Abstract
Third-generation cephalosporin resistance of Salmonella and commensal Escherichia coli isolates from cattle in the United States is predominantly conferred by the cephamycinase CMY-2, which inactivates β-lactam antimicrobial drugs used to treat a wide variety of infections, including pediatric salmonellosis. The emergence and dissemination of blaCMY-2--bearing plasmids followed and may in part be the result of selection pressure imposed by the widespread utilization of ceftiofur, a third-generation veterinary cephalosporin. This study assessed the potential effects of ceftiofur on blaCMY-2 transfer and dissemination by (i) an in vivo experimental study in which calves were inoculated with competent blaCMY-2-bearing plasmid donors and susceptible recipients and then subjected to ceftiofur selection and (ii) an observational study to determine whether ceftiofur use in dairy herds is associated with the occurrence and frequency of cephalosporin resistance in Salmonella and commensal E. coli. The first study revealed blaCMY-2 plasmid transfer in both ceftiofur-treated and untreated calves but detected no enhancement of plasmid transfer associated with ceftiofur treatment. The second study detected no association (P = 0.22) between ceftiofur use and either the occurrence of ceftiofur-resistant salmonellosis or the frequency of cephalosporin resistance in commensal E. coli. However, herds with a history of salmonellosis (including both ceftiofur-resistant and ceftiofur-susceptible Salmonella isolates) used more ceftiofur than herds with no history of salmonellosis (P = 0.03) These findings fail to support a major role for ceftiofur use in the maintenance and dissemination of blaCMY-2-bearing plasmid mediated cephalosporin resistance in commensal E. coli and in pathogenic Salmonella in these dairy cattle populations.
The major mechanism of third-generation cephalosporin resistance among U.S. human and veterinary clinical isolates of Salmonella enterica subsp. enterica is the beta-lactamase CMY-2 (12, 17, 43, 44, 46). blaCMY-2, which likely originated from the chromosomal AmpC locus of Citrobacter freundii, is disseminated among a group of similar plasmids harbored by diverse Enterobacteriaceae species (1, 2, 20, 26, 30, 31, 42, 45). In Salmonella, blaCMY-2-bearing plasmids have been observed in more than 30 serovars, notably including serovar Newport, which has gained specific attention from public health officials as a rapidly emerging threat (2, 6, 31).
Commensal Escherichia coli frequently harbors blaCMY-2-bearing plasmids (15, 33, 44), and these plasmids may be transferable to pathogens, since blaCMY-2 plasmids isolated from E. coli and S. enterica share extensive sequence similarity in addition to the blaCMY-2 open reading frame (5, 12, 42, 44). This transfer may occur in the gastrointestinal tracts of cattle, where these bacterial species periodically coexist and where transconjugants may be subjected to specific antimicrobial selection pressure. In fact, in vivo transfer of blaCMY-2 in the gastrointestinal tract has been reported between a Klebsiella pneumoniae blaCMY-2 plasmid donor and a Salmonella enterica serovar Typhimurium isolate in cattle and goats (29).
Ceftiofur is the only third-generation cephalosporin antimicrobial drug that is used in cattle production systems and is labeled for the treatment of pneumonia, postpartum metritis, necrotizing pododermatitis, and mastitis. Two ceftiofur preparations, ceftiofur sodium (Naxcel) and ceftiofur hydrochloride (Excenel) (Pfizer Animal Health, New York, NY), are unique in the veterinary pharmacopeia because they require no withholding and discard of milk collected from treated cows, making them frequent therapeutic choices in lactating animals (19, 35). Ceftiofur was licensed in 1988 (41) and its resistance in Salmonella spp. isolated from U.S. cattle, presumably conferred by blaCMY-2, was first documented in 1998 (6).
The effects of ceftiofur use on selection of blaCMY-2-bearing commensal E. coli has been examined for cattle both epidemiologically and experimentally. Tragesser et al. studied 18 Ohio dairy herds and determined that the 11 herds that used ceftiofur in any capacity (labeled indications and/or extralabel use) were 25 times more likely to have E. coli with reduced susceptibility to ceftriaxone (an expected blaCMY-2 phenotype) than the seven herds that reported no ceftiofur use (40). Interestingly, however, within eight herds that had detailed treatment records, no association was detected between the prevalence of E. coli with reduced susceptibility to ceftriaxone and use of ceftiofur on an individual-animal basis (40). In an experimental study by Jiang et al., ceftiofur administered to dairy calves was correlated with a 14% increase in ceftriaxone-resistant fecal E. coli compared to untreated controls (21). Together, these studies show a correlation between selection pressure within the gastrointestinal tracts at the individual-animal level and show that ceftiofur use may promote the dissemination of resistance in commensal E. coli at the whole-herd level.
Whether or not ceftiofur treatment directly affects in vivo horizontal transfer of blaCMY-2-bearing elements among E. coli and Salmonella has yet to be addressed. The diversity of blaCMY-2 plasmid-bearing bacterial hosts is consistent with wide dissemination of this genetic element. One hypothesis that could explain this wide dissemination is that ceftiofur may itself promote the in vivo horizontal transfer of blaCMY-2-bearing plasmids. Specifically, due to the relatively slow bactericidal activity of aminothiazolyl cephalosporins such as ceftiofur, it has been suggested that exposure to these compounds promotes filament formation in gram-negative bacteria prior to cell death that may increase the surface area and increase receptiveness of the cells for resistance plasmids (11).
Because blaCMY-2 may be disseminated by horizontal transfer of R plasmids and/or clonal expansion of individual strains, we examined the effect of ceftiofur use on these processes with two approaches; the first approach specifically considered the issue of horizontal transfer in an experimental in vivo calf model, while the second approach, a field study, assessed the overall relationship between ceftiofur use and blaCMY-2 prevalence in the primary agricultural animal niche where it is used.
MATERIALS AND METHODS
Bacterial strains.
Strains used and their plasmid content are described in Table 1 and Fig. 1, respectively. E. coli strains used were originally isolated from healthy cattle from different Pacific Northwest dairy herds by plating feces directly onto VRB-MUG agar (Accumedia, Lansing, MI) and incubating at 37°C overnight. Lactose-fermenting, beta-glucuronidase-positive colonies were subsequently streaked for isolation on Columbia agar supplemented with 5% sheep blood (Hardy Diagnostics, Santa Maria, CA), and isolated colonies were stored in brain heart infusion broth (BBL, Franklin Lakes, NJ) with 10% glycerol at −86°C. S. enterica strains were provided by the Washington Animal Disease Diagnostic Laboratory and the New York State Department of Health. Selectable markers were obtained by plating turbid broth cultures on MacConkey agar supplemented with either rifampin (75 μg/ml; Sigma, St. Louis, MO) or nalidixic acid (20 μg/ml; Sigma) followed by overnight incubation at 35°C. Colonies were picked and streaked onto the selective plates again and incubated overnight at 35°C to obtain isolated colonies to be used as the calf inocula.
TABLE 1.
Plasmid donor strains and in vitro-competent recipient strains
| Strain identificationa | Source | Yr isolated | Resistance phenotypeb |
|---|---|---|---|
| Plasmid donors | |||
| E793 | Healthy calf feces | 2002 | AMP CAZ CHL KAN SXT TET SSS STR (RIF) |
| E2699 | Healthy calf feces | 2002 | AMP CAZ CHL KAN SSS SXT GEN TET STR (RIF) |
| Recipients | |||
| E3679 | Healthy cow feces | 2002 | (NAL) |
| E4069 | Healthy cow feces | 2003 | (NAL) |
| E4079 | Healthy cow feces | 2003 | (NAL) |
| E4126 | Healthy cow feces | 2003 | (NAL) |
| S10801 | Mesenteric lymph nodec | 2005 | AMP CHL SSS STR TET (NAL) |
| S13812 | Human (clinical)d | 2004 | (NAL) |
The “E” prefix indicates E. coli; the “S” prefix indicates S. enterica
Abbreviations in boldface denote phenotypes correlated with the presence of the blaCMY-2 bearing plasmid characterized by Daniels et al. (12). AMP, ampicillin; CAZ, ceftazidime, CHL, chloramphenicol; GEN, gentamicin; KAN, kanamycin; NAL, nalidixic acid; SSS, triple sulfa (BD); TET, tetracycline; STR, streptomycin. Parentheses indicate phenotypes used for selectable markers.
Serotype Typhimurium, isolated from a calf with sepsis.
Serotype Newport, courtesy of the New York State Department of Health.
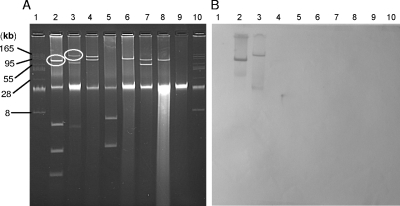
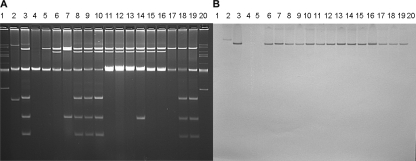
FIG. 1.
(A) Plasmid content of donor and recipient bacteria listed in Table 1. blaCMY-2 plasmids from the donors are circled. Lanes: 1 and 10, BacTracker supercoiled DNA ladder; 2, E793; 3, E2699; 4, E3679; 5, E4069; 6, E4079, 7, E4126; 8, S10801; 9, S13812. (B) Southern hybridization of the gel from panel A with a full-length blaCMY-2 probe.
The six recipient strains were determined to be capable of receiving the blaCMY-2-containing resistance plasmids from both of the donor strains in filter mating experiments. Briefly, 10 μl of turbid broth cultures of each donor/recipient combination was mixed and spotted onto 1-cm2 sterile nitrocellulose pieces overlying Columbia agar plus 5% sheep blood (Hardy) and incubated overnight at 35°C. Nitrocellulose pieces were suspended in 3 ml sterile 1× phosphate-buffered saline, and 0.3-ml volumes of the suspensions were plated onto LB agar supplemented with nalidixic acid (20 μg/ml) and ceftazidime (8 μg/ml). Growth was compared with donor-only and recipient-only negative controls.
Stocks used for calf inocula were prepared from turbid LB broth overnight cultures by adding 50% (vol/vol) sterile glycerol (Sigma) to obtain a final glycerol concentration of 10%. The bacterial/glycerol suspensions were then aliquoted into individual 1-ml doses (E. coli, ca. 109 CFU) or 10-ml doses (S. enterica, ca. 1010 CFU) and stored at −80°C until the day of use. Representative aliquots of each stock were thawed, serially diluted, and spiral plated using a WASP II spiral plater (Don Whatley Scientific, West Yorkshire, United Kingdom) onto MacConkey agar for quantification.
Plasmid detection was accomplished using the alkaline lysis method described by Kado and Liu (22). Aliquots (20 μl) of lysates were loaded into a 1× Tris-acetate-EDTA gel containing 1.1% agarose (Genepure LE; ISC BioExpress, Kaysville, UT) and run 3 h at 3 V/cm. A supercoiled DNA ladder (BacTracker; Epicentre Technologies, Madison, WI) was used to estimate plasmid size.
Animal subjects: in vivo experimental determination of blaCMY-2 plasmid transfer in calves.
Ten male Holstein (n = 6) or Jersey (n = 4) calves between 3 and 4 months of age that received no antimicrobial agents either parenterally or orally administered since birth were used in this study. Calves were obtained as age-matched pairs to permit random allocation into ceftiofur and control treatment groups. Animals were isolated in individual temperature-controlled 120-ft2 rooms upon arrival and were fed alfalfa hay and water ad libitum for the duration of the study. Calf grain (Animax; Purina, St. Louis, MO) was also provided once daily to the calves per the manufacturer's recommendations. Disinfectant footbaths containing 1% VirkonS (Antec, Suffolk, United Kingdom) were used by all personnel entering or exiting each animal room. Disposable gloves and room-specific coveralls and full-length boot covers were used when handling animals to prevent fomite-mediated transmission between rooms. Animal manipulations were divided into three phases: (i) pretreatment phase (days −7 to −1), (ii) treatment phase (days 0 to 8), and (iii) posttreatment phase (days 9 to 14). Fecal samples for bacteriologic analyses were obtained daily from each calf through all three phases. All studies for each pair of calves were completed and the isolation rooms were cleaned and disinfected prior to acquisition of the next pair of calves.
Pretreatment phase.
Upon arrival at housing units, one dose of intramuscular ceftiofur (ceftiofur sodium; Pfizer, NY; 2.2 mg/kg) was administered (in accordance with label instructions) to each calf to increase the likelihood of detection of resident ceftiofur-resistant bacteria that could potentially act as plasmid donors (although data obtained subsequently indicated that this treatment probably did not alter this likelihood) (38). Feces were sampled and cultured daily for seven days to detect preexisting ceftiofur-, nalidixic acid-, or rifampin-resistant flora using culture methods that are described below.
Treatment phase.
On days when recipient strains were administered to the calves, all six strains were given to each animal as a mixture of the thawed glycerol stocks; likewise, on days when donor strains were administered to the calves, both donor strains were given to each animal as a mixture. Donor and recipient strains were always administered on different days in order to decrease the chance of the occurrence of bacterial conjugation in the mouths of the animals.
Calves were inoculated with recipient (days 0, 2, 4, and 6) and donor (days 1, 3, and 5) stocks by oral drench. As the recipient stock included Salmonella enterica in high numbers, calves were monitored for clinical signs consistent with salmonellosis, including diarrhea, inappetence, and dehydration.
Ceftiofur (ceftiofur sodium; 2.2 mg/kg) was administered intramuscularly to the calf assigned to the treatment group beginning on treatment day 2 and subsequently every 24 h for 5 doses. Thus, calves had received both donor and recipient strains prior to administration of the first dose of ceftiofur.
Posttreatment phase.
Alternate-day inoculations with donor and recipient stocks were continued for five additional days (days 9 to 13). This phase served as a within-calf control period during which no further exposure to ceftiofur occurred.
Animal subjects: herd-level association of ceftiofur use with prevalence of ceftiofur resistance.
Feces were collected from lactating cows of 42 dairy herds in Washington State between June 2006 and September 2007. The average number of lactating cows in the study herds was 1,490 (range, 200 to 7,150). Feces (ca. 20 g) were combined into pools of 10 samples in sterile plastic bags (Whirl-pac; Nasco, Ft. Atkinson, WI) and manually mixed prior to spiral plating using the dilution scheme described later.
Confirmed cases of clinical salmonellosis involving ceftiofur-resistant strains in the study herds between January 2003 and September 2007 were identified through the records of the Washington Animal Disease Diagnostic Laboratory (Pullman, WA) with the permission of producers. Similarly, with producers' permission, purchase records of all ceftiofur formulations were obtained via on-farm receipts, computer records, and/or supply-house facsimiles of products dispensed. Ceftiofur use was expressed as milligrams ceftiofur/lactating cow/month for each study herd.
Culture methods.
Spiral plating (WASP II spiral plater) was used for all quantitative bacteriologic cultures. Two primary dilutions of feces in 1× phosphate-buffered saline (1:10 and 1:600) were used for enumeration of colonies between 3 × 102 CFU/g (the detection limit) and 2.4 × 108 CFU/g, using the 200-μl log and 50-μl log settings on the spiral plater, respectively. The 1:10 dilution was poured through a glass fiber filter (catalog no. A75448; Labconco Corp., Kansas City, MO) fitted to a sterile Osmonics filter funnel (GE Water and Process Technologies) atop a 125-ml sterile vacuum filter flask to remove large particulates prior to analysis. Results from two sector pairs on the polar counting grid were averaged and used for each count. When fewer than 35 colonies were detected in a sector, the entire plate area was counted.
Total E. coli was enumerated on VRB-MUG agar, using a long-wavelength UV light source to identify fluorescent (beta-glucuronidase-positive) colonies. Throughout this study, ceftazidime was used for selection of blaCMY-2-expressing isolates (3). For detection of resistant strains, antimicrobial agents were added to MacConkey agar (Hardy) in the following concentrations: ceftazidime, 8 μg/ml (Sigma); nalidixic acid, 20 μg/ml (Sigma); and rifampin, 75 μg/ml (Sigma). Ceftazidime was combined with either rifampin or nalidixic acid in the plating media to detect donor and transconjugant strains, respectively, in the calf inoculation study. Concentrated rifampin stock was prepared in dimethyl sulfoxide (Fisher, Pittsburgh, PA) and stored at 4°C (24). Cultures were incubated at 37°C overnight.
For the herd-level study only, up to ten lactose-fermenting ceftazidime-resistant colonies from each plate were transferred to EC-MUG agar (Hardy) to estimate the proportion of presumptive E. coli based on β-glucuronidase activity (25). Fifty MUG-negative lactose-fermenting colonies, each originating from a different fecal sample, were analyzed using the API20E system (Biomerieux, Hazelwood, MO) to estimate the proportion of MUG-negative colonies that were E. coli.
Salmonella spp. were detected by enrichment cultures of the same pooled fecal specimens used for the E. coli enumeration. Briefly, 10 g of feces was inoculated into 100 ml tetrathionate broth (Hardy), incubated at 37°C for 24 h, subcultured to XLD agar plates, and incubated at 37°C overnight (Accumedia). Identification of Salmonella spp. was confirmed by appropriate reactions with lysine iron agar (Accumedia), triple sugar iron agar (Accumedia), urea agar (Hardy), oxidase test (Hardy), and agglutination in poly-O antisera (Hardy). Salmonella serotyping was performed by the National Veterinary Service Laboratory (Ames, IA). For the calf study only, poly-B and poly-C2 antisera (Hardy) were used to distinguish between the two Salmonella recipient strains listed in Table 1.
Antimicrobial susceptibility testing.
Antimicrobial susceptibility testing was performed using the Kirby-Bauer agar diffusion method in accordance with CLSI guidelines and interpretive criteria (8). Briefly, a 0.5 McFarland turbidity suspension of a pure culture in brain heart infusion broth (Hardy) was spread onto a 150-mm petri plate containing Mueller-Hinton agar (Hardy) and the following antimicrobial-containing discs were applied: ampicillin (10 μg), amoxicillin-clavulanate (20 μg/10 μg), chloramphenicol (30 μg), gentamicin (10 μg), kanamycin (30 μg), sulfamethoxazole-trimethoprim (23.75 μg/1.25 μg), tetracycline (30 μg), ceftazidime (30 μg), ceftazidime-clavulanate (30 μg/10 μg), nalidixic acid (30 μg), streptomycin (10 μg), and triple sulfa (0.25 mg) (BD, Sparks, MD). Susceptibility testing of clinical Salmonella isolates by the Washington Animal Disease Diagnostic Laboratory was performed with broth microdilution with the BOP06F Sensititre plate (Trek Systems, Cleveland, OH) in accordance with manufacturer's instructions and CLSI guidelines (7).
Detection of blaCMY-2.
A colony representing each positive plate from the field study was propagated in 200 μl LB broth containing ceftazidime (8 μg/ml) in 96-well plates. A 96-pin replicator (Boekel Scientific, Feasterville, PA) was used to subculture to 150-mm-diameter LB agar plates along with positive and negative control strains (E. coli strains E2699 [12] and C600 [ATCC 23724], respectively) and incubated overnight at 37°C. Bacterial colony DNA was then immobilized on MagnaLift nylon membranes (GE Water Process Technologies) using a standard procedure (36).
Plasmid profile gels of representative putative in vivo transconjugants from the calf study were transferred to MagnaLift nylon membranes using a standard procedure (36).
A digoxigenin-labeled full-length (1,146-bp) blaCMY-2 probe was generated with the primers cmy-F (5-GACAGCCTCTTTCTCCACA-3) and cmy-R (5-TGGAACGAAGGCTACGTA-3) (46) using the DIG-PCR kit (Roche, Basel, Switzerland) following the manufacturer's instructions. A gel-purified blaCMY-2 PCR product generated from total DNA isolated from E. coli strain W1559 (23) was used as a template in the PCR for probe generation, ensuring that only blaCMY-2 sequence was amplified. Reaction conditions were as follows: denaturation 95°C (2 min) followed by 35 cycles of 95°C (30 s) to 48°C (30 s) to 72°C (45 s) in an iCycler (Bio-Rad, La Jolla, CA).
Nylon membranes were hybridized with the digoxigenin-labeled blaCMY-2 probe using Easy-hyb buffer (Roche) overnight at 42°C followed by stringent washes per the manufacturer's instructions. For the colony lifts, CDP-Star (Roche) was used for probe detection, followed by a 1-min exposure to radiography film (Hyperfilm, Amersham, Buckinghamshire, United Kingdom). For the membranes containing DNA transferred from the plasmid profile gels, probe detection was facilitated using NBT/BCIP (Roche) per the manufacturer's instructions.
Discrimination of transconjugants from plasmid donor strains.
Because of the potential for spontaneous mutations conferring nalidixic acid resistance in donor strains, we applied a genetic test to confirm that dual ceftazidime-nalidixic acid-resistant colonies were true transconjugants. The recipient and donor E. coli strains selected for this study did not share any flagellar alleles based on HhaI restriction fragment length polymorphism (RFLP) of fliC PCR products (28); therefore, transconjugants should have flagellar antigens consistent with one or more of the recipient strains. Consequently, HhaI RFLP of fliC PCR products from donor strains, recipient strains, and up to 10 putative transconjugants per calf was performed according to the method described by Machado et al. (28).
Data analysis.
The frequency of transconjugant isolation was compared between treatment and control groups using a repeated-measures analysis of variance with treatment group and day as fixed effects and subject animal as a random effect (SAS 9.1; SAS Institute Inc., Cary, NC).
Separate univariate linear regression models were used to examine the relationship between ceftiofur use (expressed as milligrams/cow/month) and each of the following dependent variables: (i) proportion of E. coli with decreased susceptibility to ceftazidime, (ii) mean frequency of E. coli with decreased susceptibility to ceftazidime, and (iii) animal prevalence of E. coli with decreased susceptibility to ceftazidime, all using herd as the unit of observation (NCSS, Kaysville, UT). Separate univariate linear regression models were also used to examine the relationship between herd size (number of lactating cattle) and (i) ceftiofur use and (ii) proportion of E. coli with decreased susceptibility to ceftazidime. The Mann-Whitney test (http://elegans.swmed.edu/∼leon/stats/utest.cgi) was used to compare ceftiofur use between herds with histories of clinical or environmental ceftiofur-resistant Salmonella.
RESULTS
In vivo experimental determination of blaCMY-2 plasmid transfer in calves. (i) Colonization of experimental calves with donor and recipient inocula.
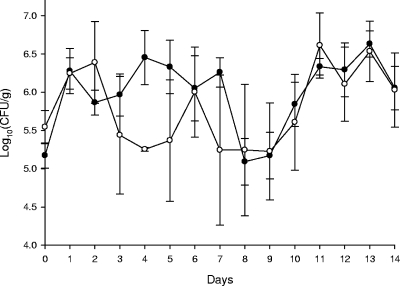
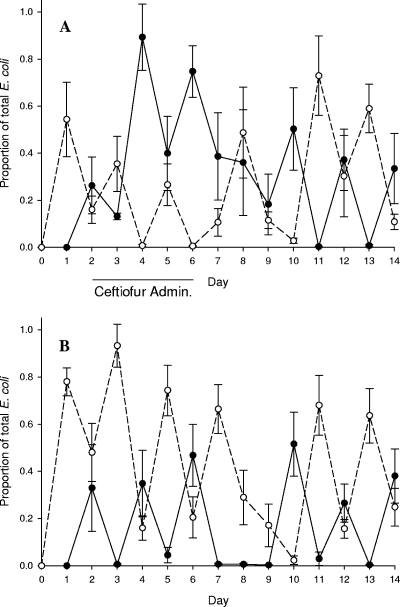
During the pretreatment phase, no coliform bacteria with the ceftazidime, rifampin, or nalidixic acid resistance characteristics of the donor, recipient, or putative transconjugants strains were recovered from any of the calves. Both donor (Rifr Cazr) and recipient (Nalr) E. coli phenotype strains were isolated from fecal specimens of all calves, beginning in every case on the day following the initial oral dose administration of these strains. During the treatment phase, calves receiving ceftiofur had decreased numbers of total fecal E. coli, reaching a minimum on day 4 (Fig. 2). Presumptive donor and recipient E. coli strains comprised substantial proportions of the E. coli fecal flora throughout the experiment in both treatment and control calves. However, ceftiofur treatment transiently favored the donor strains, while in the control nontreated calves, the inoculum strains administered the previous day predominated (Fig. 3A and B).
FIG. 2.
Mean fecal coliform concentrations in ceftiofur-treated and control (nontreated) calves. Bars represent one standard error of the mean. Open circles denote treatment group animals. Ceftiofur was administered on days 2, 3, 4, 5, and 6.
FIG. 3.
Mean proportions of donor (Rifr Cazr; closed circles) and recipient (Nalr; open circles) E. coli strains relative to total fecal coliforms in ceftiofur-treated calves (A) and control (nontreated) calves (B). Bars indicate one standard error of the mean.
Non-lactose-fermenting Nalr (presumptive Salmonella recipient strain) colonies were cultured from all calves' fecal specimens on at least one and up to six days. When present, non-lactose-fermenting Nalr colonies never exceeded 1% of the total lactose-fermenting colony counts from the same fecal specimens. No calves showed signs of clinical salmonellosis during these experiments.
(ii) In vivo transconjugants.
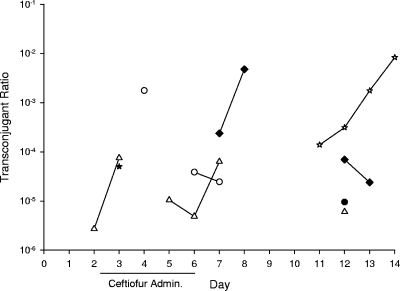
Lactose-fermenting Nalr Cazr colonies (presumptive E. coli transconjugants) were isolated from three calves each in the treatment and control groups, and there was no significant difference in average transconjugant frequencies between the groups (F = 0.42, P = 0.54). No tendency for increased detection of transconjugants was observed during the ceftiofur treatment phase. Non-lactose-fermenting Nalr Cazr colonies (presumptive Salmonella transconjugants) were cultured from one calf each from treatment and control groups. The rates of plasmid transfer from donors to recipient strains in vivo were estimated by average and maximum transconjugant ratios (quotient of Nalr Cazr lactose-fermenting colonies and total E. coli), as reported in Table 2 and Fig. 4. Representative putative E. coli transconjugants (n = 39) and S. enterica transconjugants (n = 15) contained plasmids indistinguishable in size from the donor strain E793 plasmid (Fig. 5).
TABLE 2.
In vivo generation of blaCMY-2 transconjugant E. coli in experimental calves
| Calfa | Resistance ratiob
|
Day of first detection | Duration of detection (days) | |
|---|---|---|---|---|
| Mean | Maximum | |||
| C1 | 7.26E−07 | 9.43E−06 | 12 | 1 |
| T1 | 1.41E−04 | 1.76E−03 | 4 | 3 |
| C2 | 3.92E−04 | 4.76E−03 | 7 | 4 |
| T2 | 0 | 0 | NAc | NA |
| C3 | 0 | 0 | NA | NA |
| T3 | 0 | 0 | NA | NA |
| C4 | 3.85E−06 | 5.00E−05 | 3 | 1 |
| T4 | 8.11E−04 | 8.33E−03 | 11 | 4 |
| C5 | 0 | 0 | NA | NA |
| T5 | 1.25E−05 | 7.50E−05 | 2 | 6 |
| C (mean) | 7.93E−05 | 9.64E−04 | NA | 2 |
| T (mean) | 1.93E−04 | 2.03E−03 | NA | 4.3 |
A “T” prefix indicates the treatment group; a “C” prefix indicates the control group.
Resistance ratio = transconjugants (CFU/g)/total fecal E. coli (CFU/g).
NA, not applicable.
FIG. 4.
Transconjugant ratios (Nalr Cazr/total coliforms) for individual calves. Open symbols signify treatment group animals.
FIG. 5.
(A) Plasmid content of donor strains, two recipient negative controls for Southern hybridization and representative transconjugants. Lanes: 1 and 20, BacTracker supercoiled DNA ladder; 2, E2699 (donor); 3, E793 (donor); 4, S13812 (recipient); 5, S10801 (recipient); transconjugant lanes: 6 to 9, 15, and 16, E. coli from animal T1; 10 to 14, E. coli from animal C2; 17 poly-B-positive S. enterica from animal T5; 18 and 19, poly-B-positive S. enterica from animal T4. (B) Southern hybridization of gel from panel A with full-length blaCMY-2 probe.
HhaI RFLP patterns of fliC PCR products generated from E. coli transconjugants in all cases corresponded to those of one of the recipient strains, indicating that colonies with resistance to both nalidixic acid and ceftazidime were not spontaneous nalidixic acid-resistant mutants of the donor strains but instead represented true transconjugants. Non-lactose-fermenting Nalr Cazr isolates demonstrated agglutination in poly-B antiserum, consistent with the S. Typhimurium recipient.
Herd-level association of ceftiofur use with prevalence of ceftiofur resistance: density of E. coli with decreased susceptibility to ceftazidime in dairy herds and blaCMY-2 prevalence.
A mean of 28 fecal pools per herd (range, 10 to 44) were collected on a mean of 4.5 collection dates (range, 3 to 6) for determination of the prevalence and concentration of ceftiofur-resistant E. coli on the dairy farms in this study. Coliforms with decreased susceptibility to ceftazidime were isolated from 41 of the 42 study herds. Coliforms with decreased susceptibility to ceftazidime across all herds comprised 0.7% (standard deviation = 1.2%) of the total coliform flora. Among 367 colonies with decreased susceptibility to ceftazidime, one colony representing each selective plate with growth, 328 (89%) were positive for blaCMY-2 by colony lift hybridization. The correlation between lactose fermentation and positive MUG reaction was high (r = 0.97), as expected (25). Among the 50 MUG-negative colonies selected for API20E biochemical analysis, 35 were identified as E. coli (API confidence level ≥ 85.5%), 13 had low confidence identifications, and 2 were identified as Klebsiella oxytoca (confidence level of 97.1%). All 35 of the API20E-identified MUG-negative E. coli isolates fermented sorbitol and therefore were unlikely to be serotype O157:H7.
Ceftiofur use varied widely among farms, ranging from 0 to 980 mg/lactating cow/month (approximately 0 to 0.5 dose/animal/month). Four farms reported no ceftiofur use, including three organic farms that used no antimicrobial drugs of any kind on their premises. Ceftiofur use was not correlated with the frequency of E. coli isolates with decreased susceptibility to ceftazidime, whether expressed as CFU/g feces (r = 0.035, P = 0.827) or as the proportion of resistant E. coli colonies (r = 0.145, P = 0.360). The prevalence of cattle from which fecal E. coli with reduced susceptibility to ceftazidime was detectable was also uncorrelated with ceftiofur use (r = 0.07). Finally, neither the intensity of ceftiofur use nor the proportion of E. coli with decreased susceptibility to ceftazidime was associated with herd size (r = 0.018 and r = 0.107, respectively).
Fifteen of the 39 study herds with accessible diagnostic records had a history of clinical salmonellosis due to ceftiofur-resistant (MIC ≥ 8 μg/ml) Salmonella serovar Dublin, Newport, or Typhimurium since January 2003. These 15 herds with a history of clinical salmonellosis due to ceftiofur-resistant Salmonella tended to have higher ceftiofur use than herds with no history of ceftiofur-resistant salmonellosis, although the pattern was not statistically significant (Mann-Whitney U = 223, two-tailed P = 0.22). Eight of the fifteen herds with a history of clinical ceftiofur-resistant salmonellosis also had experienced clinical salmonellosis due to ceftiofur-susceptible strains, whereas only four of the twenty-four herds with no clinical history of ceftiofur-resistant Salmonella had diagnosed salmonellosis due to ceftiofur-susceptible strains (P = 0.02, Fisher's exact test). Herds with any history of salmonellosis (including either ceftiofur-resistant or ceftiofur-susceptible; n = 20) used more ceftiofur than herds with no history of salmonellosis (n = 19) (Mann-Whitney U = 268, two-tailed P = 0.03).
Enrichment cultures of pooled fecal specimens from 20 of the 42 study dairies yielded Salmonella spp. with decreased susceptibility to ceftazidime (zone of inhibition ≤ 17 mm). Ceftiofur use among these 20 herds did not differ significantly from use on the other 22 study herds (Mann-Whitney U = 273, two-tailed P = 0.18).
DISCUSSION
The goal of this study was to examine the impact of ceftiofur, the antimicrobial agent most widely used in dairy cattle (19), on the dissemination of an antimicrobial resistance gene, blaCMY-2, among Salmonella (zoonotic pathogens of significant public health importance) and commensal E. coli (a putative reservoir of blaCMY-2 resistance plasmids) (12, 44, 46). Two complementary approaches were employed: an in vivo experimental inoculation model and a multiple-herd-based field study. The in vivo study focused on the horizontal dissemination and subsequent selection of novel transconjugants bearing blaCMY-2 resistance elements, while the field study focused on the dissemination and selection of these resistance elements in a broader sense, determining whether the blaCMY-2--expressing bacterial populations in real-world dairy production settings varies with the amount of ceftiofur used.
In vivo horizontal transfer of blaCMY-2 plasmids among E. coli and Salmonella was detected, albeit at a low frequency and with no evidence of enhancement of transfer under ceftiofur selection pressure. In addition, total fecal E. coli in ceftiofur-treated calves was transiently suppressed by ceftiofur treatment with a corresponding transient increase in the proportion of the E. coli flora with decreased resistance to ceftazidime. This observation was consistent with observations by other investigators (21, 27, 38). These changes in the composition of the fecal flora clearly illustrate selective pressure on the gastrointestinal flora due to the parenteral use of ceftiofur administered in accordance with its labeled dosage.
One remarkable finding of the current study that may be indirectly relevant to the epidemiology of third-generation cephalosporin resistance in the cattle enteric commensal flora is the dramatic impact of single oral doses of the defined blaCMY-2 donors and cephalosporin-susceptible recipient E. coli strains on the composition of the fecal enteric flora of the calves on the following day. For example, the recipient strains accounted for greater than 50% of the total fecal E. coli population on day 1 of the study, and the donor strains accounted for greater than 20% on day 2. Similar dominant effects on the fecal flora composition following oral E. coli inoculation have been seen previously when the inoculated strain was serotype O157:H7 (10, 39). The amplification of these E. coli strains suggests that introduction of these strain types by animal movements (for example, following cattle purchase or wildlife contacts) or by purchase of contaminated feeds in commerce (13) may be sufficient to disseminate and amplify potentially problematic clones. If so, the resultant regional or national dissemination could make it much more difficult to demonstrate the effects of drug usage on an individual farm basis, as we attempted to do in this study. The epidemiology of the fecal E. coli flora of cattle may be critical to a full understanding of the dissemination of antimicrobial drug resistance.
In contrast to the high colonization rates of the calves' gastrointestinal tracts with donor and recipient E. coli strains, the Salmonella recipient strains were detected only periodically and in low numbers, despite administration of a 10-fold-larger inocula compared to E. coli. This is consistent with previous observations in 10-week-old dairy calves inoculated with 109 CFU Salmonella Typhimurium phage type DT-104 (J. Gay, unpublished data). Younger calves, less than 2 weeks of age, are readily infected with doses 10,000-fold lower than those utilized here (16, 34). The colonization resistance that we observed is likely due to the fact that our calves were healthy and unstressed and had functional rumens.
The field study revealed no direct effect of ceftiofur use on the blaCMY-2-bearing population of commensal E. coli at the herd level: neither resistance ratio nor cow prevalence of E. coli with decreased susceptibility to ceftazidime was correlated with ceftiofur use. The lack of an association between the amount of ceftiofur use and the prevalence of cattle colonized by E. coli with reduced susceptibility to ceftazidime observed here in 42 modern large dairy farms confirms and extends the observation in a previous report of this relationship in 8 smaller (mean, 86 lactating cows per herd) Ohio dairy herds (40). Interestingly, the only herd in this study in which no E. coli with reduced ceftazidime susceptibility was detected was one of the three organic herds in which no antimicrobial drugs are used. Surprisingly, the frequencies of ceftazidime-resistant E. coli in the other two organic herds were comparable to those of the nonorganic herds in this study, implying that ceftiofur or indeed any other antimicrobial drug use is unnecessary for maintenance of blaCMY-2-bearing E. coli on individual dairy farms.
R plasmids encoding blaCMY-2 frequently encode resistance determinants for additional classes of antimicrobial agents (12, 17), and it is possible that the use of antimicrobial drugs other than ceftiofur could select for organisms carrying blaCMY-2 plasmids. As we have no information about the magnitude of other beta-lactam or other nonprescription drug classes on our study dairies, we cannot rule out possible confounding effects caused by this factor. However, there are two reasons to believe that such an effect is not very likely. (i) Ceftiofur is among the most common antimicrobial agents used on dairy farms (32, 35, 47). (ii) Among adult cattle, the relatively high prevalence of E. coli strains susceptible to ceftiofur but resistant to tetracyclines and/or sulfonamides (i.e., those resistances most frequently coencoded on blaCMY-2 plasmids) renders those drugs less likely to specifically coselect for blaCMY-2 carriage (4, 14, 37).
Consistent with the lack of an association between the magnitude of ceftiofur use on farms and the frequency of resistance in the commensal E. coli population, no significant association (P = 0.22) was observed between historical occurrence of clinical salmonellosis with ceftiofur-resistant strain(s) and the magnitude of ceftiofur use on dairy farms in this study. Interestingly, herds with a history of ceftiofur-resistant Salmonella infection were significantly more likely to have also experienced infections with ceftiofur-susceptible Salmonella, and herds experiencing any salmonellosis, regardless of ceftiofur resistance phenotype, used significantly more ceftiofur than herds with no clinical salmonella history. These observations suggest that some dairy farms engage in management practices that predispose the cows to salmonellosis in general and that increased ceftiofur use may be a frequent therapeutic response to suspected salmonellosis in these herds.
Previous studies have identified management factors that are significantly associated with salmonellosis. Cobbold et al. found that blaCMY-2-bearing Salmonella Newport was significantly more prevalent in a herd with a combined hospital-maternity pen than in a herd with separate facilities for sick animals (9). A large multistate study found that Salmonella shedding by cows was significantly associated with a wide variety of management factors, including absence of monensin use, storage of feeds in nonenclosed buildings, and cattle access to surface water sources (18). It is therefore plausible that one reason to increase ceftiofur use is a result of increased clinical problems resulting from poor management or biosecurity practices, rather than being a root cause of ceftiofur-resistant Salmonella problems in herds.
E. coli is considered a reservoir of resistance determinants (including blaCMY-2) that are potentially transferable to pathogens. Therefore, development of effective measures to prevent or reduce resistance to third-generation cephalosporins among E. coli strains, particularly those in the enteric flora of food-producing animals like cattle, would be highly desirable. The results of this study raise important questions about the specific role of ceftiofur use in promoting or maintaining third-generation cephalosporin resistance in the gastrointestinal flora of cattle. The in vivo calf study provided no evidence that ceftiofur use affected the generation or dissemination of blaCMY-2 plasmids. Only a relatively weak and transient effect on selection of ceftiofur-resistant strains among commensal E. coli populations was detected at the individual-animal level. The herd-based study failed to demonstrate any significant effect of the magnitude of ceftiofur use on the frequency of commensal E. coli or pathogenic Salmonella resistant to third-generation cephalosporins. Together, these findings suggest that a focus on reduction of ceftiofur use in individual animals or on individual farms is very unlikely to significantly affect the prevalence of ceftiofur resistance in the bovine enteric flora.
Acknowledgments
We thank Russell McClanahan, Lisa Jones, and Yubei Zhang for their technical assistance.
This work was funded in part by USDA-NRI Epidemiological Approaches to Food Safety grant 2005-01373, NIAID NIH contract N01-AI-30055, and the Agricultural Animal Health Program, WSU College of Veterinary Medicine, Pullman, WA.
Footnotes
Published ahead of print on 17 April 2009.
REFERENCES
- 1.Barlow, M., and B. G. Hall. 2002. Origin and evolution of the AmpC beta-lactamases of Citrobacter freundii. Antimicrob. Agents Chemother. 46:1190-1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bauernfeind, A., Y. Chong, and K. Lee. 1998. Plasmid-encoded AmpC beta-lactamases: how far have we gone 10 years after the discovery? Yonsei Med. J. 39:520-525. [DOI] [PubMed] [Google Scholar]
- 3.Bauernfeind, A., I. Stemplinger, R. Jungwirth, and H. Giamarellou. 1996. Characterization of the plasmidic beta-lactamase CMY-2, which is responsible for cephamycin resistance. Antimicrob. Agents Chemother. 40:221-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berge, A. C., S. C. Champagne, R. M. Finger, and W. M. Sischo. 2007. The use of bulk tank milk samples to monitor trends in antimicrobial resistance on dairy farms. Foodborne Pathog. Dis. 4:397-407. [DOI] [PubMed] [Google Scholar]
- 5.Call, D. R., M. S. Kang, J. Daniels, and T. E. Besser. 2006. Assessing genetic diversity in plasmids from Escherichia coli and Salmonella enterica using a mixed-plasmid microarray. J. Appl. Microbiol. 100:15-28. [DOI] [PubMed] [Google Scholar]
- 6.CDC. 2003. National Antimicrobial Resistance Monitoring System-Enteric Bacteria (NARMS): 2003 executive report. U.S. Department of Health and Human Services, U.S. FDA, Silver Spring, MD.
- 7.CLSI. 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, vol. 26. Clinical Laboratory Standards Institute, Wayne, PA.
- 8.CLSI. 2006. Performance standards for antimicrobial disk susceptibility tests; approved standard, 9th ed. Clinical and Laboratory Standards Institute document M2-A9. Clinical and Laboratory Standards Institute, Wayne, PA.
- 9.Cobbold, R. N., D. H. Rice, M. A. Davis, T. E. Besser, and D. D. Hancock. 2006. Long-term persistence of multi-drug-resistant Salmonella enterica serovar Newport in two dairy herds. J. Am. Vet. Med. Assoc. 228:585-591. [DOI] [PubMed] [Google Scholar]
- 10.Cray, W. C., Jr., and H. W. Moon. 1995. Experimental infection of calves and adult cattle with Escherichia coli O157:H7. Appl. Environ. Microbiol. 61:1586-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dancer, S. J. 2001. The problem with cephalosporins. J. Antimicrob. Chemother. 48:463-478. [DOI] [PubMed] [Google Scholar]
- 12.Daniels, J. B., D. R. Call, and T. E. Besser. 2007. Molecular epidemiology of blaCMY-2 plasmids carried by Salmonella enterica and Escherichia coli isolates from cattle in the Pacific Northwest. Appl. Environ. Microbiol. 73:8005-8011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davis, M. A., D. D. Hancock, D. H. Rice, D. R. Call, R. DiGiacomo, M. Samadpour, and T. E. Besser. 2003. Feedstuffs as a vehicle of cattle exposure to Escherichia coli O157:H7 and Salmonella enterica. Vet. Microbiol. 95:199-210. [DOI] [PubMed] [Google Scholar]
- 14.DeFrancesco, K. A., R. N. Cobbold, D. H. Rice, T. E. Besser, and D. D. Hancock. 2004. Antimicrobial resistance of commensal Escherichia coli from dairy cattle associated with recent multi-resistant salmonellosis outbreaks. Vet. Microbiol. 98:55-61. [DOI] [PubMed] [Google Scholar]
- 15.Donaldson, S. C., B. A. Straley, N. V. Hegde, A. A. Sawant, C. DebRoy, and B. M. Jayarao. 2006. Molecular epidemiology of ceftiofur-resistant Escherichia coli isolates from dairy calves. Appl. Environ. Microbiol. 72:3940-3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fecteau, M. E., J. K. House, S. F. Kotarski, N. S. Tankersley, M. M. Ontiveros, C. R. Alcantar, and B. P. Smith. 2003. Efficacy of ceftiofur for treatment of experimental salmonellosis in neonatal calves. Am. J. Vet. Res. 64:918-925. [DOI] [PubMed] [Google Scholar]
- 17.Fey, P. D., T. J. Safranek, M. E. Rupp, E. F. Dunne, E. Ribot, P. C. Iwen, P. A. Bradford, F. J. Angulo, and S. H. Hinrichs. 2000. Ceftriaxone-resistant Salmonella infection acquired by a child from cattle. N. Engl. J. Med. 342:1242-1249. [DOI] [PubMed] [Google Scholar]
- 18.Fossler, C. P., S. J. Wells, J. B. Kaneene, P. L. Ruegg, L. D. Warnick, J. B. Bender, L. E. Eberly, S. M. Godden, and L. W. Halbert. 2005. Herd-level factors associated with isolation of Salmonella in a multi-state study of conventional and organic dairy farms I. Salmonella shedding in cows. Prev. Vet. Med. 70:257-277. [DOI] [PubMed] [Google Scholar]
- 19.Hales, J. R. C. 2005. Antibacterials in the animal health industry: current markets and future prospects. PJB Publications, London, United Kingdom. http://www.pjbpubs.com/cms.asp?pageid=1897.
- 20.Huang, I. F., C. H. Chiu, M. H. Wang, C. Y. Wu, K. S. Hsieh, and C. C. Chiou. 2005. Outbreak of dysentery associated with ceftriaxone-resistant Shigella sonnei: first report of plasmid-mediated CMY-2-type AmpC beta-lactamase resistance in S. sonnei. J. Clin. Microbiol. 43:2608-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang, X., H. Yang, B. Dettman, and M. P. Doyle. 2006. Analysis of fecal microbial flora for antibiotic resistance in ceftiofur-treated calves. Foodborne Pathog. Dis. 3:355-365. [DOI] [PubMed] [Google Scholar]
- 22.Kado, C. I., and S. T. Liu. 1981. Rapid procedure for detection and isolation of large and small plasmids. J. Bacteriol. 145:1365-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kang, M. S., T. E. Besser, and D. R. Call. 2006. Variability in the region downstream of the blaCMY-2 beta-lactamase gene in Escherichia coli and Salmonella enterica plasmids. Antimicrob. Agents Chemother. 50:1590-1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karlson, A. G., and J. A. Ulrich. 1969. Stability of rifampin in dimethylsulfoxide. Appl. Microbiol. 18:692-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kilian, M., and P. Bulow. 1976. Rapid diagnosis of Enterobacteriaceae. I. Detection of bacterial glycosidases. Acta Pathol. Microbiol. Scand. Sect. B 84B:245-251. [DOI] [PubMed] [Google Scholar]
- 26.Koeck, J. L., G. Arlet, A. Philippon, S. Basmaciogullari, H. V. Thien, Y. Buisson, and J. D. Cavallo. 1997. A plasmid-mediated CMY-2 beta-lactamase from an Algerian clinical isolate of Salmonella Senftenberg. FEMS Microbiol. Lett. 152:255-260. [DOI] [PubMed] [Google Scholar]
- 27.Lowrance, T. C., G. H. Loneragan, D. J. Kunze, T. M. Platt, S. E. Ives, H. M. Scott, B. Norby, A. Echeverry, and M. M. Brashears. 2007. Changes in antimicrobial susceptibility in a population of Escherichia coli isolated from feedlot cattle administered ceftiofur crystalline-free acid. Am. J. Vet. Res. 68:501-507. [DOI] [PubMed] [Google Scholar]
- 28.Machado, J., F. Grimont, and P. A. Grimont. 2000. Identification of Escherichia coli flagellar types by restriction of the amplified fliC gene. Res. Microbiol. 151:535-546. [DOI] [PubMed] [Google Scholar]
- 29.McCuddin, Z. P., S. A. Carlson, M. A. Rasmussen, and S. K. Franklin. 2006. Klebsiella to Salmonella gene transfer within rumen protozoa: implications for antibiotic resistance and rumen defaunation. Vet. Microbiol. 114:275-284. [DOI] [PubMed] [Google Scholar]
- 30.Odeh, R., S. Kelkar, A. M. Hujer, R. A. Bonomo, P. C. Schreckenberger, and J. P. Quinn. 2002. Broad resistance due to plasmid-mediated AmpC beta-lactamases in clinical isolates of Escherichia coli. Clin. Infect. Dis. 35:140-145. [DOI] [PubMed] [Google Scholar]
- 31.Philippon, A., G. Arlet, and G. A. Jacoby. 2002. Plasmid-determined AmpC-type beta-lactamases. Antimicrob. Agents Chemother. 46:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pol, M., and P. L. Ruegg. 2007. Treatment practices and quantification of antimicrobial drug usage in conventional and organic dairy farms in Wisconsin. J. Dairy Sci. 90:249-261. [DOI] [PubMed] [Google Scholar]
- 33.Pruiett, C. S. 2004. Identification and characterization of Escherichia coli with reduced susceptibility to ceftazidime from Washington and Idaho cattle herds. M.S. thesis. Washington State University, Pullman.
- 34.Rankin, J. D., and R. J. Taylor. 1966. The estimation of doses of Salmonella Typhimurium suitable for the experimental production of disease in calves. Vet. Rec. 78:706-707. [DOI] [PubMed] [Google Scholar]
- 35.Raymond, M. J., R. D. Wohrle, and D. R. Call. 2006. Assessment and promotion of judicious antibiotic use on dairy farms in Washington State. J. Dairy Sci. 89:3228-3240. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 37.Sawant, A. A., N. V. Hegde, B. A. Straley, S. C. Donaldson, B. C. Love, S. J. Knabel, and B. M. Jayarao. 2007. Antimicrobial-resistant enteric bacteria from dairy cattle. Appl. Environ. Microbiol. 73:156-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Singer, R. S., S. K. Patterson, and R. L. Wallace. 2008. Effects of therapeutic ceftiofur administration to dairy cattle on Escherichia coli dynamics in the intestinal tract. Appl. Environ. Microbiol. 74:6956-6962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tkalcic, S., T. Zhao, B. G. Harmon, M. P. Doyle, C. A. Brown, and P. Zhao. 2003. Fecal shedding of enterohemorrhagic Escherichia coli in weaned calves following treatment with probiotic Escherichia coli. J. Food Prot. 66:1184-1189. [DOI] [PubMed] [Google Scholar]
- 40.Tragesser, L. A., T. E. Wittum, J. A. Funk, P. L. Winokur, and P. J. Rajala-Schultz. 2006. Association between ceftiofur use and isolation of Escherichia coli with reduced susceptibility to ceftriaxone from fecal samples of dairy cows. Am. J. Vet. Res. 67:1696-700. [DOI] [PubMed] [Google Scholar]
- 41.Upjohn Company. 1988. New animal drug application 140-338. Upjohn Company, Kalamazoo, MI.
- 42.Welch, T. J., W. F. Fricke, P. F. McDermott, D. G. White, M. L. Rosso, D. A. Rasko, M. K. Mammel, M. Eppinger, M. J. Rosovitz, D. Wagner, L. Rahalison, J. E. Leclerc, J. M. Hinshaw, L. E. Lindler, T. A. Cebula, E. Carniel, and J. Ravel. 2007. Multiple antimicrobial resistance in plague: an emerging public health risk. PLoS ONE 2:e309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Winokur, P. L., A. Brueggemann, D. L. DeSalvo, L. Hoffmann, M. D. Apley, E. K. Uhlenhopp, M. A. Pfaller, and G. V. Doern. 2000. Animal and human multidrug-resistant, cephalosporin-resistant Salmonella isolates expressing a plasmid-mediated CMY-2 AmpC beta-lactamase. Antimicrob. Agents Chemother. 44:2777-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Winokur, P. L., D. L. Vonstein, L. J. Hoffman, E. K. Uhlenhopp, and G. V. Doern. 2001. Evidence for transfer of CMY-2 AmpC beta-lactamase plasmids between Escherichia coli and Salmonella isolates from food animals and humans. Antimicrob. Agents Chemother. 45:2716-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu, S. W., K. Dornbusch, G. Kronvall, and M. Norgren. 1999. Characterization and nucleotide sequence of a Klebsiella oxytoca cryptic plasmid encoding a CMY-type beta-lactamase: confirmation that the plasmid-mediated cephamycinase originated from the Citrobacter freundii AmpC beta-lactamase. Antimicrob. Agents Chemother. 43:1350-1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao, S., D. G. White, P. F. McDermott, S. Friedman, L. English, S. Ayers, J. Meng, J. J. Maurer, R. Holland, and R. D. Walker. 2001. Identification and expression of cephamycinase blaCMY genes in Escherichia coli and Salmonella isolates from food animals and ground meat. Antimicrob. Agents Chemother. 45:3647-3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zwald, A. G., P. L. Ruegg, J. B. Kaneene, L. D. Warnick, S. J. Wells, C. Fossler, and L. W. Halbert. 2004. Management practices and reported antimicrobial usage on conventional and organic dairy farms. J. Dairy Sci. 87:191-201. [DOI] [PubMed] [Google Scholar]