Abstract
Phoslactomycins (PLMs) and related leustroducsins (LSNs) have been isolated from a variety of bacteria based on antifungal, anticancer, and other biological assays. Streptomyces sp. strain HK 803 produces five PLM analogs (PLM A and PLMs C to F) in which the C-18 hydroxyl substituent is esterified with a range of branched, short-alkyl-chain carboxylic acids. The proposed pathway intermediate, PLM G, in which the hydroxyl residue is not esterified has not been observed at any significant level in fermentation, and the only route to this potentially useful intermediate has been an enzymatic deacylation of other PLMs and LSNs. We report that deletion of plmS3 from the PLM biosynthetic cluster gives rise to a mutant which accumulates the PLM G intermediate. The 921-bp plmS3 open reading frame was cloned and expressed as an N-terminally polyhistidine-tagged protein in Escherichia coli and shown to be an 18-O acyltransferase, catalyzing conversion of PLM G to PLM A, PLM C, and PLM E using isobutyryl coenzyme A (CoA), 3-methylbutyryl-CoA, and cyclohexylcarbonyl-CoA, respectively. The efficiency of this process (kcat of 28 ± 3 min−1 and Km of 88 ± 16 μM) represents a one-step chemoenzymatic alternative to a multistep synthetic process for selective chemical esterification of the C-18 hydroxy residue of PLM G. PlmS3 was shown to catalyze esterification of PLM G with CoA and N-acetylcysteamine thioesters of various saturated, unsaturated, and aromatic carboxylic acids and thus also to provide an efficient chemoenzymatic route to new PLM analogs.
Attachment of either short (C2 to C6) or medium (C8 to C12) acyl chains to both amine and alcohol moieties on polyketide and polypeptide natural products can represent a key step in generating the final biologically active molecule. This step is often, but not always, one of the later biosynthetic steps and is catalyzed by an acyltransferase. The corresponding gene is typically associated with the polyketide or polypeptide biosynthetic gene cluster. Despite the importance of this step, a relatively small number of these acyltransferases from actinomycetes have been identified, and very few have been fully characterized (2, 10, 15, 18).
Studies of biosynthetic processes where there is an acylation of a polyketide chain, have indicated that the enzymes have various degrees of promiscuity for the carboxylic acid substrates. For instance, genetic evidence has shown that mdmB and acyA encode 3-O-acyltransferases which transfer either acetyl or propionyl groups to position 3 in 16-membered macrolides such as midecamycin, spiramycin, carbomycin, and tylosin (3, 10). The asm19 gene product has been identified as the 3-O-acyltransferase which catalyzes the attachment of the biologically essential acyl group in the macrocyclic ansamitocins (18). The asm19 mutant accumulates an N-demethyl-4,5-desepoxymytansinol, indicating that acylation of the macrocycle precedes N methylation and epoxidation. Escherichia coli cell extracts containing a recombinant Asm19 protein have been shown to catalyze acylation of this mytansinol intermediate using a range of short straight- and branched-chain acyl coenzyme A (CoA) thioesters (C2 to C5). Finally, LovD, which catalyzes the acylation of the C-8 hydroxyl group of monacolin J to yield the natural product lovastatin, a pharmaceutically important fungal polyketide product produced by Aspergillus nidulans, has been characterized (30). This enzyme is able to utilize a wide range of different acyl donors activated as CoA, N-acetylcysteamine (NAC), or methyl thioglycolate esters and thus offers an economically attractive route for generating novel lovasotatin analogs for treatment of hypercholesterolemia.
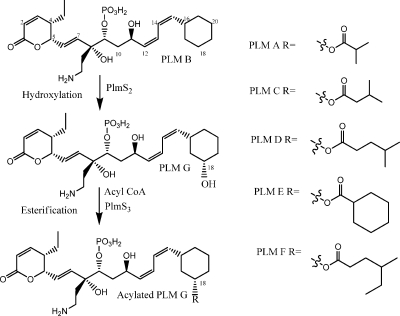
One or multiple O-acyltransferases have been implied to be involved in the post-polyketide synthase tailoring steps leading to a series of natural products known as the phoslactomycins (PLMs) (Fig. 1) (6). These compounds (also known as leustroducsins [LSNs], phospholines, and phosphazomycins) have been isolated from various actinomycetes, and their structures are all identical with the exception of the acyl substituent at C-18 (4, 5, 12, 13, 20, 27, 28). Streptomyces sp. strain HK 803 produces at least five such acylated analogs (PLM A and PLMs C to F) (Fig. 1) as well as PLM B, in which the C-18 hydroxyl substituent is absent (4, 5, 27).
FIG. 1.
Proposed biosynthetic relationship between PLM products made by Streptomyces sp. strain HK 803. A cytochrome P450 monooxygenase (PlmS2) catalyzes C-18 hydroxylation of PLM B to generate PLM G, which is subsequently 18-O acylated by PlmS3.
These natural products have been isolated based on their potent activity (as low as 0.008 μg/ml) against some phytopathogenic fungi (27, 28). The compounds also have relatively weak antitumor activity (50% inhibitory concentration of 2 to 3 μg/ml against L1210, P38,8, and El-4 cell lines) (19) which may arise from their activity as selective inhibitors of protein phosphatase 2A. (26). These natural products also show induction of a colony-stimulating factor (12) via NF-κB activation and thrombopoiesis (14). This array of promising biological activities has stimulated research into the field of PLMs for treatments of various diseases. Low yields and the presence of multiple acylated products from fermentations have provided a barrier to this work, and circuitous routes to obtaining individual compounds have been described. For instance, there have been no reports of any actinomycetes which produce PLM G (LSN H) (Fig. 1), in which the C-18 hydroxyl residue is present but not acylated, and this compound has been reported to be obtained only by cleavage of the acyl groups of a mixture of other PLMs and LSNs using porcine liver esterase (24). A multistep synthetic route to selectively acylate PLM G with 6-methyloctanoic acid (producing LSN B) has also been described (17).
Recently the entire 75-kbp Plm biosynthetic gene cluster has been cloned, sequenced, and analyzed (21) and has provided an opportunity to study the enzymatic hydroxylation and acylation processes which give rise to the range of PLM products. Deletion of the plmS2 open reading frame (ORF), showing high sequence similarity to bacterial cytochrome P450 monooxygenases, has resulted in an NP1 mutant producing only PLM B (Fig. 1) (21). The plmS2 ORF has been expressed as an N-terminally polyhistidine-tagged protein in Streptomyces coelicolor, and the purified protein has been shown to catalyze conversion of PLM B to PLM G (7). This work in conjunction with other studies (1, 23) has led to a proposal that the final two biosynthetic steps involve hydroxylation of PLM B (to give PLM G) and subsequent acylation with a broad range of acyl-CoA substrates (to give PLM A and PLMs C to F). The acylation step is required for potent antifungal activity of the PLMs. Initial analysis of the PLM biosynthetic gene cluster (21) did not reveal a candidate gene or genes whose products might be responsible for this acylation.
Here we identify the plmS3 gene product as the singular acyltransferase in Streptomyces sp. strain HK 803 responsible for C-18 acylation of PLM G. Generation of a plmS3 deletion mutant results in selective production of PLM G, supporting the proposed role of this gene product and providing the first direct fermentation method to access this intermediate. The 921-bp plmS3 ORF was cloned and expressed as an N-terminally polyhistidine-tagged protein in E. coli, and the recombinant purified protein was shown to catalyze acylation of PLM G with isobutyryl-CoA, 3-methylbutyryl-CoA, and cyclohexylcarbonyl-CoA to give PLM A, PLM C, and PLM E, respectively. This efficient one-step enzymatic process offers an attractive alternative to the multistep synthetic process for selective acylation of PLM G. PlmS3 was also shown to catalyze esterification of PLM G with CoA and NAC thioesters with a remarkably wide range of various saturated, unsaturated, and aromatic carboxylic acids and thus provides an efficient chemoenzymatic route to new PLM analogs.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. The wild-type Streptomyces sp. strain HK 803 and its deletion mutants NP8 and NP8/pMSG1′ were maintained on SY agar (1% soluble starch, 0.1% yeast extract, 0.1% N-Z amine type A), and the PLM production was performed as described previously (21). Escherichia coli cultures were grown at 37°C in Luria-Bertani medium supplemented with ampicillin or chloramphenicol (50 μg/ml) when necessary (22).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Reference or source |
|---|---|---|
| Streptomyces strains | ||
| HK 803 | Wild type; produces PLMs A through F | Riken, Japan |
| NP8 | plmS2-plmS3 deletion mutant; Amr; produces only PLM B | This work |
| NP8/pMSG1' | plmS3 deletion mutant; Amr Spr; produces only PLM G | This work |
| E. coli strains | ||
| TG2 | supE hsdΔ5 thi Δ(lac-proAB) Δ(srl-recA)306::Tn10(Tetr) F′[traD36 proAB+lacIqlacZΔM15] | 22 |
| BW 25113/pIJ790 | Recombination host; Chlr | 8 |
| ET12567/pUZ8002 | Nonmethylating strain harboring nontransmissible pUZ8002 (plasmid capable of mobilizing oriT-containing vectors) | 8 |
| BL21-Codon Plus(DE3) | E. coli expression host; Chlr | Stratagene |
| Plasmids | ||
| pET15b | E. coli expression vector; Apr | Novagen |
| pMSG1 | pGF200 with 0.7-kbp NdeI-BglI fragment replaced by 1.2-kbp NdeI-BamHI plmS2; Apr | 7 |
| pMSG1′ | pMSG1 with aac(3)IV replaced by aadA; Spr | This work |
| pMSG4 | pET15b with 921-bp NdeI-BamHI plmS3 ORF; Apr | This work |
| pIJ773 | Contains apramycin cassette for gene disruption; Apr Amr | 8 |
| pIJ778 | Contains streptomycin and spectinomycin cassette for gene disruption; Spr Amr | 8 |
| Cosmid clone 3A11 | Supercos 1 (Stratagene) derived; Apr Kanr | 21 |
Apr, ampicillin resistance; Amr, apramycin resistance; Chlr, chloramphenicol resistance; Kanr, kanamycin resistance; Spr, spectinomycin resistance.
Targeted disruption of plmS2-plmS3.
The plmS2-plmS3 genes are adjacent in the PLM biosynthetic gene cluster and were replaced with aac(3)IV (the apramycin resistance gene) using a PCR-targeted Streptomyces gene replacement method (8). The apramycin cassette for gene disruption from pIJ773 was amplified using the forward primer CCGTGCCGGGCCACCGGCGGATTGGAGAACGCAACCATGATTCCGGGGATCCGTCGACC and reverse primer CCCGGACGACCGGGCCGGTTCAGCGTGCGGGAACGCTCATGTAGGCTGGAGCTGCTTC (the pIJ773-homologous sequence is in bold). The resulting PCR product was used to replace plmS2-plmS3 first in a cosmid clone 3A11 (21) and then in Streptomyces sp. strain HK 803 following the established methodologies (8), using SY agar instead of MS agar (11). The allelic replacement of the plmS2-plmS3 genes in the NP8 mutant was confirmed by PCR amplification and sequencing.
Genetic complementation of the NP8 mutant.
The recombinant conjugative plasmid pMSG1′ was generated from pMSG1 (Table 1) by recombinant allelic replacement of aac(3)IV with the spectinomycin resistance gene, aadA. The forward primer GATCGACTGATGTCATCAGCGGTGGAGTGCAATGTCGTGATGAGGGAAGCGGTGATCGC and reverse primer GCCCCTCCAACGTCATCTCGTTCTCCGCTCATGAGCTCATTATTTGCCGACTACCTTG (pIJ778-homologous sequence is in bold) were used to amplify aadA from pIJ778, and the resulting PCR product was used to carry out the replacement using established methodology (9). pMSG1′, conferring spectinomycin resistance, was transferred by conjugation into the NP8 mutant using standard protocols (11).
Cloning of the plmS3 gene.
General DNA manipulations were performed following standard protocols (22). The plmS3 ORF was PCR amplified using a forward primer (5′-CATATGGCCGACAGCCTTGCGGCC-3′) introducing a unique NdeI site at the 5′ end of the gene and a reverse primer (5′-CGCTCAGGTCCTAGGTCACCATGCGG-3′) introducing a unique BamHI site downstream from the TGA translational stop codon. DNA of cosmid clone 3A11 from a Streptomyces sp. strain HK 803 genomic library was used as a template (21). The 921-bp amplified DNA fragment was first cloned into the pCR2.1 TOPO TA vector (Invitrogen). The 921-bp NdeI-BamHI insert was further subcloned into the pET15b expression vector to generate pMSG4. The ORF was confirmed by DNA sequencing, and pMSG4 was transformed into E. coli BL21-Codon Plus(DE3) cells.
Expression and purification of N-terminally hexahistidine-tagged PlmS3.
E. coli BL21-Codon Plus(DE3)/pMSG4 transformants were inoculated into 30 ml LB medium containing 50 μg/ml ampicillin and 25 μg/ml chloramphenicol and were grown at 220 rpm and 37°C overnight. Each overnight culture was added to 600 ml of LB containing 50 μg/ml ampicillin and 25 μg/ml chloramphenicol, and the cultures were grown at 220 rpm and 37°C until an optical density at 600 nm of 0.6 was reached. The cultures were transferred to 25°C and were induced with 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 5 h at 200 rpm. The cells were harvested by centrifugation. Cells from 1,200 ml culture were resuspended in 75 ml lysis buffer (50 mM sodium phosphate monobasic, 300 mM sodium chloride, and 10 mM imidazole, pH 8.0) containing lysozyme (0.5 mg/ml) and incubated on ice for 30 min. The cells were further ruptured by sonication, cell debris were removed by centrifugation, and the soluble fraction was applied to Ni-nitrilotriacetic acid (NTA) agarose (Qiagen) equilibrated with lysis buffer. The column was washed with 75 ml of wash buffer (50 mM sodium phosphate monobasic, 300 mM sodium chloride, and 20 mM imidazole, pH 8.0), and the protein was eluted with buffer containing 300 mM imidazole. Fractions of 500 μl were collected, and an aliquot of 10 μl was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis to check for the presence of PlmS3. Desired fractions were pooled and dialyzed using a Slide-A-Lyser dialysis cassette (3,500-molecular-weight cutoff) against 100 mM potassium phosphate buffer, pH 7.0. The protein concentration was determined with the Bio-Rad protein assay dye reagent concentrate, using bovine serum albumin as a standard. Aliquots of protein were stored at −80°C.
PLM production and analysis.
Production, purification, and analysis of PLM G and other PLM analogs using high-pressure liquid chromatography (HPLC) and liquid chromatography-mass spectrometry (LC-MS) were carried out as described previously with some modifications (21). The mobile phases A (acetonitrile-water, 20:80) and B (acetonitrile-water, 80:20) both contained 0.05% formic acid, and gradients from 0% B to 60% B over either 40 or 70 min (at a flow rate of 1 ml/min) were used. PLM G was stored as a concentrated solution (5 mM) in methanol.
In vitro acylation of PLM G.
Enzymatic conversion of PLM G into various PLM analogs was monitored with an HPLC assay. A standard assay was carried out by incubating the Ni-NTA affinity-purified PlmS3 (40 μg) in 100 mM potassium phosphate buffer (500 μl; pH 7.0) with PLM G (22 μM), acyl-CoA, or various NAC thioesters (500 μM) at 30°C for 1.5 h. The acyl-CoA substrates were purchased from Sigma-Aldrich, whereas NAC thioesters of various carboxylic acids were synthesized as described previously (1). Simultaneous control experiments in the absence of either the enzyme or substrate were conducted. The enzymatic reaction was stopped by addition of 125 μl acetonitrile (20%), and the reaction mixture was filtered. HPLC analysis of an aliquot under conditions previously described (21) allowed the loss of the PLM G peak and the formation of an acylated PLM product peak to be monitored. The mass of the new peak was determined by LC-MS.
For kinetic determination of PLM A synthesis, the Ni-NTA affinity-purified PlmS3 (6.8 μg) was incubated with isobutyryl-CoA (500 μM) and various concentrations of PLM G (from a 4.3 mM stock solution) in 100 mM potassium phosphate buffer (pH 7.0) in a reaction volume of 500 μl at 30°C for 15 min. The reaction rate was linear during this assay, and the peak area of the PLM A determined by HPLC analysis was used to calculate the rates of product formation. One unit of the enzyme activity was defined as the amount of enzyme yielding 1 nmol of product per min under these assay conditions.
Determination of the relative efficiency of the acylation reaction with various acyl thioester substrates (100 μM) was carried out using the Ni-NTA affinity-purified PlmS3 (6.9 μg) with PLM G (22 μM) and acyl-CoA/NAC in 100 mM potassium phosphate buffer (pH 7.0) in a reaction volume of 500 μl at 30°C for 15 min. The samples were analyzed by HPLC as stated above, using the loss of PLM G to determine the percentage of PLM G acylation with each substrate.
Deacylation of PLM A and PLMs C to F.
Deacylation of a mixture of natural PLMs by PlmS3 was monitored by HPLC analysis, looking at both loss of substrate and formation of the PLM G product. For kinetic assays, the enzyme (44 μg) was incubated with various concentrations of PLM A in 100 mM potassium phosphate buffer (pH 7.0) in a reaction volume of 500 μl at 30°C for 2 h.
Thioesterase activity of PlmS3.
The thioesterase activity of PlmS3 with acyl-CoA thioesters was determined spectrophotometrically by measuring an increase in A412, reflecting formation of 5-thio-2-nitrobenzoate as a result of reaction of the SH group of the released CoA-SH product with 5,5′-dithiobis-(2-nitrobenzoic acid) (DTNB) (9). For kinetic assays, the reaction mixture (500 μl) contained 100 mM potassium phosphate buffer (pH 7.0), DTNB (0.5 mM), PlmS3 (14.6 μg), and various concentrations of isobutyryl-CoA. The samples were incubated at 30°C for 30 min, and the absorbance was measured at 412 nm. The nanomoles of CoA-SH product formed were calculated using a standard curve for reaction of various concentrations of CoA-SH with DTNB.
RESULTS
In-frame deletion of plmS2-plmS3 in Streptomyces sp. strain HK 803.
The plmS3 ORF, adjacent to plmS2 in the plm biosynthetic gene cluster, encodes a protein with 25 to 30% amino acid identity to several putative hydrolytic enzymes (proteases, lipases, and esterases) which share a common alpha/beta hydrolase fold. A multiple alignment of PlmS3 and these proteins revealed that it contained the highly conserved serine (Ser106)-aspartate (Asp229)-histidine (His257) catalytic triad associated with these proteins. Only very low amino acid sequence similarity was observed between PlmS3 and other acyltransferases involved in post-polyketide synthase tailoring reactions.
Targeted disruption of the plmS2 gene has previously resulted in an NP1 mutant which produces only PLM B (21). A strain, NP8, in which both the adjacent plmS2 and plmS3 genes were replaced with the apramycin resistance gene was generated from Streptomyces sp. strain HK 803. The NP8 mutant, like the NP1 mutant, produced only PLM B, consistent with the proposal that PlmS2 and PlmS3 catalyze the final two steps in PLMs biosynthesis (Fig. 1).
Selective production of PLM G by the NP8/pMSG1′ strain.
Genetic complementation of a plmS2 deletion strain, NP2 (an NP1 derivative) by pMSG1 (a recombinant conjugative plasmid expressing the plmS2 gene under the control of the ermE* promoter) has previously resulted in an NP2/pMSG1 strain in which production of PLM A and PLMs C to F is restored (7). Plasmid pMSG1′ was generated from pMSG1 by facile allelic replacement of aac(3)IV by aadA, and the resulting spectinomycin resistance-conferring plasmid was transferred into the NP8 derivative strain by conjugation (Fig. 2). HPLC analysis of the fermentation products of the resulting strain, NP8/pMSG1′, revealed neither PLM B nor any acylated PLM products (PLM A and PLMs C to F). Rather, a new peak with a much shorter retention time was observed (Fig. 3). This new product coeluted with a PLM G standard (previously generated by PlmS2-catalyzed hydroxylation of PLM B [7]). LC-MS analysis of the fermentation broth confirmed the expected mass (529 Da) for PLM G, and nuclear magnetic resonance analyses of the purified product were consistent with the PLM G structure. As plmS3 is the only plm biosynthetic gene absent in NP8/pMSG1′, the selective production of a PLM G product clearly implicates PlmS3 as an 18-O-acyltransferase which catalyzes the final step in the biosynthesis of the PLMs.
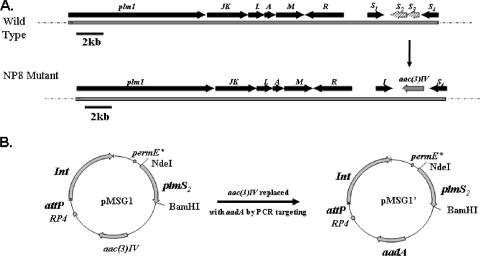
FIG. 2.
Genetic strategy for generation of the NP8/pMSG1′ derivative of Streptomyces sp. strain HK 803. (A) Replacement of plmS2-plmS3 by aac(3)IV using a PCR targeting method. (B) Replacement of aac(3)IV in the plmS2 expression plasmid (pMSG1) by aadA to generate pMSG1′.
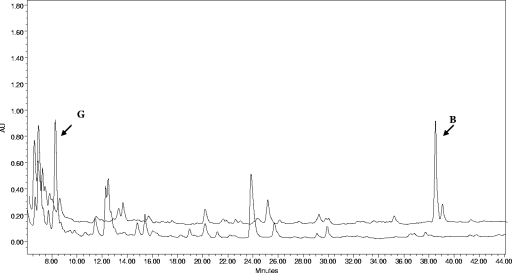
FIG. 3.
HPLC analysis demonstrating PLM B production by the NP8 derivative of Streptomyces sp. strain HK 803 (top trace) and PLM G production by NP8/pMSG1′ (plmS3 expression plasmid) (bottom trace).
Production of PLM G by NP8/pMSG1′ was typically about 6 mg/liter under standard fermentation conditions and was comparable to the yields of PLM B produced by NP8 or NP2 mutants. This mutant was used to obtain pure PLM G as a substrate for in vitro acylation assays with PlmS3.
Heterologous expression of PlmS3.
The 921-bp plmS3 ORF was cloned into prokaryotic expression vector pET15b, and PlmS3 was expressed as a soluble N-terminally polyhistidine-tagged protein in Escherichia coli BL21-Codon Plus(DE3) cells. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis of E. coli BL21(DE3)/pMSG4 cell extracts grown under IPTG induction conditions revealed the presence of relatively high levels of a 36-kDa protein (consistent with the predicted masses of PlmS3 and the 2-kDa hexahistidine tag). This expressed protein was absent in similarly prepared extracts of a control culture of E. coli/pET15b. PlmS3 was purified as a His-tagged protein by affinity chromatography on an Ni-NTA agarose column to more than 50% purity. The typical yields of purified PlmS3 were 1 to 1.5 mg/liter.
In vitro enzymatic conversions by recombinant PlmS3.
Commercially available isobutyryl-CoA and isovaleryl-CoA were incubated separately with the purified PLM G and Ni-NTA-purified PlmS3 in a potassium phosphate buffer (pH 7) at 30°C for 1.5 h. An HPLC assay which allowed the conversion of the PLM G substrate to either PLM A or PLM C clearly demonstrated the predicted acyltransferase activity of PlmS3 (Fig. 4). Coinjection of PLM A and PLM C standards and LC-MS analyses further confirmed that the predicted acylated PLMs were generated by PlmS3 incubations. The esterification of PLM G was PlmS3 dependent; no reaction in the absence of the enzyme was observed. The use of HEPES buffer instead of phosphate buffer did not alter the levels of PLM G acylation by PlmS3.
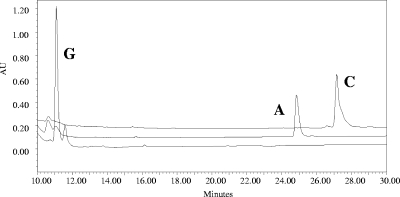
FIG. 4.
HPLC chromatogram demonstrating PlmS3-catalyzed esterification of PLM G (bottom trace) to yield PLM A (middle trace) and PLM C using isobutyryl-CoA and isovaleryl-CoA, respectively.
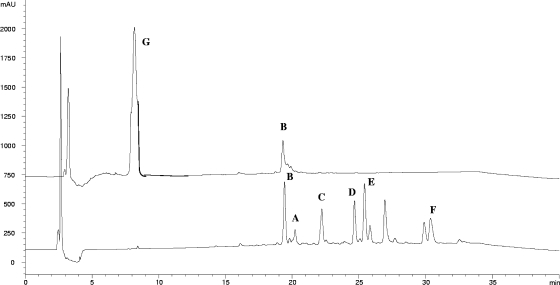
In addition to PLM G acylation activity, the recombinant PlmS3 exhibited the ability to hydrolyze both acyl thioesters and the acylated PLM products (PLM A and PLMs C to F). Prolonged incubation of a mixture of PLMs (isolated from the wild-type Streptomyces sp. strain HK 803 strain) with PlmS3 at 30°C resulted in conversion to PLM G (Fig. 5). The mixture also contained PLM B, in which the C-18 hydroxyl is absent, and this was predictably unchanged during the incubation. The recombinant PlmS3 also exhibited acyl-CoA thioesterase activity, releasing the free CoA-SH which was detected using a spectrometric DTNB assay (measuring an increase in A412 due to the formation of 5-thio-2-nitrobenzoate). This activity was observed with a range of short branched-chain acyl-CoA thioesters.
FIG. 5.
HPLC assay demonstrating PlmS3-catalyzed generation of PLM G (top trace) by hydrolysis of a mixture of acylated PLMs (bottom trace). PLM B, which does not contain an acylated C-18-hydroxyl substituent, is present at the beginning and end of the incubation.
Kinetic parameters of the reactions catalyzed by His-tagged-PlmS3.
Kinetic parameters for the acyltransferase activities of PlmS3 were established using an HPLC assay to monitor the acylation of PLM G with 500 μM isobutyryl-CoA (generation of PLM A). Kinetic parameters for the deacylation of PLM A by PlmS3 were assayed by HPLC, and those for isobutyryl-CoA hydrolysis were assayed using a DTNB spectrophotometric assay. As shown in Table 2 the PLM G acylation reaction (apparent kcat of 28 min−1) is significantly faster than the deacylation of the resulting PLM A product (apparent kcat of 0.5 min−1). Km values for PLM G (acylation) and PLM A (hydrolysis) were very similar. The rate of isobutyryl-CoA hydrolysis by PlmS3 was also low (apparent kcat of 3 min−1) relative to the acyl transfer reaction.
TABLE 2.
Apparent kinetic parameters of the catalytic activities exhibited by PlmS3
| Catalytic activity | Substrate | Mean ± SD
|
||
|---|---|---|---|---|
| Km(μM) | kcat (min−1) | kcat/Km (μM min−1) | ||
| Acyltransferase | PLM G | 88 ± 16 | 28 ± 3 | 0.32 ± 0.04 |
| Deacylation | PLM A | 92 ± 10 | 0.5 ± 0.05 | 0.005 ± 0.0005 |
| Thioesterase | Isobutyryl-CoA | 320 ± 50 | 3 ± 0.3 | 0.01 ± 0.014 |
Acyl side chain specificity and chemoenzymatic synthesis of new PLM analogs.
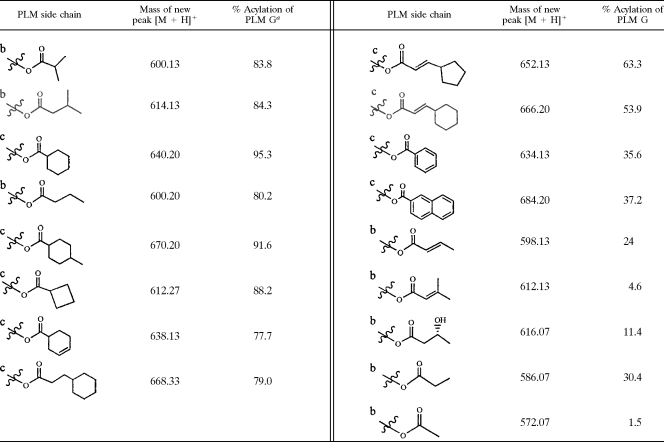
The absence of a single gene, plmS3, led to a mutant that was blocked in formation of all acylated PLMs. This observation and other properties of the recombinant enzyme (slow hydrolysis of all natural acylated PLMs) were consistent with a biosynthetic role of this enzyme in catalyzing acylation of PLM G with a range of acyl-CoA thioesters. This role indicates promiscuity with regard to the acyl thioester substrate, and we tested the extent of this using CoA-SH and NAC thioesters. As shown in Table 3, PlmS3 has a preference for generating PLM products with short, branched and aromatic side chains at C-18. Under a given set of conditions, comparable levels of new and known PLMs were generated using branched and cyclic acyl thioesters (Table 3, left). The use of aromatic, unsaturated, or even hydroxylated substrates was also possible (Table 3, right), even though the amounts of these PLM products were less under these same conditions. The use of more PlmS3 or prolonged incubation allowed us to increase the conversion rate and obtain good quantities of these new PLM products. Two products with identical m/z values were observed using dl-3-hydroxybutyryl-CoAs, indicating that both enantiomers were used as substrates by PlmS3, giving rise to two diastereomeric products. Long-chain acyl-CoAs were not substrates, indicating that strains which produce LSNs with longer acyl side chains must have a PlmS3 homolog with different substrate specificity. Thioesters of heterocyclic carboxylic acids were also not substrates for PlmS3.
TABLE 3.
Generation of 14 novel PLM compounds and 3 known PLM compounds by PlmS3-catalyzed acylation of PLM G
Calculated by determining loss of PLM G peak relative to control over a standard 500-ml assay mixture (PlmS3 [6.8 μg], PLM G [22 μM], or acyl-CoA [b] or NAC derivative [c] [100 μM] at 30°C for 15 min in 100 mM potassium phosphate buffer at pH 7.0).
DISCUSSION
The accumulation of PLM G by the NP8/pMSG1′ strain, together with the observation of PlmS3-catalyzed acylation of PLM G using various acyl thioester substrates, has provided unequivocal evidence that this enzyme alone is the 18-O-acyltransferase proposed to catalyze the final step in the PLM biosynthetic pathway. The yields of PLM G produced by this mutant are comparable to those for PLM B production by the NP8 mutant (5 to 10 mg/liter), and thus deletion of plmS3 in PLM-producing strains provides for a direct and effective fermentation route to access this PLM intermediate. Previously there have been only less-direct routes to PLM G, i.e., enzymatic hydroxylation of purified PLM B using the recombinant PlmS2 P450 monooxygenase (7) or enzyme-catalyzed deacylation of a mixture of acylated PLMs/LSNs using pig liver esterase (24).
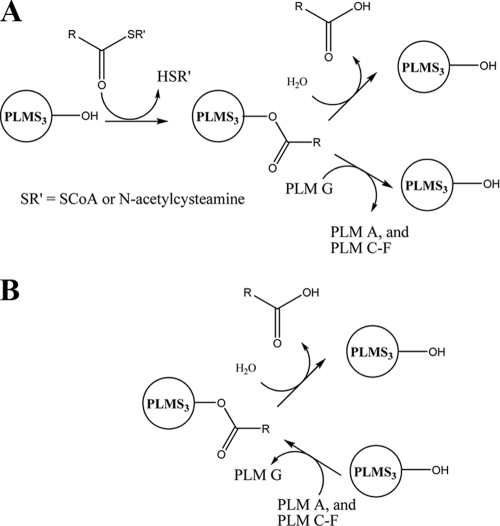
PlmS3 was most efficient in the ability to catalyze acylation of PLM G with the proposed physiological substrates, such as isobutyryl-CoA. Nonetheless, it also can catalyze hydrolysis of either acyl-CoA thioesters or a mixture of PLM A and PLMs C to F (providing PLM G). The predicted amino acid sequence revealed homology to the alpha/beta hydrolase fold superfamily of enzymes and the canonical catalytic triad associated with this group of enzymes. We envision that the acylation reaction proceeds via a ping-pong mechanism (Fig. 6A) in which the first step is formation of an acyl-enzyme intermediate using the conserved nucleophilic serine and the appropriate acyl-CoA substrate (releasing CoA as the first product). Binding of the second substrate, PLM G, then permits the acyl transfer generating the final PLM product and restoration of PlmS3 to its initial state. LovD, which catalyzes acylation of monacolin J to generate lovastatin, has also been proposed to proceed via such a ping-pong mechanism, and this has been supported by mutational and kinetic studies (30). In the case of PlmS3, the slow acyl-CoA hydrolysis reaction presumably involves the same initial formation of the acyl-enzyme intermediate. In this case the acyl group is released by reaction with water rather than the C-18 hydroxyl group of PLM G (Fig. 6A). The reversibility of the PlmS3-catalyzed reaction also means that the same hydrolytically susceptible acyl-enzyme intermediate can be generated by reacting an acylated PLM with PLM S3 (Fig. 6B), leading to the observed PlmS3-catalyzed deacylation (generation of PLM G) of PLM A and PLMs C to F. The hydrolytic activity of PlmS3 was significantly smaller than the acyltransferase activity, suggesting that in the active site PLM G can position more optimally than water for reaction with the acyl-enzyme intermediate. PlmS3 represents an alternate to pig liver esterase (24) for catalyzing formation of PLM G by hydrolysis of acylated PLMs.
FIG. 6.
Proposed role for a common acyl-enzyme intermediate in the reactions catalyzed by PlmS3.
The enzyme PlmS3 has significant promiscuity with respect to the acyl group that can be transferred to PLM G, suggesting a rather nonspecific acyl binding pocket in the enzyme. The ability of PlmS3 to transfer this acyl group to an alcohol, other than that at C-18 of PLM G, has not been evaluated due to a lack of appropriate PLM G analogs. Streptomyces sp. strain HK 803 and Streptomyces nigrescens SC273 have both been shown to produce five different acylated PLM products (4, 23). The structures of these suggest isobutyryl-CoA (PLM A), 3-methylbutyryl-CoA (PLM C), 4-methylbutyryl-CoA (PLM D), cyclohexylcarbonyl-CoA (PLM E), and 4-methylhexanoyl-CoA (PLM F) as the PlmS3 substrates. These acyl-CoAs are presumably provided by primary metabolic processes such as branched-chain amino acid catabolism and fatty acid biosynthesis/degradation. The exception is the cyclohexylcarbonyl-CoA which is generated by the PLM biosynthetic process and used to both initiate biosynthesis of the PLM core structure and convert PLM G to PLM E (21, 23). The demonstrated ability of the recombinant PlmS3 to preferentially use branched-chain acyl-CoA substrates is consistent with the range of PLM products generated in a fermentation. It is predicted that the ratio of these products is determined by the relative pools of acyl-CoA precursors and the substrate specificity of PlmS3. Furthermore, there may be lower levels of other naturally occurring acylated PLM products which have not yet been identified from these strains. These PLMs may be generated by PlmS3-catalyzed acylation of PLM G with other branched-chain acyl-CoAs or straight-chain acyl-CoA substrates (butyryl-CoA and crotonyl-CoA are both known primary metabolites [16] and substrates for this enzyme).
The promiscuity of PlmS3 toward acyl-CoA substrates and the resulting mixture of fermentation products create issues with generating adequate amounts of a specific acylated product. For instance, LSN B (PLM G acylated with a 6-methyloctanoic acid) has a number of interesting and potentially useful biological activities (17, 24, 25). The natural product is produced as a mixture of LSNs in Streptomyces platensis SANK 60191 and could be obtained only at low levels (<10 mg) by large-scale fermentation (60 liters) and considerable purification efforts. An alternative approach has been to isolate the mixture of LSNs, convert them to PLM G using an enzymatic hydrolysis, and then carry out chemical esterification of the C-18 by the Yamaguchi method (17). This 11-step process occurs in less than 2% overall yield and requires temporary removal of the C-9 phosphate monoester (by enzymatic hydrolysis) and a series of protection and deprotection steps with the other alcohol groups of PLM G. A one-step, potentially quantitative enzymatically catalyzed acylation of PLM G (using PlmS3 or the presumed homolog from the LSN B producer S. platensis) represents an attractive chemoenzymatic approach to generating specific acylated known PLMs.
Finally, PlmS3 offers a facile route to access new acylated PLMs. To date, all biological studies have focused on the known naturally occurring C-18-acylated PLMs. An initial analyses demonstrated comparable protein phosphatase 2A-inhibitory activity (29) and antifungal activity (5). However, we have shown recently that the acyl group is required for PLM antifungal activity against Rhodotorula glutinis and, in this case at least, plays an important role (7). PlmS3 can catalyze the attachment of a broader range of acyl substrates (including both aromatic and hydroxycarboxylic acids) than those found in the isolated PLMs. This enzymatic esterification, alongside established chemical methods for N acylation of PLM G (24) and other methods to alter the product of the PLM biosynthetic process, now offers new possibilities to generate libraries of PLM analogs which can be tested and evaluated for a range of biological activities and potential pharmaceutical or agricultural applications.
Acknowledgments
This work was supported by grant AI51629 from the National Institutes of Health.
The original Streptomyces sp. strain HK 803 was kindly provided by Hiroyuki Osada (Riken, Japan).
Footnotes
Published ahead of print on 20 March 2009.
REFERENCES
- 1.Alhamadsheh, M. M., N. Palaniappan, S. Daschouduri, and K. A. Reynolds. 2007. Modular polyketide synthases and cis double bond formation: establishment of activated cis-3-cyclohexylpropenoic acid as the diketide intermediate in phoslactomycin biosynthesis. J. Am. Chem. Soc. 129:1910-1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arisawa, A., N. Kawamura, T. Narita, I. Kojima, K. Okamura, H. Tsunekawa, T. Yoshioka, and R. Okamoto. 1996. Direct fermentative production of acyltylosins by genetically-engineered strains of Streptomyces fradiae. J. Antibiot. (Tokyo) 49:349-354. [DOI] [PubMed] [Google Scholar]
- 3.Arisawa, A., N. Kawamura, K. Takeda, H. Tsunekawa, K. Okamura, and R. Okamoto. 1994. Cloning of the macrolide antibiotic biosynthesis gene acyA, which encodes 3-O-acyltransferase, from Streptomyces thermotolerans and its use for direct fermentative production of a hybrid macrolide antibiotic. Appl. Environ. Microbiol. 60:2657-2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fushimi, S., K. Furihata, and H. Seto. 1989. Studies on new phosphate ester antifungal antibiotics phoslactomycins. II. Structure elucidation of phoslactomycins A to F. J. Antibiot. (Tokyo) 42:1026-1036. [DOI] [PubMed] [Google Scholar]
- 5.Fushimi, S., S. Nishikawa, A. Shimazu, and H. Seto. 1989. Studies on new phosphate ester antifungal antibiotics phoslactomycins. I. Taxonomy, fermentation, purification and biological activities. J. Antibiot. (Tokyo) 42:1019-1025. [DOI] [PubMed] [Google Scholar]
- 6.Ghatge, M., N. Palaniappan, S. Das Choudhuri, and K. Reynolds. 2006. Genetic manipulation of the biosynthetic process leading to phoslactomycins, potent protein phosphatase 2A inhibitors. J. Ind. Microbiol. Biotechnol. 33:589-599. [DOI] [PubMed] [Google Scholar]
- 7.Ghatge, M. S., and K. A. Reynolds. 2005. The plmS2-encoded cytochrome P450 monooxygenase mediates hydroxylation of phoslactomycin B in Streptomyces sp. strain HK803. J. Bacteriol. 187:7970-7976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gust, B., G. L. Challis, K. Fowler, T. Kieser, and K. F. Chater. 2002. REDIRECT technology: PCR-targeting system in Streptomyces coelicolor. John Innes Center, Norwich, United Kingdom.
- 9.Habeeb, A. F. S. A. 1972. Reaction of protein sulfhydryl groups with Ellman's reagent. Methods Enzymol. 25:457-464. [DOI] [PubMed] [Google Scholar]
- 10.Hara, O., and C. R. Hutchinson. 1992. A macrolide 3-O-acyltransferase gene from the midecamycin-producing species Streptomyces mycarofaciens. J. Bacteriol. 174:5141-5144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, United Kingdom.
- 12.Kohama, T., R. Enokita, T. Okazaki, H. Miyaoka, A. Torikata, M. Inukai, I. Kaneko, T. Kagasaki, Y. Sakaida, A. Satoh, et al. 1993. Novel microbial metabolites of the phoslactomycins family induce production of colony-stimulating factors by bone marrow stromal cells. I. Taxonomy, fermentation and biological properties. J. Antibiot. 46:1503-1511. [DOI] [PubMed] [Google Scholar]
- 13.Kohama, T., T. Nakamura, T. Kinoshita, I. Kaneko, and A. Shiraishi. 1993. Novel microbial metabolites of the phoslactomycins family induce production of colony-stimulating factors by bone marrow stromal cells. II. Isolation, physico-chemical properties and structure determination. J. Antibiot. 46:1512-1519. [DOI] [PubMed] [Google Scholar]
- 14.Koishi, R., C. Yoshimura, T. Kohama, and N. Serizawa. 2002. Leustroducsin B activates nuclear factor-κB via the acidic sphingomyelinase pathway in human bone marrow-derived stromal cell line KM-102. J. Interferon Cytokine Res. 22:343-350. [DOI] [PubMed] [Google Scholar]
- 15.Kruger, R., W. Lu, M. Oberthur, J. Tao, D. Kahne, and C. T. Walsh. 2005. Tailoring of glycopeptide scaffolds by the acyltransferases from the teicoplanin and A-40,926 biosynthetic operons. Chem. Biol. 12:131-140. [DOI] [PubMed] [Google Scholar]
- 16.Li, C., G. Florova, K. Akopiants, and K. A. Reynolds. 2004. Crotonyl-coenzyme A reductase provides methylmalonyl-CoA precursors for monensin biosynthesis by Streptomyces cinnamonensis in an oil-based extended fermentation. Microbiology 150:3463-3472. [DOI] [PubMed] [Google Scholar]
- 17.Matsuhashi, M., and K. Shimada. 2002. Chemical transformation of leustroducsins: synthesis of leustroducsin B. Tetrahedron Lett. 58:5619-5626. [Google Scholar]
- 18.Moss, S., L. Bai, S. Toelzer, B. J. Carroll, T. Mahmud, T. W. Yu, and H. G. Floss. 2002. Identification of asm19 as an acyltransferase attaching the biologically essential ester side chain of ansamitocins using N-desmethyl-4,5-desepoxymaytansinol, not maytansinol, as its substrate. J. Am. Chem. Soc. 124:6544-6545. [DOI] [PubMed] [Google Scholar]
- 19.Ozasa, T., K. Suzuki, M. Sasamata, K. Tanaka, M. Kobori, S. Kadota, K. Nagai, T. Saito, S. Watanabe, and M. Iwanami. 1989. Novel antitumor antibiotic phospholine. 1. Production, isolation and characterization. J. Antibiot. (Tokyo) 42:1331-1338. [DOI] [PubMed] [Google Scholar]
- 20.Ozasa, T., K. Tanaka, M. Sasamata, H. Kaniwa, M. Shimizu, H. Matsumoto, and M. Iwanami. 1989. Novel antitumor antibiotic phospholine. 2. Structure determination. J. Antibiot. (Tokyo) 42:1339-1343. [DOI] [PubMed] [Google Scholar]
- 21.Palaniappan, N., B. S. Kim, Y. Sekiyama, H. Osada, and K. A. Reynolds. 2003. Enhancement and selective production of phoslactomycin B, a protein phosphatase IIa inhibitor, through identification and engineering of the corresponding biosynthetic gene cluster. J. Biol. Chem. 278:35552-35557. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 23.Sekiyama, Y., N. Palaniappan, K. A. Reynolds, and H. Osada. 2003. Biosynthesis of phoslactomycins: cyclohexanecarboxylic acid as the starter unit. Tetrahedron Lett. 59:7465-7471. [Google Scholar]
- 24.Shibata, T., S. Kurihara, T. Oikawa, N. Ohkawa, N. Shimazaki, K. Sasagawa, T. Kobayashi, T. Kohama, F. Asai, A. Shiraishi, et al. 1995. Preparation of leustroducsin H and the structure-activity relationship of its derivatives. J. Antibiot. 48:1518-1520. [DOI] [PubMed] [Google Scholar]
- 25.Sugimura, Y., T. Shibata, K. Tamaki, K. Kurihara, T. Kohama, T. Shiraishi, T. Kobayashi, K. Sasagawa, and N. Shimazaki. April 1995. Leustroducsin H, its preparation and its therapeutic use. U.S. patent 5,409,912.
- 26.Teruya, T., S. Simizu, N. Kanoh, and H. Osada. 2005. Phoslactomycin targets cysteine-269 of the protein phosphatase 2A catalytic subunit in cells. FEBS Lett. 579:2463-2468. [DOI] [PubMed] [Google Scholar]
- 27.Tomiya, T., M. Uramoto, and K. Isono. 1990. Isolation and structure of phosphazomycin C. J. Antibiot. (Tokyo) 43:118-121. [DOI] [PubMed] [Google Scholar]
- 28.Uramoto, M., Y. C. Shen, N. Takizawa, H. Kusakabe, and K. Isono. 1985. A new antifungal antibiotic, phosphazomycin A. J. Antibiot. 38:665-668. [DOI] [PubMed] [Google Scholar]
- 29.Usui, T., G. Marriott, M. Inagaki, G. Swarup, and H. Osada. 1999. Protein phosphatase 2A inhibitors, phoslactomycins. Effects on the cytoskeleton in NIH/3T3 cells. J. Biochem. 125:960-965. [DOI] [PubMed] [Google Scholar]
- 30.Xie, X., K. Watanabe, W. A. Wojcicki, C. C. Wang, and Y. Tang. 2006. Biosynthesis of lovastatin analogs with a broadly specific acyltransferase. Chem. Biol. 13:1161-1169. [DOI] [PubMed] [Google Scholar]