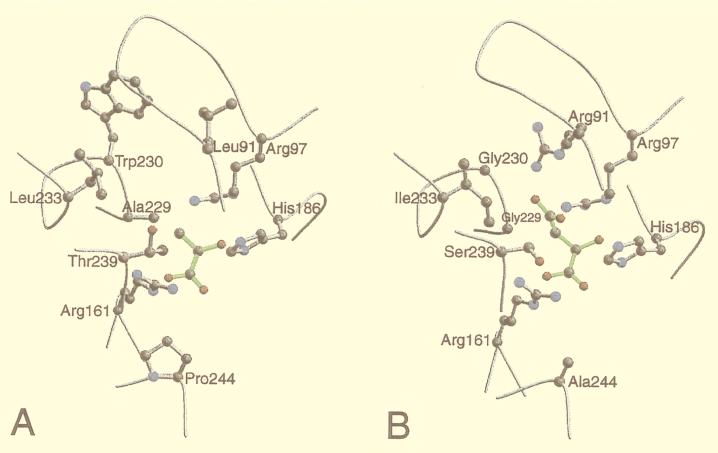
Several insights into the reasons for the differences in substrate specificity of the highly similar TvLDH and TvMDH were obtained by comparing the active site environments in their 3D models (Fig. 3). The most notable difference was the switch from Arg91[102] in TvMDH to Leu91 in TvLDH. This change is explained by the role of Arg91 that coordinates the carboxyl group of the malate substrate in MDHs (7, 8). A further significant difference was the change from Gly230[237] in TvMDH to Trp230 in TvLDH. This difference also was rationalized by the 3D models: A simultaneous presence of both Arg91 and Trp230 would create a steric clash. Thus, a change of Trp230 in the TvMDH structure to a smaller amino acid in TvLDH is necessary if similar backbone conformation is to be maintained. This feature is shared with other dehydrogenases of known structure. At position 230[237], the LDH structures generally have a large aromatic residue (Trp or Tyr) whereas the MDH structures always have Ser. In the 5-Å environment, there are two additional differences between TvMDH and TvLDH: Gly229Ala[236] and Ser239Thr[246]. These residues may play an important role in substrate recognition because they point directly to the variable parts of the two substrates. These amino acid mutations also can be understood with the aid of the models because they result in a more hydrophobic environment for the more hydrophobic lactate/pyruvate substrate.
Figure 3.
Plots of the active site environments in the 3D models of T. vaginalis lactate (A) and malate (B) dehydrogenases. The active site environment includes all of the residues that have at least one atom within 5 Å of at least one substrate atom. The backbone is traced in gray. The conserved Arg161 and His186 residues, presumed to be crucial in catalysis, and the residues that differ in type between the two sequences are shown in the ball-and-stick representation. The substrate molecules are shown in green. The plots were drawn with the programs molscript (41) and raster3d (42).
Expression Constructs.
Sequencing the constructs confirmed the identity of the inserted ORFs with the wild-type genes Tvldh and Tvmdh. The constructs were modified by introducing a heptamer His tag immediately downstream of the initiator ATG. Single nucleotide changes produced the desired amino acid replacements Leu91Arg in TvLDH and Arg91Leu in TvMDH. Sequencing of these mutated constructs verified that the planned replacements indeed occurred.
Expression and Biochemical Characterization.
Total extracts of induced bacteria that carried the various constructs contained a high amount of the corresponding heterologous protein. The histidine-tagged recombinant TvLDH1 and TvMDH were purified by nickel-agarose affinity chromatography. Each purified enzyme corresponded to a single band on SDS/PAGE, with molecular masses of 37.5 kDa for TvLDH1 and 36.5 kDa for TvMDH, in good agreement with the sizes estimated from the derived amino acid sequences. The products had the expected substrate specificities (Table 1). The wild-type TvLDH also showed significant MDH activity with oxalacetate as substrate. The results confirm the identification of the two ORFs as Tvldh and Tvmdh. The single-point mutation of TvLDH, Leu91Arg, made it active with the oxalacetate substrate, although it markedly decreased its activity toward pyruvate. TvMDH, in contrast, exhibited no measurable activity with pyruvate. The reverse mutant, Arg91Leu, remained a less efficient MDH and did not acquire detectable activity with pyruvate (Table 1). Attempts were made to make TvMDH active against pyruvate by introducing additional point mutations, as suggested by comparison of the 3D models of the active site clefts (Fig. 3): Gly230Trp, Arg91Leu/Gly229Ala/Gly230Trp, Arg91Leu/Gly230Trp/Ser239Thr, and Arg91Leu/Gly229Ala/Gly230Try/Ser239Thr. None of the mutants gained any LDH activity. MDH activity was somewhat increased in the Gly230Trp mutant whereas all others lost this activity (data not shown).
Table 1.
Kinetic characterization of the enzymatic activities with oxalacetate and pyruvate of T. vaginalis cell extract and genes expressed in E. coli
| Enzyme sample | Activity
|
|||||
|---|---|---|---|---|---|---|
| Oxalacetate
|
Pyruvate
|
|||||
| pH | Km* | Vmax† | pH | Km* | Vmax† | |
| Homogenate | 7.6 | 67 | 23 | 7.2 | 175 | 5 |
| TvLDH1 | 5.9 | 40 | 31 | 6.1 | 45 | 400 |
| Leu91Arg | 6.3 | 425 | 1,430 | 6.1 | 85 | 123 |
| TvMDH | 7.6 | 33 | 3,076 | ND | ND | ND |
| Arg91Leu | 6.5 | 222 | 400 | ND | ND | ND |
ND, not detectable.
μM.
μmol⋅min−1⋅mg protein−1.
DISCUSSION
Our studies revealed that LDH (27, 28) of the amitochondriate eukaryote T. vaginalis is exceptional among its functional analogs. The sequence of this enzyme is more closely related to the cMDH of the same species and to other members of the cMDH subfamily than to any other sequenced LDH. Phylogenetic reconstruction clearly included it in the cMDH clade and placed it outside of both the mMDH group and the family containing all other known LDH molecules. The close relationship of TvLDH to cMDHs suggests that the gene duplication leading to this enzyme was independent from the primary separation of the MDH and LDH families of 2-ketoacid:NAD(P) oxidoreductases (1).
The high activity of the expressed TvLDH1 on oxalacetate is an unusual property for an LDH. For instance, Bacillus stearothermophilus LDH shows a 1,000-fold higher catalytic efficiency toward pyruvate than oxalacetate (10). This suggests that the second, independent invention of an LDH by convergent evolution, as seen in TvLDH, did not achieve the same substrate selectivity as did the first, more ancient divergence. A shift of specificity in TvLDH toward oxalacetate could be achieved easily by a single Leu91Arg substitution, not a surprising result in view of the effect of the similar Gln102Arg substitution produced by site-directed mutagenesis in other organisms (1, 10). That the reverse conversion could not be achieved by either a single Arg91Leu substitution or by multiple substitutions corresponding to differences in the active site clefts of the two enzymes probably reflects the weaker binding of the LDH substrate, pyruvate (7), and shows that other differences between the sequences of TvLDH and TvMDh are necessary for the switch in specificity.
The high similarity of LDH and MDH (29) in T. vaginalis indicates that this conversion occurred relatively recently, possibly in the parabasalid lineage. The nested position of Tvldh among the cMDHs suggests that the change occurred in the MDH to LDH direction. The available limited taxon sampling, especially the lack of published cMDH sequences for other protists, with the exception of G. lamblia MDH (15), prevented us from testing this evolutionary scenario. However, such close pairing of MDH and LDH molecules has not been noted in other organisms. In fact, phylogenetic reconstructions suggest that LDH and MDH separated in an early gene duplication, followed by functional diversification and divergent evolution (1, 5, 13).
The present results are of interest also in regard to the comparative biochemistry of the Parabasala. The few species of this group so far explored reveal significant metabolic differences. Lactate is formed also by other Trichomonas species (21) but not by a less closely related species, Tritrichomonas foetus from cattle, which has no LDH and produces succinate as a major reduced end product (43). These species belong to the two major suborders of the order Trichomonadida, Trichomonadina and Tritrichomonadina, respectively (19). A study of additional representatives of these groups will be necessary to test whether these metabolic differences are characteristic of the two suborders and, of more interest, to seek an answer to the question of whether LDH arose after the separation of the suborders or has been present already in their common ancestor and was lost in the Tritrichomonadina lineage. Enzymes of the monophyletic LDH subfamily, derived from an early gene duplication (1, 5, 13), are used widely by other organisms to reoxidize NADH and keep glycolysis flowing. Our data indicate that essentially the same mechanism was discovered independently and more recently by an ancestor of Trichomonas species.
Acknowledgments
Earlier studies by Dr. Anton Markoš (26, 29) in our laboratory provided a starting point of this research. His contributions are gratefully acknowledged. We thank Drs. Patricia J. Johnson for the T. vaginalis gDNA library, Sally Harmich for sharing her results, Hervé Philippe for the must package, and Lidya B. Sánchez and Jennifer A. Lee for help and advice. Oligonucleotide synthesis and peptide and DNA sequencing were performed by the Nucleic Acid Sequencing Facility of the Rockefeller University. A.F. is a Burroughs Wellcome Fellow. A.Š. is a Sinsheimer Scholar and an Alfred P. Sloan Research Fellow. This research was supported by U.S. Public Health Service National Institutes of Health Grants AI11942 to M.M. and GM 54762 to A.Š. as well as National Science Foundation Grant BIR-9601845 to A.Š.
ABBREVIATIONS
- cMDH
cytosolic-type malate dehydrogenase
- chlMDH
NADP-dependent chloroplast malate dehydrogenase
- gDNA
genomic DNA
- LDH
lactate dehydrogenase
- MDH
malate dehydrogenase
- mMDH
mitochondrial-type MDH
- PDB
Protein Data Bank
- 3D
three-dimensional
- TvLDH
Trichomonas vaginalis lactate dehydrogenase
- TvMDH
Trichomonas vaginalis malate dehydrogenase
Footnotes
References
- 1.Golding G B, Dean A M. Mol Biol Evol. 1998;15:355–369. doi: 10.1093/oxfordjournals.molbev.a025932. [DOI] [PubMed] [Google Scholar]
- 2.Gietl C. Biochim Biophys Acta. 1992;1100:217–234. doi: 10.1016/0167-4838(92)90476-t. [DOI] [PubMed] [Google Scholar]
- 3.Goward C R, Nicholls D J. Protein Sci. 1994;3:1883–1888. doi: 10.1002/pro.5560031027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Birktoft J J, Rhodes G, Banaszak L J. Biochemistry. 1989;28:6065–6081. doi: 10.1021/bi00440a051. [DOI] [PubMed] [Google Scholar]
- 5.McAlister-Henn L. Trends Biochem Sci. 1988;13:178–181. doi: 10.1016/0968-0004(88)90146-6. [DOI] [PubMed] [Google Scholar]
- 6.Cordwell S J, Basseal D J, Pollack J D, Humphrey-Smith J. Gene. 1997;195:113–120. doi: 10.1016/s0378-1119(97)00063-2. [DOI] [PubMed] [Google Scholar]
- 7.Cunningham M A, Ho L L, Nguyen D T, Gillilan R E, Bash P A. Biochemistry. 1997;36:4800–4816. doi: 10.1021/bi962734n. [DOI] [PubMed] [Google Scholar]
- 8.Nicholls D J, Miller J, Scawen M D, Clarke A R, Holbrook J J, Atkinson T, Goward C R. Biochem Biophys Res Commun. 1992;189:1057–1062. doi: 10.1016/0006-291x(92)92311-k. [DOI] [PubMed] [Google Scholar]
- 9.Clarke A R, Wigley D B, Chia W N, Barstow D, Atkinson T, Holbrook J J. Nature (London) 1986;324:699–702. doi: 10.1038/324699a0. [DOI] [PubMed] [Google Scholar]
- 10.Wilks H M, Hart K W, Feeney R, Dunn C R, Muirhead H, Chia W N, Barstow D A, Atkinson T, Clarke A R, Holbrook J J. Science. 1988;242:1541–1544. doi: 10.1126/science.3201242. [DOI] [PubMed] [Google Scholar]
- 11.Cendrin F, Chroboczek J, Zaccai G, Eisenberg H, Mevarech M. Biochemistry. 1993;32:4308–4313. doi: 10.1021/bi00067a020. [DOI] [PubMed] [Google Scholar]
- 12.Boernke W E, Sanville Millard C, Wilkins Stevens P, Kakar S N, Stevens F J, Donnelly M L. Arch Biochem Biophys. 1995;322:43–52. doi: 10.1006/abbi.1995.1434. [DOI] [PubMed] [Google Scholar]
- 13.Iwabe N, Kuma K-i, Kishino H, Hasegawa M, Miyata T. J Mol Evol. 1990;31:205–210. doi: 10.1007/BF02109497. [DOI] [PubMed] [Google Scholar]
- 14.Berkemeyer M, Scheibe R, Ocheretina O. J Biol Chem. 1998;273:27927–27933. doi: 10.1074/jbc.273.43.27927. [DOI] [PubMed] [Google Scholar]
- 15.Roger, A. J., Morrison, H. G. & Sogin, M. L. (1999) J. Mol. Evol., in press. [DOI] [PubMed]
- 16.Ocheretina O, Scheibe R. Gene. 1997;199:145–148. doi: 10.1016/s0378-1119(97)00361-2. [DOI] [PubMed] [Google Scholar]
- 17.Synstad B, Emmerhoff O, Sirevåg R. Arch Microbiol. 1996;165:346–353. doi: 10.1007/s002030050337. [DOI] [PubMed] [Google Scholar]
- 18.Charnock C. J Bacteriol. 1997;179:4066–4070. doi: 10.1128/jb.179.12.4066-4070.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brugerolle G. J Eukaryotic Microbiol. 1993;40:616–618. doi: 10.1111/j.1550-7408.1993.tb04935.x. [DOI] [PubMed] [Google Scholar]
- 20.Coombs G H, Müller M. In: Biochemistry and Molecular Biology of Parasites. Marr J J, Müller M, editors. London: Academic; 1995. pp. 33–47. [Google Scholar]
- 21.Müller M. Annu Rev Microbiol. 1988;42:465–488. doi: 10.1146/annurev.mi.42.100188.002341. [DOI] [PubMed] [Google Scholar]
- 22.Mack S R, Müller M. Comp Biochem Physiol B Comp Biochem. 1980;67:213–216. [Google Scholar]
- 23.Müller M, Gorrell T E. Antimicrob Agents Chemother. 1983;24:667–673. doi: 10.1128/aac.24.5.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.ter Kuile B H. Microbiology. 1996;142:3337–3345. doi: 10.1099/13500872-142-12-3337. [DOI] [PubMed] [Google Scholar]
- 25.Kulda J, Tachezy J, Čerkasovová A. J Eukaryotic Microbiol. 1993;40:262–269. doi: 10.1111/j.1550-7408.1993.tb04915.x. [DOI] [PubMed] [Google Scholar]
- 26.Markoš A, Miretsky A, Müller M. J Mol Evol. 1993;37:631–643. doi: 10.1007/BF00182749. [DOI] [PubMed] [Google Scholar]
- 27.Baernstein H D. J Parasitol. 1959;45:491–498. [PubMed] [Google Scholar]
- 28.Harmych S E, Sidawy E, Komuniecki R. Comp Biochem Physiol B Biochem Mol Biol. 1996;115:405–409. [Google Scholar]
- 29.Markoš A, Morris A, Rozario C, Müller M. FEMS Microbiol Lett. 1996;135:259–264. doi: 10.1111/j.1574-6968.1996.tb07998.x. [DOI] [PubMed] [Google Scholar]
- 30.Eventoff W, Rossman M G, Taylor S S, Torff H-J, Meyer H, Keil W, Kiltz H H. Proc Natl Acad Sci USA. 1977;74:2677–2681. doi: 10.1073/pnas.74.7.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Šali A, Blundell T L. J Mol Biol. 1998;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 32.Sánchez, R. & Šali, A. (1997) Proteins, Suppl. 1, 50–58. [DOI] [PubMed]
- 33.Sippl M J. Proteins. 1993;17:355–362. doi: 10.1002/prot.340170404. [DOI] [PubMed] [Google Scholar]
- 34.Laskowski R A, McArthur M W, Moss D S, Thornton J M. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 35.Hall M D, Banaszak L J. J Mol Biol. 1993;232:213–222. doi: 10.1006/jmbi.1993.1377. [DOI] [PubMed] [Google Scholar]
- 36.Philippe H. Nucleic Acids Res. 1993;21:5264–5272. doi: 10.1093/nar/21.22.5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Adachi J, Hasegawa M. molphy 2.3: Programs for Molecular Phylogenetics Based on Maximum Likelihood. Tokyo: Institute of Statistical Mathematics; 1996. pp. 1–150. [Google Scholar]
- 38.Jones D T, Taylor W R, Thornton J M. Comput Appl Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 39.Livingstone C D, Barton G J. Comput Appl Biosci. 1993;6:745–756. doi: 10.1093/bioinformatics/9.6.745. [DOI] [PubMed] [Google Scholar]
- 40.Barton G J. Protein Eng. 1993;6:37–40. doi: 10.1093/protein/6.1.37. [DOI] [PubMed] [Google Scholar]
- 41.Kraulis P. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 42.Merritt E A, Murphy M E P. Acta Crystallogr D. 1994;50:869–873. doi: 10.1107/S0907444994006396. [DOI] [PubMed] [Google Scholar]
- 43.Ryley J F. Biochem J. 1955;59:361–369. doi: 10.1042/bj0590361. [DOI] [PMC free article] [PubMed] [Google Scholar]