Abstract
Asobara tabida wasps are fly endoparasitoids that naturally harbor three Wolbachia strains, which induce cytoplasmic incompatibility and control oogenesis. To investigate whether other bacteria play a role in wasp biology, we surveyed the bacterial communities of wild A. tabida populations originating from different regions of France and of laboratory colonies using PCR-denaturing gradient gel electrophoresis and culture methods. Proteobacteria and Firmicutes were found to be the main phyla represented in these populations. Among these were several cultured and uncultured representatives of the genera Acetobacter, Acidomonas, Bacillus, Brevibacillus, Duganella, Herbaspirillum, Pseudomonas, Staphylococcus, and Streptococcus. In addition to Wolbachia, wild individuals harbored Rickettsia, which tended to be lost when insects were reared in the laboratory. The antibiotic treatment used to generate wasp sublines singly infected with Wolbachia also affected the overall bacterial composition, with most fingerprint sequences being characteristic of the family Enterobacteriaceae. We also screened for potentially heritable endosymbionts by PCR and fluorescence in situ hybridization in stable laboratory lines, with only Wolbachia being consistently found in wasp ovaries.
Bacteria associated with insects play a crucial role in host development, survival, and reproduction (13). Many insects harbor bacterial endosymbionts, which establish close relationships, like the mutualistic interaction between aphids and their primary endosymbiont of the genus Buchnera; the bacterium uses the host as a habitat to which it supplies essential amino acids, facilitating insect growth when the diet of plant phloem sap is insufficient (5, 23). Aphids host many other nonessential bacteria as secondary or facultative symbionts. However, aphids with secondary symbionts can gain a fitness advantage in terms of diet, regimen plant host range, heat tolerance, or resistance to pathogens and parasitoids (reviewed in references 45 and 48). Multiple infections are costly to hosts and are perhaps maintained because of the benefits they confer. Recently, Wolbachia has been shown to protect the host Drosophila melanogaster from viral damage (37). However, investigating the evolutionary significance of interspecific symbioses in bacterial communities in invertebrates is challenging in that the majority of bacteria are not yet cultivable outside host cells.
Here we analyzed the main bacterial populations of Asobara tabida (Hymenoptera: Braconidae), endoparasitoids of Drosophila species and related genera (14). Usually, members of A. tabida are naturally multiply infected with bacteria of the genus Wolbachia, obligate intracellular Alphaproteobacteria of the order Rickettsiales (2, 25, 51), found in association with numerous arthropods, mainly insects, and certain nematodes, where they are mostly vertically transmitted from mother to progeny (74). The interaction between Wolbachia spp. and their hosts is very complex and ranges from parasitism to mutualism. In filarial nematodes, Wolbachia organisms are required in the host's biology (4), but Wolbachia spp. are mostly parasites that affect arthropod reproduction, such as by inducing parthenogenesis in some parasitoid wasps (63), feminizing genetic males in isopods (9), and inducing male killing and cytoplasmic incompatibility in many insects (39, 64). The wasp A. tabida harbors three Wolbachia strains; strains wAtab1 and wAtab2 induce cytoplasmic incompatibility, whereas wAtab3 is necessary for the completion of oogenesis (19, 20, 73). The involvement of Wolbachia strains in wasp reproduction was discovered when wasps were treated with antibiotics to generate lines harboring subsets of Wolbachia or aposymbiotic lines (19, 21). Only oogenesis was affected by curing Wolbachia wAtab3; other traits, such as insect size, weight, locomotion, and behavior, were unchanged (19). While antibiotic treatment has been used to determine biological roles of symbionts in this way, their effect on the overall composition of bacterial populations has not been investigated.
To investigate the potential role of bacterial endosymbionts in the biology of A. tabida, we studied the bacterial communities in insect populations originating from different regions of France using culture and nonculture methods and fluorescence in situ hybridization (FISH). We also examined whether antibiotherapy to generate lines with a subset of Wolbachia strains altered the composition or density of the bacterial communities.
MATERIALS AND METHODS
Insect collection and maintenance.
Wasps of the genus Asobara (Hymenoptera: Braconidae) are endoparasitoids of Drosophila. Female wasps lay their eggs in fly larvae, where the parasitic wasp larvae feed and develop (14). A. tabida individuals were collected from six sites in France—Condrieu (Co), Igé (Ig), Sainte Foy-Lès-Lyon (Sfl), Saint Laurent d'Agny (Sla), and Verpillère (Vp)—and reared in the laboratory (except those from Vp). Four lines originating from Montpellier (Mo), Pierrefeu (Pi), Sfl, and Sla that had been maintained in an insectarium for six months to four years were also used. Co, Sfl, Sla, and Vp are located in Rhône-Alpes region (southwest of France), around 40 km from the city of Lyon. Igé is situated in region of Bourgogne, 80 km from Lyon. Mo (304 km from Lyon) and Pi (500 km from Lyon) are in Languedoc-Roussillon and Provence-Alpes Côte d'Azur, respectively, both in the south.
The wasps were reared on a Wolbachia-free Drosophila melanogaster strain, originating from Sfl, at 20°C with a 12-h-12-h light-dark cycle and 70% relative humidity. Members of A. tabida are usually naturally infected with three Wolbachia strains, named wAtab1, wAtab2, and wAtab3 (73). An A. tabida line, Pi(3), infected with only wAtab3 was obtained from Pi(123), a triply infected line, using a moderate antibiotic treatment (19). After antibiotherapy, sublines were reared without antibiotics for seven generations (new line) or for four years (old line) before analysis.
Isolation of bacteria from insect ovaries.
Emerging A. tabida adults were anesthetized at 4°C, surface sterilized in 70% ethanol for 5 min, and rinsed five times in sterile water. Ovaries from anesthetized females were dissected with needles in SPG buffer (218 mM sucrose, 7.2 mM K2HPO4, 3.8 mM KH2PO4, 4.9 mM l-glutamate; pH 7.2) under a binocular microscope and were surface sterilized as described above. A total of 100 ovaries were crushed in 50 μl IPL41 medium (Gibco, Invitrogen, France). Directly plating the homogenate gave rise to only a few very small colonies that did not grow further either aerobically or anaerobically. To enrich the homogenate, it was diluted and inoculated (final optical density at 540 nm of 0.01) into three Venoject tubes containing 3 ml of different media: IPL41 (Gibco), modified Luria Bertani (10 g liter−1 Bacto-tryptone, 5 g liter−1 yeast extract, 5 g liter−1 NaCl), and PYC (5 g liter−1 peptone, 3 g liter−1 yeast extract, 6 mM CaCl2·2H2O; pH 7.0). After five days' incubation at 26°C without agitation, cultures were reinoculated in both 5-ml liquid cultures and on agar plates of the respective medium. Single colonies were streaked out to check for purity and then screened using standard bacteriological techniques to determine the dilution factor and colony morphology (size, shape, margin, color, opacity, and consistency). Purified isolates were cultured in liquid medium and stored in 25% glycerol at −80°C until use.
Genomic and plasmid DNA extractions.
Genomic DNA was isolated using a DNeasy tissue kit (Qiagen, Courtaboeuf, France) with the following modifications. Pools of 5 to 15 emerging adults or 100 dissected ovaries, prepared and disinfected as described above, were homogenized in 180 μl of DNeasy ATL buffer. The mixture was treated with 2 mg ml−1 lysozyme (Eurogentec, Angers, France) for 3 h at 37°C and then with proteinase K (Qiagen) for 12 h at 56°C. The mixture was centrifuged twice at 12,000 × g for 1 min. The supernatant was transferred to a new tube and treated with RNase A (final concentration, 1 mg ml−1) for 2 min at room temperature. Then 200 μl of DNeasy AL buffer was added and incubated for 10 min at 70°C. After the addition of 200 μl absolute ethanol, the sample was vortexed and pipetted into the DNeasy mini-spin column. DNA was eluted with 30 μl of TE buffer (10 mM Tris [pH 7.5], 0.1 mM EDTA [pH 8]), quantified using an ND-1000 spectrophotometer (NanoDrop Technologies, Inc.), and stored at −20°C until use.
Genomic and plasmid DNA (recombinant vector [TOPO 2.1] containing cloned DNA fragments for sequencing) were extracted from bacteria using Nucleospin tissue or plasmid kits (Macherey-Nagel, Düren, Germany) following the manufacturer's instructions.
Diagnostic and quantitative PCR.
General PCR conditions are given in Table 1. A PCR specific for Wolbachia using primers (Table 1) targeting 16S rRNA genes and wsp loci was as previously reported (44). A LightCycler LC480 apparatus (Roche) was used for real-time quantitative PCR. The 20-μl reaction mixture contained 1× LightCycler DNA master SYBR green I (Roche), each primer at 300 nM, and 20 ng of template DNA. Amplification conditions were 10 min at 95°C; 40 cycles of 15 s at 95°C and 1 min at 60°C or 63°C for wsp and rrs, respectively; and 72°C for 30 s. Standard curves were drawn for amplification from a DNA plasmid containing wsp and 16S rRNA gene fragments, respectively (Table 1).
TABLE 1.
Primers used in this study
| Sample | Gene | Primer | Primer sequence (5′-3′) | Amplicon size (bp) and Tm (°C) | Reference |
|---|---|---|---|---|---|
| Eubacteria | rrs + IGa | REUB | 5′ GCCAAGGCATCCACC 3′ | Variable; 55 | 29 |
| rrs | pA | 5′ AGAGTTTGATCCTGGCTCAG 3′ | 1,500; 55 | 12 | |
| pH | 5′ AAGGAGGTGATCCAGCCGCA 3′ | ||||
| V3 region rrs | 16S (V3) 338F | 5′GCCGCCCGCCGCGCGCGGCGGGCGGGGCGGGGGCACGGGGGGACTCCTACGGGAGGCAGCAG 3′ | Variable; 55 | 49 | |
| 16S (V3) 520R | 5′ ATTACCGCGGCTGCTGG 3′ | ||||
| Wolbachia | rrs | 99F | 5′ TTGTAGCCTGCTATGGTATAACT 3′ | 864; 52 | 51 |
| 994R | 5′ GAATAGGTATGATTTTCATGT 3′ | ||||
| wsp | 81F | 5′ TGGTCCAATAAGTGTATGAAGAAAC 3′ | 600; 55 | 10 | |
| 165F | 5′ AAAGGGGACTGATGATGT 3′ | 526; 52 | 79 | ||
| 172F | 5′ TGGTCCAATAAGTGATGAAGAAAC 3′ | 519; 52 | 79 | ||
| Aso3F | 5′ AAAAATTAAACGCTACTCCA 3′ | 515; 52 | 73 | ||
| 691R | 5′ AAAGGGGACTGATGATGT 3′ | 79 | |||
| Bacillus | rrs | BK-1F | 5′ TCACAAGGCRACGATGG 3′ | 1,100; 58 | 77 |
| BK-1R | 5′ CGTATTCACCGCGGCATG 3′ | ||||
| Brevibacillus | rrs | F-1 | 5′ GCYTAAYACATGCAAGTCGARCG 3′ | 318; 68 | 32 |
| R-1 | 5′ ACTGCTGCCTCCCGTAGGAGT 3′ | ||||
| Pseudomonas | rrs | Ps For | 5′ GGTCTGAGAGGATGATCAGT 3′ | 990; 52 | 76 |
| Ps Rev | 5′ TTAGCTCCACCTCGCGGC 3′ | ||||
| Rickettsia | rrs | Rb-F | 5′ GCTCAGAACGAACGCTATC 3′ | 963; 58 | 33 |
| Rb-R | 5′ GAAGGAAAGCATCTCTGC 3′ | ||||
| Staphylococcus | rrs | 16S3 up | 5′ ATGCAAGTCGAGCGAAC 3′ | 265; 57 | 61 |
| 16S3 down | 5′ TGTCTCAGTTCCAGTGTGGC 3′ | ||||
| TOPO 2.1 | M13F | 5′ GTAAAACGACGGCCAG 3′ | Variable; variable | ||
| M13R | 5′ CAGGAAACAGCTATGAC 3′ | ||||
| pQuantAlb | wsp A group | QAtdir1 | 5′ GRGTTGATRTTGAAGGRS 3′ | 264; 60 | This study |
| QArev2 | 5′ CACCAGCTTTTACTTGACC 3′ | 68 | |||
| pQuantAlb16S | rrs fragment | 519F | 5′ CAGCMGCCGCGGTAANWC 3′ | 407; 63 | 65 |
| 907R | 5′ CCGTCAATTCMTTTRAGTT 3′ |
IG, intergenic gene between rrs and the 23S gene.
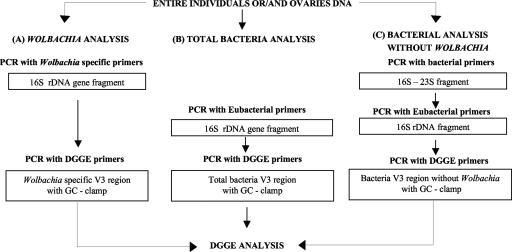
To analyze the total bacterial populations while excluding Wolbachia amplicons, we first targeted the 16S-23S region and then 16S alone, as all Wolbachia strains identified so far harbor unlinked 16S and 23S genes (7) (Fig. 1). This strategy also allowed us to disregard other bacteria, such as Buchnera, having a similar split rRNA gene organization (58). For this, 16S and 23S regions were amplified with standard universal primers (Table 1) as follows: 95°C for 3 min; 35 cycles of 94°C for 30 s, 55°C for 40 s, and 72°C for 3 min; and 72°C for 10 min. The 16S rRNA gene was amplified with the same conditions except that the final elongation step was for 1 min 30 s. Amplification reaction mixtures (25 μl) contained the genomic DNA template (60 ng) in 1× polymerase reaction buffer (Clontech), 200 μM each deoxynucleoside triphosphate, 200 nM each primer, and 1 U Titanium Taq polymerase (Clontech) and used a T-gradient thermocycler (Biometra). PCR products were electrophoresed on 0.8% agarose gel, stained with ethidium bromide, and photographed under UV with the Fisher Bioblock scientific system (Fisher, Ilkirch, France).
FIG. 1.
Schematic view of PCR-DGGE strategies used to analyze the composition of bacterial communities in whole individuals and ovaries of A. tabida. (A and B) Two-step nested PCR strategy for amplification of the rrs V3 region from Wolbachia (A) and the total bacterial community (B). (C) Three-step nested-PCR strategy to amplify the rrs V3 region from the bacterial community while excluding Wolbachia amplicons.
For DGGE, 16S rRNA gene V3 regions were amplified in 50-μl reaction mixtures containing 1 μl of rrs amplicons in 1× polymerase reaction buffer (Invitrogen), 1.5 mM MgCl2, 250 μM each deoxynucleoside triphosphate, 0.5 μM each primer, and 0.4 U of Taq polymerase (Invitrogen). Amplification conditions were 94°C for 2 min; 7 cycles of 94°C for 30 S, 55°C for 30 s, and 72°C for 1 min; 21 cycles of 92°C for 30 S, 55°C for 30 s, and 72°C for 1.21 min; and 72°C for 10 min.
DGGE.
Bio-Rad Dcode or Ingeny PhorU (Apollo Instruments, Compiègne, France) systems were used for DGGE analysis of the V3 PCR products. The 6% acrylamide gels contained a linear chemical gradient of urea and formamide from 35% to 65%. PCR products (5 μg per well) were run in TAE buffer (40 mM Tris [pH 8.0], 20 mM acetic acid, 1 mM EDTA) at 60°C for 16 h at 75 V (Dcode) or for 17 h at 100 V (Ingeny PhorU). After electrophoresis, gels were immersed in SYBR green for 30 min at 4°C, rinsed in water, and photographed. PCR-DGGE replicates made with the same DNA sample generated identical patterns, indicating that the amplification was reproducible. When required, bands were excised, transferred to Eppendorf tubes, and washed five times with sterilized water. After all trace of liquid had been removed, 30 μl of water was added to tubes that were heated at 60°C for 30 min and kept overnight at 4°C; 2 μl of solution was used for reamplification, and products were cloned and sequenced.
Cloning and sequencing.
PCR products were purified using a QIAquick PCR purification kit (Qiagen) and cloned into chemically competent Escherichia coli cells (DH5α or TOP10) according to the TOPO TA 2.1 cloning kit protocol (Invitrogen). Transformants containing DNA inserts were selected for sequencing by Genoscreen (Lille, France). Sequences were analyzed with the NCBI Blastn program (http://www.ncbi.nlm.nih.gov/).
FISH.
Ovaries and oocytes were fixed for 20 min in freshly prepared 4% formaldehyde in PBS buffer (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4 [pH 7.4]). Samples were hybridized overnight at 37°C in hybridization buffer [50% formamide, 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 200 g liter−1 dextran sulfate, 250 μg ml−1 poly(A), 250 μg ml−1 salmon sperm DNA, 250 μg ml−1 tRNA, 0.1 M dithiothreitol (DTT), 0.5× Denhardt's solution] containing 200 ng of each probe. Fluor-labeled oligonucleotide probes (Invitrogen) were used: two Wolbachia probes, W2 (5′-CTTCTGTGAGTACCGTCATTATC-3′) (36) and Wol3 (5′-TCCTCTATCCTCTTTCAATC-3′) (60), 5′-end labeled with rhodamine; two Pseudomonas probes, PSEUD (5′-ACCTAGGCTCATCTGATAGCGCAAGG-3′) (59) and PSEUG (5′-GATCCGGACTACGATCGGTTT-3′) (34), 5′-end labeled with Alexa 488; and two Brevibacillus probes, Brevigenus Brbac2 (5′-CATCTCCCAGTGACAGCCGA-3′) and Breviagri Brbac6 (FITC-CCCAGTGATAGCGAAAAGC) (M. Kyselková, J. Kofecky, M. Frapolli, G. Défago, M. Ságová-Marécková, G. Grundmann, and Y. Moënne-Loccoz, unpublished data), 5′-end labeled with FITC. Samples were washed twice in 1× SSC, 10 mM DTT and twice in 0.5× SSC, 10 mM DTT at 55°C for 15 min, rinsed in PBS, mounted in glycerol on a glass slide, and viewed with fluorescent (Axio Imager.Z1; Zeiss) and confocal (LSM510; Zeiss) microscopes. Fixed samples were stained with 1 μg ml−1 DAPI in glycerol.
Nucleotide sequence accession numbers.
Sequences determined in this study were deposited in the GenBank database and are listed in Table 2.
TABLE 2.
Phylogenetic affiliation of sequences obtained in this study
| Analysis and sample type | Band(s), name of clone, or fragment | Size (bp) | Accession no. | Phylogenetic affiliation | Closest relative organism | Accession no. | No. with identity/total (%) |
|---|---|---|---|---|---|---|---|
| DGGE | |||||||
| Entire individuals | 1; 2; 7; 8; 12; 13; 27; 28 | 169 | FJ481970 | Alphaproteobacteria | Wolbachia endosymbiont of D. melanogaster wMelPop | AB360385.1 | 169/169 (100) |
| 3a | 171 | FJ562342 | Alphaproteobacteria | Rickettsia sp. strain Brugge | AF322443.1 | 170/171 (99) | |
| 3b | 193 | FJ562343 | Betaproteobacteria | Uncultured Duganella sp. | AM946212 | 186/193 (96) | |
| 4 | 171 | FJ562342 | Alphaproteobacteria | Rickettsia sp. strain Brugge | AF322443.1 | 170/171 (99) | |
| 5a | 175 | FJ562344 | Uncultured bacterium, clone COL[lowem]aai14h01 | EU460112.1 | 166/175 (94) | ||
| 5b | 194 | FJ562345 | Gammaproteobacteria | Pseudomonas sp. | EU308476 | 194/194 (100) | |
| 6 | 194 | FJ562346 | Betaproteobacteria | Herbaspirillum sp. strain IEH 4430 | FJ267649 | 190/194 (97) | |
| 9; 16; 22-26 | 194 | FJ481973 | Gammaproteobacteria | Enterobacteriaceae | AM940408.1 | 191/194 (98) | |
| 10-14-17 | 169 | FJ481971 | Alphaproteobacteria | Acetobacter pasteurianus bh12 | FJ227313.1 | 168/169 (99) | |
| 11-15-18 | 169 | FJ481972 | Alphaproteobacteria | Acidomonas methanolica | AF127398.1 | 168/169 (99) | |
| 19-21 | 195 | FJ481974 | Gammaproteobacteria | Enterobacteriaceae | AM940408.1 | 193/195 (98) | |
| Ovaries | 29-30 | 194 | FJ481975 | Gammaproteobacteria | Pseudomonas sp. R-35722 strain Z57b | AM886087.1 | 194/194 (100) |
| 31-32 | 194 | FJ481969 | Gammaproteobacteria | Endosymbiont of Petrasma sp. | FM213449.1 | 194/194 (100) | |
| 33 | 194 | FJ481976 | Firmicutes | Uncultured Staphylococcus sp., clone WLB24 | FJ405254.1 | 194/194 (100) | |
| 34 | 195 | FJ481977 | Firmicutes | Streptococcus sp. strain F1 | FJ405281.1 | 194/194 (100) | |
| 35 | 194 | FJ481978 | Firmicutes | Staphylococcus pasteuri NJ-1 | FJ392804.1 | 193/194 (99) | |
| Bacterial isolates from ovaries | |||||||
| Tri-infected A. tabida | KZ17a | 1528 | FJ481959 | Firmicutes | Brevibacillus agri NCHU 1002 | AY319301.1 | 1516/1530 (99) |
| Mono-infected A. tabida | KZ17b | 1528 | FJ481959 | Firmicutes | Brevibacillus agri NCHU 1002 | AY319301.1 | 1516/1530 (99) |
| KZ2 | 1542 | FJ481960 | Firmicutes | Staphylococcus epidermis strain SR1, clone step.1051c07 | AF270147.1 | 1539/1543 (99) | |
| KZ3 | 1543 | FJ481961 | Firmicutes | Bacillus pumilus SAFR-032 | CP000813.1 | 1537/1544 (99) | |
| Genus specific amplification | |||||||
| Tri-infected A. tabida | Brevibacillus | 321 | FJ481962 | Firmicutes | Bacillus sp. OS1 | EF428970.1 | 320/321 (99) |
| Pseudomonas | 990 | FJ481964 | Gammaproteobacteria | Uncultured bacterium, clone nbt05e04 | EU535862.1 | 990/990 (100) | |
| Staphylococcus | 282 | FJ481965 | Firmicutes | Uncultured Staphylococcus sp., clone IS034B20 | AY807427.1 | 280/283 (98) | |
| Staphylococcus | 282 | FJ481966 | Firmicutes | Staphylococcus pasteuri ELA-9 | FJ195007.1 | 271/282 (96) | |
| Rickettsia | 967 | FJ603467 | Alphaproteobacteria | Rickettsia sp. strain Brugge | AF322443.1 | 955/967 (99) | |
| Mono-infected A. tabida | Brevibacillus | 299 | FJ481963 | Firmicutes | Bacillus pumilus Sua-BAC003 | EU870500.1 | 298/299 (99) |
| Pseudomonas | 990 | FJ481964 | Gammaproteobacteria | Uncultured bacterium, clone nbt05e04 | EU535862.1 | 990/990 (100) | |
| Staphylococcus | 282 | FJ481967 | Firmicutes | Uncultured bacterium, clone AGB17 | FM211024.1 | 281/282 (99) | |
| Staphylococcus | 282 | FJ481968 | Firmicutes | Uncultured bacterium, clone nbt221d06 | EU536773.1 | 278/282 (98) |
RESULTS
Whole-insect bacterial profiling by PCR-DGGE.
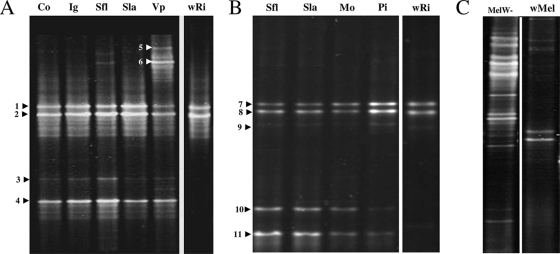
To explore the bacterial community of A. tabida, PCR-DGGE fingerprints of samples from whole insects were produced using primers (Table 1) specific to rrs genes and the corresponding hypervariable V3 regions, which has been shown to be useful for analyzing bacterial communities (15, 78). The genomic DNA was extracted from insects collected from different sites in France and from individuals reared in the laboratory. For each population, three pools of five individuals were tested independently. Whole-insect-body samples from females caught in the field had similar DGGE profiles (Fig. 2A), except for those from Vp. There were three intense and some faint bands in the profiles of all individuals, whereas another two bands were found only in individuals from Vp. Bands were excised from the gel, and some were cloned and sequenced (Table 2). The two major bands in the middle of the gel were considered to be from the genus Wolbachia, as they were present in the control V3 fragments amplified from Wolbachia strain wRi purified from infected Drosophila simulans. One faint band gave rise to two distinct sequences characteristic of the uncultivable genera Duganella and Rickettsia. The intense and lowest band also corresponded to Rickettsia. Amplification with specific rrs primers (Table 1) and sequencing of the 967-bp PCR products confirmed the band was from a Rickettsia sp., closely related (99% identity) to Rickettsia strain Brugge (Table 2). The upper bands exclusively found in individuals from Vp generated three sequences closely related to sequences from Herbaspirillum, Pseudomonas, and an unknown uncultivable bacterium. In some experiments, Duganella, Herbaspirillum, and Pseudomonas have been reported as contaminants (66). However, clonable PCR products were not generated with V3 primers in controls without a DNA template under the conditions used here, suggesting that there was no contamination.
FIG. 2.
DGGE profiles of hypervariable rrs V3 segments of A. tabida in native (A) and laboratory (B) populations. wRi, Wolbachia strain purified from D. simulans Riverside (44). Numbers correspond to cloned and sequenced bands (Table 2). (C) Profile of D. melanogaster used to rear A. tabida lines. wMel, Wolbachia strain purified from infected D. melanogaster.
Of the natural A. tabida populations, only lines from Co, Ig, Sfl, and Sla could be maintained in an insectarium. F2 females and males of these recent laboratory-reared lines mostly had DGGE patterns similar to those of their F0 parents, except for an additional band in Sfl F2 insects and the absence of fragments corresponding to Duganella and Rickettsia in Co F2 insects (see Fig. S1 in the supplemental material). In contrast, the long-term laboratory colonies (Sfl, Sla, Mo, and Pi) gave identical patterns (Fig. 2B) that differed from those of both natural and laboratory F2 populations. Although the two bands corresponding to Wolbachia were detected, bands indicative of Duganella and Rickettsia were absent. Three new bands found were indicative of the family Enterobacteriaceae and the genera Acetobacter and Acidomonas (Table 2). As laboratory rearing clearly influences the composition of the bacterial community, we examined whether the bacteria detected in the wasps could be acquired from fly larvae during parasitic development. DGGE fingerprints (Fig. 2C) of the nursery Wolbachia-free D. melanogaster had many bands, which were clearly distinct from those found in A. tabida. DGGE profiles were similar in adult insects at emergence and at one week after emergence.
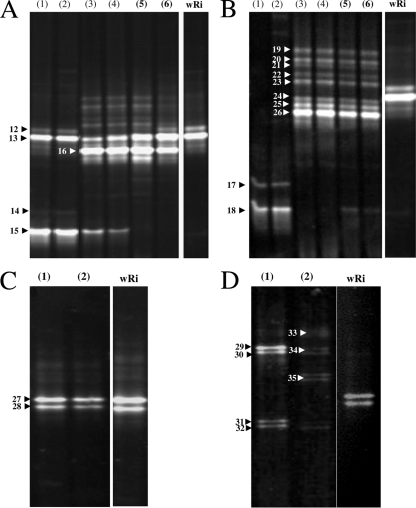
DGGE fingerprints of antibiotic-treated individuals.
Usually, antibiotics are applied to host tissues to eliminate Wolbachia in order to analyze a particular phenotype ascribed to this symbiont. To investigate whether antibiotic treatment affects the bacterial composition of A. tabida, we used the Pi(3) line, generated from the triply infected line Pi(123) after moderate tetracycline treatment (19), and individuals were stably maintained in the laboratory. Pi(3) is singly infected with Wolbachia strain wAtab3, which is required for wasp oogenesis. The absence or presence of wAtab1 and wAtab2 was confirmed by PCR specific for wsp alleles (data not shown). PCR-DGGE amplification of Pi(3) yielded one intense band corresponding to Wolbachia (Fig. 3A, lanes 3 to 6), compared to the V3 fragments from Pi(123) (lanes 1 and 2) and Wolbachia strain wRi, used as a control. Additional faint bands were consistently seen in both Pi(3) males and females. Moreover, the DGGE profiles of Pi(3) individuals newly generated by antibiotic treatment (Fig. 3A, lanes 3 and 4) showed one of the bands attributed to Acidomonas in Pi(123), whereas individuals reared for a long period in the laboratory had lost this band (Fig. 3A, lanes 5 and 6).
FIG. 3.
Examples of rrs V3 DGGE profiles from untreated Pi(123) and tetracycline-treated Pi(3) A. tabida. Amplification with (A and C) and without (B and D) Wolbachia sequences is depicted in Fig. 1. (A and B) Whole triply infected female (lane 1) and male (lane 2) and whole singly infected female (lanes 3 and 5) and male (lanes 4 and 6). (C and D) Ovaries from triply infected (lane 1) and singly infected (lane 2) females. wRi, Wolbachia strain purified from D. simulans Riverside. Numbers correspond to cloned and sequenced bands (Table 2).
To determine whether the additional bands observed in Pi(3) are a genuine consequence of tetracycline treatment or an artifact where the high density of Wolbachia sequences in Pi(123) affects the detection of other bacterial sequences, a fingerprinting strategy was used that excluded Wolbachia rrs gene sequences from the profile (see Materials and Methods) (Fig. 1). Fingerprints were obtained for both old and new generations of Pi(3) and of Pi(123) in this way. A positive control containing the rrs gene of Wolbachia strain wRi as the template was included. All the DGGE profiles (Fig. 3B) lacked the Wolbachia bands, as expected. Pi(123) female (lane 1) and male (lane 2) samples had only the two major fragments previously attributed to Acetobacter and Acidomonas, suggesting that the multiple rrs genes of the three Wolbachia strains did not impair amplification of the V3 regions of potential coinfecting bacteria in triply infected individuals. Again, Pi(3) had the most bands (Fig. 3B, lanes 3 to 6). Except for the absence of Wolbachia bands, the DGGE patterns were mostly similar to that of Pi(3) Wolbachia sequences (Fig. 3A), albeit with some bands being more intense. Several of these V3 fragments amplified from Pi(3) were cloned. All 16 cloned fragments (8 from males and 8 from females) were sequences characteristic of members of the Enterobacteriaceae (Table 2). Overall, these results indicate that experimental exclusion of Wolbachia V3 fragments did not significantly affect the detection of coinfecting bacteria. Therefore, tetracycline treatment affected the composition of the bacterial community of A. tabida, probably by eliminating other tetracycline-sensitive bacteria and/or allowing tetracycline-resistant bacteria to occupy free niches.
Wolbachia and total bacterial density.
As antibiotic treatment influences the composition of the bacterial community, we counted Wolbachia bacteria and all bacteria in whole bodies of untreated Pi(123) and treated Pi(3) females. There were significantly more Wolbachia organisms per wasp in triply infected (12 × 103 ± 0.02 × 103) than in singly infected (2 × 103 ± 0.001 × 103) individuals (Kruskal-Wallis test; P < 0.0003), with cell numbers being similar to those previously reported (47). Pi(3) individuals also harbored fewer bacteria, 5 × 105 ± 0.001 × 105 in total, compared to 8.4 ± 0.009 × 105 in Pi(123). Overall, these findings demonstrate that when tetracycline was applied to generate singly infected Pi(3), it greatly reduced Wolbachia density but only slightly reduced the total number of bacteria, indicating that some bacterial populations increased after antibiotic treatment.
Bacteria in A. tabida ovaries.
To search for potential maternally transmitted endosymbionts, PCR-DGGE V3 profiles of dissected insect ovaries (DIO) from Pi(123) and Pi(3) lines were obtained, and the two major bands were exclusively assigned to Wolbachia (Fig. 3C). Interestingly, PCR exclusion of the Wolbachia rrs gene fraction in DIO allowed new bands to be detected in both Pi(123) (Fig. 3D, lane 1) and Pi(3) (lane 2). A total of five doublets were excised from the gel, cloned, and sequenced. BLAST analysis (Table 2) indicated that sequences from Pi(123) were closely related to sequences from the Gammaproteobacteria (a Pseudomonas sp. and an endosymbiont of the bivalve Petrasma sp.), whereas those from Pi(3) were attributed to the group of Firmicutes (two Staphylococcus strains and one Streptococcus strain).
Because DGGE fingerprinting revealed the presence of bacterial genera that have potentially cultivable relatives, a culture-dependent analysis was performed. Liquid cultures of DIO homogenates were plated on rich media. Two colony types were recovered from Pi(123) and three from Pi(3). For each colony type, two clones were selected for complete sequencing of the rrs gene. The rrs gene sequences (Table 2) of two isolates from Pi(123) and one isolate from Pi(3) corresponded to Brevibacillus agri. Two other isolates from Pi(3) were identified as Bacillus pumilus and Staphylococcus epidermidis. For all isolates, sequence similarities were up to 99% with respect to the rrs sequences of the type strains reported in databases.
To further investigate the occurrence of the Firmicutes and the Gammaproteobacteria in A. tabida ovaries, available genus-specific primers targeting Brevibacillus, Staphylococcus, and Pseudomonas were used in PCR on Pi(123) and Pi(3) DIO samples. In all cases, a PCR product of the expected size was obtained (data not shown). Cloning and sequencing the fragments confirmed the identity of the targeted genus (Table 2) and hence its occurrence in wasp ovaries.
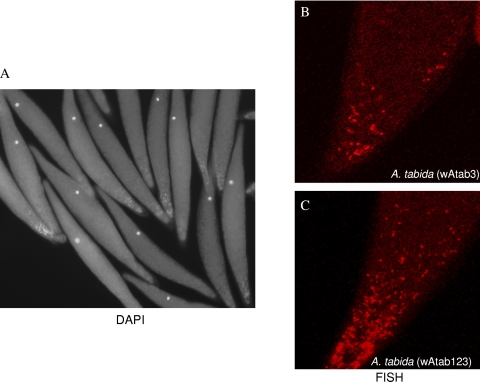
Visualization of oocyte-inhabiting bacteria by FISH.
To localize potential vertically transmitted bacterial symbionts, we used FISH for the first time in A. tabida using genus-specific fluor-labeled oligonucleotide probes on fixed oocytes from Pi(3) and Pi(123). DAPI staining showed both cell nuclei and dots which correspond to bacteria (Fig. 4A). Specific signals for Wolbachia were detected in the cytoplasm of posterior oocytes (Fig. 4B and C). The signals were more intense in Pi(123) oocytes (Fig. 4B), extending to the subpole area, than in Pi(3) oocytes (Fig. 4C), and the location was usual for Wolbachia strains in these lines (19, 57). No fluorescent signals were detected in ovaries with probes targeting the genera Brevibacillus and Pseudomonas, although fluorescent dots were observed in cultures from both bacteria (not shown).
FIG. 4.
Fluorescent confocal microscopy of A. tabida oocytes infected with Wolbachia. (A) DAPI staining of A. tabida oocytes containing Wolbachia strain wAtab3. (B and C) Specific probe for Wolbachia in singly (B) and triply (C) infected oocytes.
DISCUSSION
Bacterial endosymbionts of the wasp A. tabida were investigated by culture and nonculture methods. Members of the Alphaproteobacteria, Betaproteobacteria, Gammaproteobacteria, and Firmicutes were found, with some variation in the species recorded, depending on the method used. These results were in accordance with previous reports of microbes associated with insects, including Drosophila (17), mosquitoes (56), and termites (27). Although the main bacterial phyla are shared with many other arthropod lineages, we found that native A. tabida populations harbor a microbial community dominated by the genera Wolbachia and Rickettsia, in the order Rickettsiales. Two genera, Duganella and Herbaspirillum, were also found in some native A. tabida populations.
The multiple infection of Asobara species by Wolbachia is well documented (20, 47, 73); these bacteria are transmitted transovarially, and two strains induce cytoplasmic incompatibility, whereas one strain controls oogenesis through a cell death process (21, 52, 73). Before this study, no Asobara species had been found to be associated with Rickettsia. The latter genus is usually described as the causative agent of rickettsiosis in vertebrates transmitted by blood-feeding arthropods (3). However, in recent years, members of Rickettsia have been found in association with nonhematophagous arthropods, in which they induce some changes in reproduction, as does the related genus Wolbachia (54). The Rickettsia strain found here was similar to the obligate symbiont necessary for the development of oocytes and egg laying in parthenogenetic booklice (55). Its role in A. tabida is yet to be established.
The coexistence of heritable Wolbachia and Rickettsia with other symbionts has been reported for the hematophagous tick Ixodes scapularis (8), the aphid Cinera cedri (31), and the whitefly Bemisia tabaci (33). Wolbachia may infect an estimated 20% to 70% of insect species (38, 75). In some natural host populations, the prevalence of Wolbachia within species can be high, up to 97% incidence for Drosophila simulans infected with strain wRi (69) and almost 100% for mosquito Aedes albopictus infected with wAlbB (41). However, between 20% and 100% of Bemisia tabaci whiteflies from the north to the south of Israel were infected with Rickettsia, Wolbachia, and three other bacterial genera (33). Overall, these data indicated that multiple infections with members of the Rickettsiales occur in several invertebrate lineages.
When bacterial populations in laboratory-reared A. tabida were surveyed, Wolbachia was consistently found in all individuals, whereas Rickettsia was not detectable in the F2 generation of one of the four native populations recently maintained in the insectarium. Moreover, Rickettsia was never found in stock lines that had been reared in the laboratory for a long period; two of these lines (Sfl and Sla) originated from regions where recently caught individuals harbored Rickettsia, suggesting that this bacterium could become dispensable for the wasp under laboratory conditions. Loss of Rickettsia was accompanied by the appearance of Acetobacter, Acidomonas, and members of the family Enterobacteriaceae. The evolutionary behavior governing the dynamics of such cohabitation is mostly unknown and is under investigation.
The composition of bacterial populations was consistently modified by the antibiotic treatment applied to generate a subline of A. tabida singly infected by Wolbachia strain wAtab3. There were more bands in the DGGE patterns of the tetracycline-treated Pi(3) line than in those of the parental triply infected Pi(123) line. It is possible that the tetracycline-sensitive fraction of the bacterial population was eliminated, allowing an increase in the number of resistant bacteria. No significant difference in the overall density of bacteria was found between treated and untreated lines. However, multi-infected individuals harbored a significantly higher proportion of Wolbachia, consistent with previous data confirming that the strain wAtab3 does not proliferate to occupy the niche liberated by the other two strains (wAtab1 and wAtab2). Surprisingly, all the bands detected in the tetracycline-treated line were characteristic of the family Enterobacteriaceae. This may represent diverse genera, as the rrs-based phylogeny of Enterobacteriaceae is not clear (50, 62). Members of the Enterobacteriaceae are regularly found in invertebrates, including Photorhabdus and Xenorhabdus in Steinernema or Heterorhabditis (28), Klebsiella in Drosophila (17), Pantoea in mosquitoes (56), and Rhanella in pine beetles (71). Furthermore, several insect lineages, such as aphids, psyllids, whiteflies, weevils, and wasps, harbor members of the Rickettsiales together with some heritable Enterobacteriaceae, notably Arsenophonus, Buchnera, Hamiltonella, Regiella, Serratia, Sodalis, and Wigglesworthia (1, 5, 30, 35, 45, 46, 67). However, none of the sequences recorded here was unequivocally matched to these genera; this will require further taxonomic study.
To look for potential maternally inherited symbionts, nonculture methods, exclusion of Wolbachia sequences from DGGE analysis, and culture were used to identify members of the Gammaproteobacteria and Firmicutes in ovary homogenates. In addition to Wolbachia, DGGE rrs V3 sequences came from a Pseudomonas sp. (an endosymbiont of the bivalve Petrasma sp.), Streptococcus, and Staphylococcus. Isolated bacteria were Bacillus pumilus, Brevibacillus agri, and Staphylococcus epidermis, all members of the Firmicutes. Mostly known as being associated with vertebrates (26), members of Firmicutes have also been described in insects, including D. melanogaster (18), gypsy moths (11), and the mosquito Culex quinquefasciatus (57). However, only Wolbachia was detected in ovaries by FISH, a technique applied for the first time in A. tabida. The location of Wolbachia in oocyte poles is in agreement with published data (19, 57). Probes specific for Brevibacillus and Pseudomonas did not produce clear signals in entire ovaries, suggesting that PCR products or isolates obtained from homogenates might be dissection contaminants from other organs of the insect body, such as the gut, which is known to contain such bacteria (72).
Multiple bacterial infections, commonly found in members of the Arthropoda, may create competition or cooperation between symbionts and the host immune defense (13, 16, 24, 43). To infect and persist in a host population, symbiotic bacteria usually increase host fitness or manipulate host reproduction, maximizing their transmission from one generation to the next. In A. tabida, three Wolbachia strains coexist and are maintained either by inducing cytoplasmic incompatibility or by controlling oogenesis. The biological roles of other bacteria discovered in the wasp are under investigation. Many of these bacteria are commonly found in the environment, including soil and plants. For instance, members of the Enterobacteriaceae such as Herbaspirillum can evolve as endophytes able to fix atmospheric nitrogen in planta (53). As insects grow on diets with extremely high carbon-to-nitrogen (C:N) ratios, bacterial nitrogen fixation may provide an additional nitrogen source (6). Commensal Firmicutes in the digestive tracts of some insects contribute to the degradation of polymers, such as chitin and cellulose, and of aromatic compounds that can be toxic to microorganisms (42). Acetic acid bacteria such as Acetobacter and Acidomonas use sugar fermentation to produce ethanol, which in turn can be oxidized into acetic acid, which acidifies their habitats, notably insect guts (40). Some bacteria detected here, assigned to known or unknown groups, are not yet cultivable and may have roles in A. tabida biology. When such potential symbiotic roles are studied, antibiotics should be used in the knowledge that they may affect both the targeted and nontargeted bacteria, as shown here, and thus influence the outcomes of the biological interactions, as recently found in different animal taxa (22, 70).
Supplementary Material
Acknowledgments
We thank Fabrice Vavre, Roland Allemand, and Natacha Kremer for the use of insect rearing facilities, as well as three anonymous reviewers for helpful advice.
K.Z. was supported by a Ph.D. fellowship from the French Ministère de l'Education Nationale, de la Recherche et des Nouvelles Technologies. D.V. was a postdoctoral researcher supported by Agence Nationale de Recherche grant ANR-06-BLAN-0316. This work was funded partly by grant ANR-06-SEST07.
Footnotes
Published ahead of print on 17 April 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Akman, L., A. Yamashita, H. Watanabe, K. Oshima, T. Shita, M. Hattori, and S. Aksoy. 2002. Genome sequence of the endocellular obligate symbiont of tsetse flies, Wigglesworthia glossinidia. Nat. Genet. 32:402-407. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, C. L., and T. L. Karr. 2001. Wolbachia: evolutionary novelty in rickettsial bacteria. BMC Evol. Biol. 1:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Azad, A. F., and C. B. Beard. 1998. Rickettsial pathogens and their arthropod vectors. Emerg. Infect. Dis. 4:170-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bandi, C., J. W. McCall, C. Genchi, S. Corona, L. Venco, and L. Sachhi. 1999. Effects of tetracycline on the filarial worms Brugia pahangi and Dirofilaria immitis and their bacterial endosymbionts. Int. J. Parasitol. 29:357-364. [DOI] [PubMed] [Google Scholar]
- 5.Baumann, L., and P. Baumann. 2005. Cospeciation between the primary endosymbiont of mealybugs and their hosts. Curr. Microbiol. 50:84-87. [DOI] [PubMed] [Google Scholar]
- 6.Behar, A., B. Yuval, and E. Jurkevitch. 2005. Enterobacteria-mediated nitrogen fixation in natural populations of the fruit fly Ceratitis capitata. Mol. Ecol. 14:2637-2643. [DOI] [PubMed] [Google Scholar]
- 7.Bensaadi-Merchermek, N., J. C. Cagnon, K. S. Solange, and C. Mouchès. 1995. Characterization of the unlinked 16S rDNA and 23S-5S rRNA operon of Wolbachia pipientis, a prokaryotic parasite of insect gonads. Gene 165:81-86. [DOI] [PubMed] [Google Scholar]
- 8.Benson, M. J., J. D. Gawronski, D. E. Eveleigh, and D. R. Benson. 2004. Intracellular symbionts and other bacteria associated with deer ticks (Ixodes scapularis) from Nantucket and Wellfleet, Cape Cod, Massachusetts. Appl. Environ. Microbiol. 70:616-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bouchon, D., T. Rigaud, and P. Juchault. 1998. Evidence for widespread Wolbachia infection in isopod crustaceans: molecular identification and host feminization. Proc. R. Soc. Lond. B 265:1081-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Braig, H. R., W. Zhou, S. L. Dobson, and S. L. O'Neill. 1998. Cloning and characterization of a gene encoding the major surface protein of the bacterial endosymbiont Wolbachia pipientis. J. Bacteriol. 180:2373-2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Broderick, N. A., K. F. Raffa, R. M. Goodman, and J. Handelsman. 2004. Census of the bacterial community of the gypsy moth larval midgut using culturing and culture-independent methods. Appl. Environ. Microbiol. 70:293-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bruce, K. D., W. D. Hiorns, J. L. Hobman, A. M. Osborn, P. Strike, and D. A. Ritchie. 1992. Amplification of DNA from native populations of soil bacteria by using the polymerase chain reaction. Appl. Environ. Microbiol. 58:3413-3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buchner, P. 1965. Endosymbiosis of animals with plant microorganisms. Interscience, New York, NY.
- 14.Carton, Y., Y. M. Boulétreau, J. J. M. van Alphen, and J. C. van Lenteren. 1986. The Drosophila parasitic wasp, p. 347-394. In M. Ashburner, H. L. Carson, and J. N. Thompson (ed.), The genetics and biology of Drosophila. Academic Press Inc., London, United Kingdom.
- 15.Chakravorty, S., D. Helb, M. Burday, N. Connell, and D. Alland. 2007. A detailed analysis of 16S ribosomal gene segments for the diagnosis of pathogenic bacteria. J. Microbiol. Methods 69:330-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cherry, S., and N. Silverman. 2006. Host-pathogen interactions in Drosophila: new tricks from an old friend. Nat. Immunol. 7:911-917. [DOI] [PubMed] [Google Scholar]
- 17.Corby-Harris, V., A. C. Pontaroli, L. J. Shimkets, J. L. Bennetzen, K. E. Habel, and E. L. Promislow. 2007. Geographical distribution and diversity of bacteria associated with natural populations of Drosophila melanogaster. Appl. Environ. Microbiol. 73:3470-3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cox, C. R., and M. S. Gilmore. 2007. Native microbial colonization of Drosophila melanogaster and its use as a model of Enterococcus faecalis pathogenesis. Infect. Immun. 75:1565-1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dedeine, F., F. Vavre, F. Fleury, B. Loppin, M. E. Hochberg, and M. Bouletreau. 2001. Removing symbiotic Wolbachia bacteria specifically inhibits oogenesis in a parasitic wasp. Proc. Natl. Acad. Sci. USA 98:6247-6252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dedeine, F., F. Vavre, D. D. Shoemaker, and M. Bouletreau. 2004. Intra-individual coexistence of a Wolbachia strain required for host oogenesis with two strains inducing cytoplasmic incompatibility in the wasp Asobara tabida. Int. J. Org. Evol. 58:2167-2174. [DOI] [PubMed] [Google Scholar]
- 21.Dedeine, F., M. Bouletreau, and F. Vavre. 2005. Wolbachia requirement for oogenesis: occurrence within the genus Asobara (Hymenoptera, Braconidae) and evidence for intraspecific variation in A. tabida. Heredity 95:394-400. [DOI] [PubMed] [Google Scholar]
- 22.Dethlefsen, L., S. Huse, M. L. Sogin, and D. A. Relman. 2008. The pervasive effects of an antibiotic on the human gut microbiota. PLoS Biol. 6:e280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Douglas, A. E. 1998. Nutritional interactions in insect-microbial symbioses: aphids and their symbiotic bacteria Buchnera. Annu. Rev. Entomol. 43:17-37. [DOI] [PubMed] [Google Scholar]
- 24.Douglas, A. E. 2008. Conflict, cheats and the persistence of symbioses. New Phytol. 177:849-858. [DOI] [PubMed] [Google Scholar]
- 25.Dumler, J. S., A. F. Barbet, C. P. J. Beckker, G. A. Dasch, G. H. Palmer, S. C. Ray, Y. Rikihisa, and F. R. Rurangirwa. 2001. Reorganization of genera in the families Rickettsiaceae and Anaplasmataceae in the order Rickettsiales: unification of some species of Ehrlichia with Anamaplasma, Cowdria with Ehrlichia and Ehrlichia with Neorickettsia, description of news species combinations and designation of Ehrlichia equi and “HGE agent” as subjective synonyms of Ehrlichia phagocytophila. Int. J. Syst. Evol. Microbiol. 51:2145-2165. [DOI] [PubMed] [Google Scholar]
- 26.Eckburg, P. B., E. M. Bik, C. N. Bernstein, E. Purdom, L. Dethlefsen, M. Sargent, S. R. Gill, K. E. Nelson, and D. A. Relman. 2005. Diversity of the human intestinal microbial flora. Science 308:1635-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fall, S., J. Hamelin, F. Ndiaye, K. Assigbetse, M. Aragno, J. L. Chotte, and A. Brauman. 2007. Differences between bacterial communities in the gut of a soil-feeding termite (Cubitermes niokoloensis) and its mounds. Appl. Environ. Microbiol. 73:5199-5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Forst, S., and K. Nealson. 1996. Molecular biology of the symbiotic-pathogenic bacteria Xenorhabdus spp. and Photorhabdus spp. Microbiol. Rev. 60:21-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.García-Martínez, J., S. G. Acinas, A. I. Antón, and F. Rodríguez-Valera. 1999. Use of the 16S-23S ribosomal genes spacer region in studies of prokaryotic diversity. J. Microbiol. Methods 36:55-64. [DOI] [PubMed] [Google Scholar]
- 30.Gherna, R. L., J. H. Werren, W. Weisburg, R. Cote, C. R. Woese, L. Mandelco, and J. Benner. 1991. Arsenophonus nasoniae gen. nov., sp. nov., the causative agent of the son-killer trait in the parasitic was Nasonia vitripennis. Int. J. Syst. Bacteriol. 41:563-568. [Google Scholar]
- 31.Gómez-Valero, L., M. Soriano-Navarro, V. Pérez-Brocal, A. Heddi, A. Moya, J. M. García-Verdugo, and A. Latorre. 2004. Coexistence of Wolbachia with Buchnera aphidicola and a secondary symbiont in the aphid Cinara cedri. J. Bacteriol. 186:6626-6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goto, K., R. Fujita, Y. Kato, M. Asahara, and A. Yokota. 2004. Reclassification of Brevibacillus brevis strains NCIMB 13288 and DSM 6472 (NRRL NRS-887) as Aneurinibacillus danicus sp. nov. and Brevibacillus limnophilus sp. nov. Int. J. Syst. Evol. Microbiol. 54:419-427. [DOI] [PubMed] [Google Scholar]
- 33.Gottlieb, Y., M. Ghanim, E. Chiel, D. Gerling, V. Portnoy, S. Steinberg, G. Tzuri, A. R. Horowitz, E. Belausov, N. Mozes-Daube, S. Kontsedalov, M. Gershon, S. Gal, N. Katzir, and E. Zchori-Fein. 2006. Identification and localization of a Rickettsia sp. in Bemisia tabaci (Homoptera: Aleyrodidae). Appl. Environ. Microbiol. 72:3646-3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gunasekera, T. S., M. R. Dorsch, M. B. Slade, and D. A. Veal. 2003. Specific detection of Pseudomonas spp. in milk by fluorescence in situ hybridization using ribosomal RNA directed probes. J. Appl. Microbiol. 94:936-945. [DOI] [PubMed] [Google Scholar]
- 35.Hansen, A. K., G. Jeong, T. D. Paine, and R. Stouthamer. 2007. Frequency of secondary symbiont infection in an invasive psyllid relates to parasitism pressure on a geographic scale in California. Appl. Environ. Microbiol. 73:7531-7535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heddi, A., A. M. Grenier, C. Khatchadourian, H. Charles, and P. Nardon. 1999. Four intracellular genomes direct weevil biology: nuclear, mitochondrial, principal endosymbiont, and Wolbachia. Proc. Natl. Acad. Sci. USA 96:6814-6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hedges, L. M., J. C. Brownlie, S. L. O'Neill, and K. N. Johnson. 2008. Wolbachia and virus protection in insects. Science 322:702. [DOI] [PubMed] [Google Scholar]
- 38.Hilgenboecker, K., P. Hammerstein, P. Schlattmann, A. Telschow, and J. H. Werren. 2008. How many species are infected with Wolbachia? A statistical analysis of current data. FEMS Microbiol. Lett. 281:215-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hurst, G. D. D., F. M. Jiggins, and M. E. Majerus. 2003. Male killing bacteria in insect: mechanisms, incidence, and implications. Emerg. Infect. Dis. 6:329-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kesters, K., P. Lisdiyanti, K. Komagata, and J. Swings. 2006. The family Acetobacteraceae: the genera Acetobacter, Acidomonas, Asaia, Gluconoacetobacter, Gluconobacter, and Kozakia, p. 153-200. In M. Dworkin, S. Falkow, B. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes, 3rd ed. Springer, New York, NY.
- 41.Kittayapong, P., K. J. Baisley, R. G. Sharpe, V. Braimai, and S. L. O'Neill. 2002. Maternal transmission efficiency of Wolbachia superinfections in Aedes albopictus populations in Thailand. Am. J. Trop. Hyg. 66:103-107. [DOI] [PubMed] [Google Scholar]
- 42.König, H. 2006. Bacillus species in the intestine of termites and other soil invertebrates. J. Appl. Microbiol. 101:620-627. [DOI] [PubMed] [Google Scholar]
- 43.Massey, R. C., A. Buckling, and R. Ffrench-Constant. 2004. Interference competition and parasite virulence. Proc. R. Soc. Lond. B 271:785-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mavingui, P., V. Tran Van, E. Labeyrie, E. Rancès, F. Vavre, and P. Simonet. 2005. Efficient procedure for purification of obligate intracellular Wolbachia pipientis and representative amplification of its genome by multiple-displacement amplification. Appl. Environ. Microbiol. 71:6910-6917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moran, N. A., J. P. McCutcheon, and A. Nakabachi. 2008. Genomics and evolution of heritable bacterial symbionts. Annu. Rev. Genet. 42:165-190. [DOI] [PubMed] [Google Scholar]
- 46.Moran, N. A., J. A. Russel, R. Koga, and T. Fukatsu. 2005. Evolutionary relationships of three new species of Enterobacteriaceae living as symbionts of aphids and other insects. Appl. Environ. Microbiol. 71:3302-3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mouton, L., F. Dedeine, H. Henri, M. Boulétreau, N. Profizi, and F. Vavre. 2004. Virulence, multiple infections and regulation of symbiotic population in the Wolbachia-Asobara tabida symbiosis. Genetics 168:181-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moya, A., J. Peretó, R. Gil, and A. Latorre. 2008. Learning how to live together: genomic insights into prokaryote-animal symbioses. Nat. Rev. Genet. 9:218-229. [DOI] [PubMed] [Google Scholar]
- 49.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Naum, M., E. W. Brown, and R. J. Mason-Gamer. 2008. Is 16S rDNA a reliable phylogenetic marker to characterize relationships below the family level in the Enterobacteriaceae. J. Mol. Evol. 66:630-642. [DOI] [PubMed] [Google Scholar]
- 51.O'Neill, S. L., R. Giordano, A. M. E. Colbert, T. L. Karr, and H. M. Robertson. 1992. 16S rRNA phylogenetic analysis of the bacterial endosymbionts associated with cytoplasmic incompatibility in insects. Proc. R. Soc. Lond. B 89:2699-2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pannebakker, B. A., B. Loppin, C. P. Elemans, L. Humblot, and F. Vavre. 2007. Parasitic inhibition of cell death facilitates symbiosis. Proc. Natl. Acad. Sci. USA 104:213-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pedraza, R. O. 2008. Recent advances in nitrogen-fixing acetic acid bacteria. Int. J. Food Microbiol. 125:25-35. [DOI] [PubMed] [Google Scholar]
- 54.Perlman, S. J., M. S. Hunter, and E. Zchori-Fein. 2006. The emerging diversity of Rickettsia. Proc. R. Soc. Lond. B. 273:2097-2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Perotti, M. A., H. K. Clarke, B. D. Turner, and H. R. Braig. 2006. Rickettsia as obligate and mycetomic bacteria. FASEB J. 20:1646-1656. [DOI] [PubMed] [Google Scholar]
- 56.Pidiyar, V. J., K. Jangid, M. S. Patole, and Y. S. Shouche. 2004. Studies on cultured and uncultured microbiota of wild Culex quinquefasciatus mosquito midgut based on 16S ribosomal RNA gene analysis. Am. J. Trop. Med. Hyg. 70:597-603. [PubMed] [Google Scholar]
- 57.Rancès, E., D. Voronin, V. Tran-Van, and P. Mavingui. 2008. Genetic and functional characterization of the type IV secretion system in Wolbachia. J. Bacteriol. 190:5020-5030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rouhbakhsh, D., and P. Baumann. 1995. Characterization of a putative 23S-5S rRNA operon of Buchnera aphidicola (endosymbiont of aphids) unlinked to the 16S rRNA-encoding gene. Gene 155:107-112. [DOI] [PubMed] [Google Scholar]
- 59.Rudi, K., S. L. Flateland, J. F. Hanssen, G. Bengtsson, and H. Nissen. 2002. Development and evaluation of a 16S ribosomal DNA array-based approach for describing complex microbial communities in ready-to-eat vegetable salads packed in a modified atmosphere. Appl. Environ. Microbiol. 68:1146-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sanguin, H., A. Herrera, C. Oger-Desfeux, A. Dechesne, P. Simonet, E. Navarro, T. M. Vogel, Y. Moënne-Loccoz, X. Nesme, and G. L. Grundmann. 2006. Development and validation of a prototype 16S rRNA based taxonomic microarray for Alphaproteobacteria. Environ. Microbiol. 8:289-307. [DOI] [PubMed] [Google Scholar]
- 61.Skow, A., K. A. Mangold, M. Tajuddin, A. Huntington, B. Fritz, R. B. Thomson, Jr., and K. L. Kaul. 2005. Species-level identification of staphylococcal isolates by real-time PCR and melt curve analysis. J. Clin. Microbiol. 43:2876-2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sproer, C., U. Mendrock, J. Swiderski, E. Lang, and E. Stackebrandt. 1999. The phylogenetic position of Serratia, Buttiauxella and some other genera of the family Enterobacteriaceae. Int. J. Syst. Bacteriol. 49:1433-1438. [DOI] [PubMed] [Google Scholar]
- 63.Stouthamer, R., J. A. Breeuwer, R. F. Luck, and J. H. Werren. 1993. Molecular identification of microorganisms associated with parthenogenesis. Nature 361:66-68. [DOI] [PubMed] [Google Scholar]
- 64.Stouthamer, R., J. A. Breeuwer, and G. D. Hurst. 1999. Wolbachia pipientis: microbial manipulator of arthropod reproduction. Annu. Rev. Microbiol. 53:71-102. [DOI] [PubMed] [Google Scholar]
- 65.Stubner, S., and K. Meuser. 2000. Detection of Desulfotomaculum in an Italian rice paddy soil by 16S ribosomal nucleic acid analyses. FEMS. Microbiol. Ecol. 34:73-80. [DOI] [PubMed] [Google Scholar]
- 66.Tanner, M. A., B. Goebel, M. A. Dojka, and N. R. Pace. 1998. Specific ribosomal DNA sequences from diverse environmental settings correlate with environmental contaminants. Appl. Environ. Microbiol. 64:3110-3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Toh, H., B. L. Weiss, S. A. Perkin, A. Yamashita, K. Oshima, M. Hattori, and S. Aksoy. 2006. Massive genome erosion and functional adaptations provide insights into symbiotic lifestyle of Sodalis glossinidius in the tsetse host. Genome Res. 16:149-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tortosa, P., A. Courtiol, S. Moutailler, A. B. Failloux, and M. Weill. 2008. Chikungunya-Wolbachia interplay in Aedes albopictus. Insect. Mol. Biol. 17:677-684. [DOI] [PubMed] [Google Scholar]
- 69.Turelli, M., and A. A. Hoffmann. 1991. Rapid spread of an inherited incompatibility factor in California Drosophila. Nature 353:440-442. [DOI] [PubMed] [Google Scholar]
- 70.van der Hoeven, R., G. Betrabet, and S. Forst. 2008. Characterization of the gut bacterial community in Manduca sexta and effect of antibiotics on bacterial diversity and nematode reproduction. FEMS Microbiol. Lett. 286:149-256. [DOI] [PubMed] [Google Scholar]
- 71.Vasanthakumar, A., I. Delalibera, Jr., J. Handelsman, K. D. Klepzic, P. D. Schloss, and Raffa, K. F. 2006. Characterization of gut-associated bacteria in larvae and adults of the southern pine beetle, Dendroctonus frontalis Zimmermann. Environ. Entomol. 35:1710-1717. [Google Scholar]
- 72.Vasanthakumar, A., J. Handelsman, P. D. Schloss, L. S. Bauer, and K. F. Raffa. 2008. Gut microbiota of an invasive subcortical beetle, Agrilus planipennis Fairmaire, across various life stages. Environ. Entomol. 37:1344-1353. [DOI] [PubMed] [Google Scholar]
- 73.Vavre, F., F. Fleury, D. Lepetit, P. Fouillet, and M. Boulétreau. 1999. Phylogenetic evidence for horizontal transmission of Wolbachia in host-parasitoid associations. Mol. Biol. Evol. 16:1711-1723. [DOI] [PubMed] [Google Scholar]
- 74.Werren, J. H. 1997. Biology of Wolbachia. Annu. Rev. Entomol. 42:587-609. [DOI] [PubMed] [Google Scholar]
- 75.Werren, J. H., and D. M. Windsor. 2000. Wolbachia infection frequency in insects: evidence of a global equilibrium? Proc. R. Soc. Lond. B 267:1277-1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Widmer, F., R. J. Seidler, P. M. Gillevet, L. S. Watrud, and G. D. Di Giovanni. 1998. A highly selective PCR protocol for detecting 16S rRNA genes of the genus Pseudomonas (sensu stricto) in environmental samples. Appl. Environ. Microbiol. 64:2545-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wu, X. Y., M. J. Walker, M. Hornitzky, and J. Chin. 2006. Development of a group-specific PCR combined with ARDRA for the identification of Bacillus species of environmental significance. J. Microbiol. Methods 64:107-119. [DOI] [PubMed] [Google Scholar]
- 78.Yu, Z., and M. Morrison. 2004. Comparisons of different hypervariable regions of rrs genes for use in fingerprinting of microbial communities by PCR-denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 70:4800-4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhou, W., F. Rousset, and S. O'Neill. 1998. Phylogeny and PCR-based classification of Wolbachia strains using wsp gene sequences. Proc. Biol. Sci. 265:509-515. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.