Abstract
Salmonella enterica contamination in foods is a significant concern for public health. When DNA detection methods are used for analysis of foods, one of the major concerns is false-positive results from the detection of dead cells. To circumvent this crucial issue, a TaqMan quantitative real-time RT-PCR (qRT-PCR) assay with an RNA internal control was developed. invA RNA standards were used to determine the detection limit of this assay as well as to determine invA mRNA levels in mid-exponential-, late-exponential-, and stationary-phase cells. This assay has a detection limit of 40 copies of invA mRNA per reaction. The levels of invA mRNA in mid-exponential-, late-exponential-, and stationary-phase S. enterica cells was approximately 1 copy per 3 CFU, 1 copy per CFU, and 4 copies per 103 CFU, respectively. Spinach, tomatoes, jalapeno peppers, and serrano peppers were artificially contaminated with four different Salmonella serovars at levels of 105 and less than 10 CFU. These foods were analyzed with qRT-PCR and with the FDA's Bacteriological Analytical Manual Salmonella culture method (W. A. Andrews and T. S. Hammack, in G. J. Jackson et al., ed., Bacteriological analytical manual online, http://www.cfsan.fda.gov/∼ebam/bam-5.html, 2007). Comparable results were obtained by both methods. Only live Salmonella cells could be detected by this qRT-PCR assay, thus avoiding the dangers of false-positive results from nonviable cells. False negatives (inhibition of the PCR) were also ruled out through the use of an RNA internal control. This assay allows for the fast and accurate detection of viable Salmonella spp. in spinach, tomatoes, and in both jalapeno and serrano peppers.
Salmonella enterica contamination in various foods is a significant public health concern, domestically and internationally (22, 29, 37). Salmonella infects millions of people every year, accounting for an estimated 9.7%, 25.6%, and 30.6% of illnesses, hospitalizations, and deaths, respectively, of the total U.S. food-borne diseases caused by known food-borne pathogens (29). Consumption of fresh fruits and produce increased almost 50% between 1970 and 1994 (38). Fresh produce is vulnerable to contamination during the entire “farm-to-fork” process and may have contributed to increases in food-borne outbreaks (1), including recent Salmonella outbreaks involving jalapeno peppers (http://www.fda.gov/oc/opacom/hottopics/tomatoes.html).
A standard and accepted method for the detection of Salmonella in foods is based on a traditional microbiological method described in Chapter 5 of the Bacteriological Analytical Manual (BAM) (2, 17). It consists of a series of steps including nonselective preenrichment, selective enrichment, and selective/differential plating, followed by biochemical and serological confirmation. This method is labor-intensive and requires a minimum of 5 days for complete analysis (18). Hence, the need exists for the development of faster culture-independent screening and detection methods for this pathogen in produce.
In recent years, a plethora of new molecular methods based on Salmonella DNA detection (e.g., invA gene) either by conventional or real-time PCR have been developed (23, 27, 41). Real-time PCR (quantitative PCR [qPCR]) is faster and more sensitive than conventional PCR and provides real-time data, avoiding the use of gels as with conventional PCR (39). However, the main drawback with PCR methods is the potential detection of nonviable cells. Bacterial DNA is more stable than bacterial RNA and can persist in a sample long after the target organism has died, potentially leading to the production of false-positive results (8). If an mRNA target is chosen correctly, then it may be more suitable than DNA as a cell viability marker due to its inherent instability. The half-life of most bacterial mRNAs has been reported to range from 0.5 to 50 min (36). This implies that as long as bacteria are viable, they produce some mRNA molecules (e.g., invA mRNA, which codes for invasion protein InvA [12]). In Salmonella enterica Typhimurium grown in Luria-Bertani (LB) broth, invA mRNA was detected throughout the growth curve (24). Furthermore, several studies have shown that the invA gene and its mRNA are good candidates for detection of Salmonella spp. in environmental samples by qPCR (5, 26, 30, 31, 35, 41), quantitative reverse transcriptase real-time PCR (qRT-PCR) (10), or conventional reverse transcriptase PCR (RT-PCR) (21).
As stated by Fey et al. (10), quantification of RNA targets is a problematic issue in bacteria that can be resolved by designing appropriate RNA standards. The use of DNA standards (PCR products or plasmid or genomic DNA) will result in a broad overestimation of the RNA target molecules, largely due to different reaction efficiencies between RT-PCR and PCRs (33). Several specific RNA standards have been developed by in vitro transcription (10, 16). These reports make use of specific RNA standards that are very attractive for accurate quantification of low-level mRNAs (e.g., invA mRNA). In particular, the invA gene represents a good candidate for Salmonella detection as it is present in all pathogenic serovars described to date (6, 34). The product of this gene is essential for the organism's ability to invade mammalian cells and subsequently cause disease (13, 14).
In order to address the crucial issue of detecting viable versus nonviable Salmonella cells, a TaqMan qRT-PCR assay targeting invA mRNA was developed. A foreign internal RNA control was also incorporated into the assay with the aim of detecting potential inhibitors present in the matrices analyzed. This assay allowed for fast and accurate detection of viable S. enterica cells in four matrices (tomato, spinach, jalapeno peppers, and serrano peppers). Salmonella sp. detection in foods is usually achieved after food samples are preenriched overnight at 37°C (9). Consequently, the method described herein is intended as an initial screening of 24-h preenrichments for the presence of Salmonella in produce. In turn, this method will dramatically decrease the time and effort required during standard microbiological testing since only positive preenrichment samples will be processed.
MATERIALS AND METHODS
Bacterial strains and media.
Five strains belonging to four S. enterica serovars (serovar Enteritidis strain SE5, serovar Javiana strain 7N, serovar Newport 1240H, and serovar Saintpaul strains 1358 H and 50258), were employed in this study for artificial contamination of produce. These four strains are from the culture collection of the Food and Drug Administration (FDA), Center for Food Safety and Applied Nutrition (CFSAN), Division of Microbiology. Inclusivity of the qPCR assay for Salmonella sp. invA was demonstrated with 96 Salmonella serotypes (110 strains) from FDA's collection (see Table S1 in the supplemental material), and exclusivity was demonstrated with 32 non-Salmonella species (48 strains) from very closely related genera (Table 1). Inclusivity is the capability of the assay to detect the intended pathogen target in a wide range of strains belonging to the same bacterial species (i.e., Salmonella strains only), and exclusivity is the lack of signal or positive reaction with closely related non-Salmonella strains. Only strain SE5 was used to generate the DNA and RNA standards. Strains were grown overnight in LB medium at 35°C with shaking (250 rpm).
TABLE 1.
Organisms employed to asses the exclusivity of the invA qPCR TaqMan assay for Salmonella sp. detectiona
| Organism | No. of strains tested |
|---|---|
| Vibrio parahaemolyticus | 4b |
| Vibrio vulnificus | 1 |
| Escherichia coli | 9c |
| Enterobacter cloacae | 1 |
| Enterobacter aerogenes (ATCC 13048) | 1 |
| Cronobacter sakazakii (formerly E. sakazakii) | 1 |
| Yersinia enterocolitica | 1 |
| Yersinia pseudotuberculosis | 1 |
| Hafnia alvei | 2 |
| Morganella morganii | 1 |
| Edwardsiella tarda | 1 |
| Klebsiella pneumoniae | 1 |
| Proteus vulgaris | 1 |
| Pseudomonas aeruginosa | 1 |
| Serratia marcesans | 1 |
| Aeromonas hydrophila | 1 |
| Citrobacter freundii | 1 |
| Citrobacter koseri (ATCC 27028) | 1 |
| Staphylococcus aureus | 1 |
| Streptococcus faecalis | 1 |
| Bacillus subtilis | 1 |
| Bacillus cereus | 1 |
| Listeria monocytogenes | 1 |
| Listeria innocua | 1 |
| Shigella sonnei | 2 |
| Shigella flexneri | 2 |
| Shigella boydii | 2 |
| Shigella dysenteriae | 2 |
| Achromobacter spp. | 1 |
| Providencia stuartii (ATCC 33672) | 1 |
| Proteus mirabilis | 1 |
| Proteus hauseri (deposited as P. vulgaris) (ATCC 13315) | 1 |
| Total | 48 |
Exclusivity was demonstrated by the absence of positive signal for all strains tested; i.e., all invA qPCR results were negative.
V. parahaemolyticus pandemic strains VpHY145 and JYKVP6 and non-pandemic strains 428/00 and 357-99 (15).
Five E. coli classes (virotypes) that cause diarrheal diseases were included: strain 10009 (enterotoxigenic); strains 10010, 10015, 10016, 10017, and 10012 (enteroinvasive); strain 10023 (enterohemorrhagic); strain 10035 (enteropathogenic), and strain ATM395 (enteroaggregative).
Preparation of Salmonella inocula.
For artificial contamination, overnight cultures (109 CFU/ml) of each of the Salmonella serovars tested were diluted 100-fold and inoculated into 100 ml of LB medium at 35°C with shaking (250 rpm) for 3 h (optical density at 600 nm [OD600] of 0.5) when cells reached mid-exponential growth phase. Bacteria were harvested (3,090 × g for 10 min) and washed twice with 10 ml of Butterfield's phosphate buffer (BPB). Salmonella suspensions were serially diluted (10-fold) in BPB. Dilutions containing approximately 105 CFU/ml and less than 10 CFU/ml were used for spiking studies. CFU counts were determined by aerobic plate count as described in the BAM (3).
Sample processing and artificial contamination.
Bagged spinach, tomatoes, jalapeno, and serrano peppers (see Table 3 for a list)were obtained from local supermarkets in College Park, MD. These matrices were processed as indicated in the BAM (2). On day 1, three different 25-g portions of each food were blended individually with 225 ml of lactose broth (Difco, BD, Sparks, MD) at 10,000 to 12,000 rpm for 2 min (Waring Variable Speed Laboratory Blender; Torrington, CT). The three 25-g portions were designated A for uninoculated, B for high-level (105 CFU/ml) inoculation, and C for low-level inoculation (1 to 10 CFU/ml). The corresponding portions (B and C) were inoculated as indicated with S. Enteritidis strain SE5 and incubated for 1 h at room temperature. Portion A remained uninoculated. Subsequently, pH was measured in all three portions, and when the pH values differed from 6.8 ± 0.2, the portions were supplemented with 1 N NaOH. These three portions were later incubated at 35 ± 2°C for 24 ± 2 h. Taken together, these steps comprised the preenrichment step.
TABLE 3.
S. enterica detection by invA RNA qRT-PCR and BAM culture method in artificially contaminated produce commodities
| Food sample | Strain | Inoculation level (CFU/25 g) |
Salmonella sp. detection by the indicated method
|
DNA detection by qPCR (CT)a,b | |
|---|---|---|---|---|---|
| qRT-PCR (CT)a | BAM | ||||
| Bagged spinach | S. Enteritidis SE5 | 8 | + (27.6) | + | + (37.9) |
| S. Javiana 7N | 2 | + (28.4) | + | − | |
| Tomatoes | S. Enteritidis SE5 | 4 | + (28.7) | + | + (37.3) |
| S. Javiana 7N | 4 | + (30.1) | + | − | |
| Jalapeno peppers | S. Saintpaul 1358 H | 10 | + (33.5) | + | − |
| S. Newport 1240H | 3 | + (34.2) | + | − | |
| Serrano peppers | S. Saintpaul 50258 | 5 | + (33.9) | + | − |
| S. Newport 1240H | 3 | + (35.0) | + | − | |
CT values are given where the fluorescence signal was higher than the background.
The same RNA samples were amplified by qPCR to test for DNA contamination.
After 24 h (day 2), 1 ml was taken from each sample (A, B, and C). Two volumes (2 ml) of RNA Protect reagent (Qiagen, Valencia, CA) was added to each sample and processed as indicated by the manufacturer. This reagent is employed for RNA stabilization and protection in order to avoid the activity of endogenous RNases that can degrade RNA during the storage and extraction procedures. Samples were stored at −70°C until processed for RNA extraction.
Food samples were then analyzed as described in the BAM (2) and as depicted in Fig. 1 in Hammack et al. (18). Briefly, 0.1 ml from the preenrichment culture was added to 10 ml of Rappaport-Vassiliadis (RV) medium, and 1 ml was added to 10 ml of tetrathionate (TT) broth (Difco) and incubated for 24 ± 2 h at 42 ± 0.2 and 43 ± 0.2°C, respectively (except for tomato samples, where TT broth was incubated at 35 ± 2°C). After 24 h (day 3), tubes were vortexed, and 10-μl portions from TT and RV broths were streaked onto bismuth sulfite, xylose lysine desoxycholate, and Hektoen enteric agars. Plates were incubated at 35 ± 2°C for 24 ± 2 h. On day 4, plates were examined for the presence of typical Salmonella colonies. Typical colonies were confirmed as Salmonella as specified in the BAM (2).
FIG. 1.
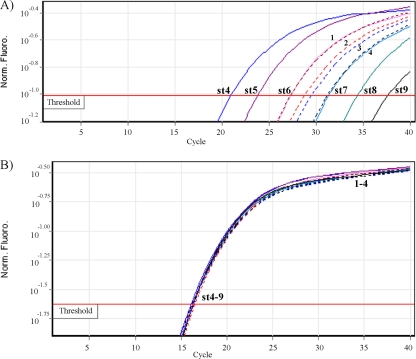
Robustness of the RNA internal control amplification in multiplexing reactions (invA qRT-PCR). Multiplex qRT-PCRs of invA RNA and the RNA internal control in bagged spinach samples artificially contaminated with different levels of Salmonella were performed. Solid lines are the invA RNA standards (st) that are identified in terms of the number of invA RNA copies per reaction mixture: st4, 1 pg (4 × 106); st5, 100 fg (4 × 105); st6, 10 fg (4 × 104); st7, 1 fg (4 × 103); st8, 0.1 fg (4 × 102); and st9, 0.01 fg (40). Dotted lines are samples inoculated with different levels of Salmonella as follows: strain SE5 at 105 CFU/25 g (line 1) and 8 CFU/25 g (line 2); strain Javiana 7N at 105 CFU/25 g (line 3) and 2 CFU/25 g (line 4). (A) qRT-PCR results for the invA RNA (ROX channel) amplification. (B) qRT-PCR results for the RNA internal control (Cy5 channel) included in the same RNA samples. Norm fluoro, normalized fluorescence.
Nucleic acid extraction.
DNA and RNA extractions were performed with the DNeasy and RNeasy Mini Kits, respectively, as recommended by the manufacturer (Qiagen). For RNA extraction, the lysis was performed with 100 μl of lysozyme (1 mg/ml) for 5 min at room temperature. DNase I (Qiagen) treatment was performed for 30 min at room temperature.
Design of primers and standards for qPCR.
All primers and probes (Table 2) employed in this study were purchased from IDT (Coralville, IA). The target for qPCR and qRT-PCR was the invA gene of S. enterica serovar Enteritidis SE5 and its corresponding mRNA. Primers and probes for the TaqMan assay were designed using Primer Express, version 1.5, using invA gene sequences available at NCBI (Applied Biosystems, Carlsbad, CA) (4). To enable an accurate quantification of the RNA targets for the establishment of the detection limit of the assay, RNA standards for invA were generated. The DNA standards for invA (865 bp) were generated as previously described (10), except using 60°C as the PCR annealing temperature. Briefly, PCR products generated with primers invA-T7-F2 and invA-T7-R1 were purified with a MinElute PCR Purification Kit (Qiagen) and subsequently transcribed in vitro with T7 polymerase using the Riboprobe System-T7 following the manufacturer's instructions (Promega, Madison, WI). This was followed by digestion with DNase I, purification using RNeasy (Qiagen), and a second DNase I digestion on the purification column. The transcripts were then analyzed by agarose gel electrophoresis (1% gels containing 0.65% formaldehyde) and quantified using a RiboGreen quantitation kit (Invitrogen, Carlsbad, CA). DNA standards were also quantified using a PicoGreen quantification kit (Invitrogen). Both measurements were conducted in a NanoDrop ND-3300 Fluorospectrometer (Thermo Fisher Scientific Inc, Waltham, MA). A second primer set (Table 2) also targeting the invA gene but with target sequences located within the sequences recognized by the first set was used to conduct the real-time quantification assays.
TABLE 2.
Primers and probes used in this study
| Function and target | Primer or probe | Sequence (5′→3′)a | Position on geneb | Product size (bp) | Accession no.c | Reference or sourced |
|---|---|---|---|---|---|---|
| Generation of standards, invA | invA-T7-F2 | TAATACGACTCACTATAGGGAACAGTGCTCGTTTACG | 1 | 885 | DQ644631 | 10 |
| invA-T7-R1 | GCAGAGTTCCCATTGAAATGGTC | 865 | 10 | |||
| PCR | ||||||
| invA | invA_176F | CAACGTTTCCTGCGGTACTGT | 175 | 116 | DQ644631 | 4 |
| invA_291R | CCCGAACGTGGCGATAATT | 291 | 4 | |||
| invA_Tx_208 | TX-CTCTTTCGTCTGGCATTATCGATCAGTACCA-BHQ2 | 207 | 4 | |||
| myIC | dd-myIC-f | CTAACCTTCGTGATGAGCAATCG | 1 | 198 | FJ357008 | CFSAN/ORS |
| dd-myIC-r | GATCAGCTACGTGAGGTCCTAC | 198 | CFSAN/ORS | |||
| dd-myIC-Cy5 | Cy5-AGCTAGTCGATGCACTCCAGTCCTCCT-Iowa BlackRQ-Sp | 62 | CFSAN/ORS |
The sequence corresponding to the T7 promoter is underlined. TX, Texas Red.
Positions on genes are given according to the accession numbers.
Accession number at the NCBI (http://www.ncbi.nih.gov/).
ORS, Office of Regulatory Science.
Calculation of copy numbers.
The numbers of copies of the qPCR and qRT-PCR standards were calculated by assuming average molecular masses of 680 Da for 1 nucleotide of double-stranded DNA and 340 Da for 1 nucleotide of single-stranded RNA. The calculation was done with the following equation: copies per nanogram = (NL × 10−9)/(n × mw), were n is the length of the standard in base pairs or nucleotides, mw is the molecular weight per nucleotide, and NL is the Avogadro constant (6.02 × 1023 molecules per mol).
Exogenous internal control.
An exogenous RNA internal control (myIC) was incorporated into the qRT-PCR assay. The myIC RNA was generated and processed as described above for invA RNA standards using as a template a modified pZErO-2 plasmid (Invitrogen) containing an inserted myIC sequence. This sequence is a synthetic construct that does not match any currently available sequence in the GenBank database (http://www.ncbi.nlm.nih.gov/). Modified pZErO-2 containing myIC sequence, primers, and probes for qPCR analysis employed for in vitro transcription were kindly provided by Deanne Deer (FDA/CFSAN) (Table 2). The myIC RNA was serially diluted to a final concentration that gave a positive signal between cycles 15.5 to 16 when the invA RNA standards were used for testing. myIC RNA was tested for robustness in qRT-PCR multiplexing reactions. Here, robustness is taken to be a reflection of the performance of the internal control across several reaction conditions (e.g., spinach samples).
invA mRNA detection in live and dead Salmonella cells.
In order to determine the feasibility of the use of invA mRNA as a marker of viability for Salmonella cells, dead (autoclaved) and live cells were spiked into 25-g bagged spinach portions. These portions were processed as indicated above (see “Sample processing and artificial contamination” above). In this case, we again employed three 25-g portions, except that portion C was inoculated with 1 ml of autoclaved Salmonella strain SE5 cells (dead cells). One ml of an inoculum containing 106 CFU/ml was added to portion B (live cells), while portion A remained uninoculated (Salmonella negative control). The procedure for preparing Salmonella cells was the same as described above (see “Preparation of Salmonella inocula” above), except that the Salmonella strain SE5 suspension was diluted 100-fold with BPB to obtain a final concentration of 106 CFU/ml rather than 105 CFU/ml (total volume, 100 ml). This suspension was separated into two 50-ml portions, one of which was autoclaved for 15 min at 121°C (portion C; dead cells) while the other remained without treatment (portion B; live cells). After 24 h, one ml was taken from each of the three preenrichment portions (A, B, and C) for RNA extraction. RNA extraction was conducted as described above. invA qRT-PCR was performed for each RNA sample extracted from each portion by triplicate.
One-step qRT-PCR and data analysis.
The qRT-PCRs were carried out using a SuperScript III Platinum One-Step Quantitative RT-PCR System kit according to the specifications of the manufacturer (Invitrogen). This kit consists of SuperScript III RT/Platinum Taq Mix, 2× Reaction Mix (a buffer containing a 0.4 mM concentration of each deoxynucleoside triphosphate, and 6 mM MgSO4), 50 mM magnesium sulfate (MgSO4), and ROX (6-carboxy-X-rhodamine) reference dye (25 μM). Reaction mixtures were scaled down to a final volume of 20 μl. Additional MgCl2 was added to the master mix to a final concentration per tube reaction mix of 5 mM. Final concentrations of primers in the qRT-PCR mix were 200 nM and 100 nM for invA and myIC, respectively. Both probes were added to a final concentration of 150 nM. qRT-PCR and data analysis were performed on a Rotor-Gene 3000 (Corbett) qPCR machine. qRT-PCR conditions were as follows: an initial cycle of 15 min at 50°C for the generation of the cDNAs; a second cycle of 2 min at 95°C to activate the hot-start Taq polymerase; and 40 cycles of denaturation at 95°C for 15 s, primer annealing at 60°C for 15 s (the acquisition of both dyes Cy5 and Texas Red were performed at the end of this cycle), and extension at 72°C for 15 s. In each reverse transcription reaction mixture, the same RNA samples were not supplemented with reverse transcriptase to detect DNA contamination. If the differences between the threshold cycle (CT) values for qRT-PCR and qPCR were greater than 4 cycles, the result was considered positive since differences in 3.1 to 3.6 cycles between samples were mostly due to differences in concentration of approximately 10-fold (if efficiency is between 90 to 110%) (http://www.stratagene.com/techtoolbox/calc/qpcr_slope_eff.aspx). For CT values that dipped below this range, a new DNase I treatment was performed. Subsequently, if similar results were obtained below the range reported, the analyzed sample was considered negative for the presence of live Salmonella cells. To generate a calibration curve, the serially diluted RNA standard (100 pg to 0.001 fg) was quantified in each qRT-PCR run. The calibration curves were generated by the Rotor-Gene software, version 6.1. For each standard, the concentration was plotted against the cycle number at which the fluorescence signal increased above the background or threshold (CT value). Reaction efficiency was calculated using the slope of the resultant calibration curve (i.e., efficiency = 10−1/slope − 1). An efficiency of 1.0 indicated a product doubling in each cycle. The number of invA mRNA copies was determined using the calibration curve generated by the Rotor-Gene software.
Nucleotide sequence accession number.
The myIC sequence was deposited in GenBank under accession number FJ357008.
RESULTS
Evaluation of the qPCR and qRT-PCR method.
The DNA standards generated by PCR from genomic DNA and the RNA standards generated by in vitro transcription of PCR products were evaluated by both qPCR and qRT-PCR. PCR primers specific for the invA gene (invA_176F and invA_291R) were used (Table 2). Linear calibration curves with a correlation coefficient (R2) of ≥0.99 and linear ranges of ≥8 orders of magnitude for both invA DNA and RNA were obtained (see Fig. S1 in the supplemental material). This corresponds to detection limits of 10 and 40 copies for invA DNA and RNA, respectively. The efficiency of the qPCR and qRT-PCR ranged from 0.93 to 0.99 and from 0.90 to 0.96, respectively. For the purposes of this study, only qRT-PCR was employed from this point forward (e.g., detection of live cells). In all qRT-PCR assays, the same RNA samples were amplified by PCR to test for DNA contamination. Most of the samples resulted in no amplification, and in some samples a very high CT value was obtained, indicative of the presence of small amounts of DNA contamination (e.g., in the standards containing 109 and 108 invA RNA copies per reaction mixture, the contaminating DNA was estimated to be 103 to 102, respectively). These data indicated that DNA contamination was negligible throughout this study.
Specificity of the invA qPCR TaqMan assay.
Supplemental Table S1 shows a panel of 110 Salmonella strains (96 serotypes) used to test for inclusivity of this assay (invA qPCR) using the primer set invA_176F and invA_291R. Salmonella sp. strains analyzed encompassed five of the six S. enterica subspecies (S. enterica subsp. I,89 serovars; S. enterica subsp. II, 3; S. enterica subsp. IIIa, 1; S. enterica subsp. IIIb, 1; and S. enterica subsp. IV, 1) and one Salmonella bongori strain. These strains represented a broad spectrum of Salmonella serovars, including many of the serotypes most frequently isolated in United States from 1968 to 1998, according to the CDC (7, 29). All Salmonella strains were identified correctly (100% inclusivity). The 48 non-Salmonella strains employed in the exclusivity tests did not give any positive signal (Table 1). These strains were chosen for exclusivity testing because many are close phylogenetic kin to the Salmonella and are known to associate with the food supply.
Evaluation of the RNA internal control.
An exogenous RNA internal control, designated myIC, that could be used to measured false-negative PCR results was generated by in vitro transcription. This myIC RNA molecule was generated from a pZErO-2 plasmid (Invitrogen) containing a DNA sequence (myIC DNA) that can be used as an internal control in DNA TaqMan assays (D. Deer, unpublished data). This plasmid also contains a T7 promoter close to the myIC sequence that allowed its in vitro transcription. The primers and probe (labeled with Cy5) employed for the amplification of myIC by TaqMan assay are described in Table 2. myIC was evaluated combined with the invA RNA qRT-PCR as a duplex TaqMan PCR assay to assess its performance across several reaction conditions. Figure 1 shows the result of the amplification by the duplex assay (invA qRT-PCR) of RNA samples extracted from 1 ml of preenrichments of bagged spinach portions (25 g) that were artificially contaminated with different levels of Salmonella (2 CFU to 105 CFU). All samples tested were positive for invA mRNA, and bagged spinach preenrichments did not inhibit the PCR. The internal control (myIC) showed approximately the same CT values for all samples analyzed.
invA mRNA quantification in pure cultures of Salmonella.
The presence of specific invA mRNA was determined in Salmonella strain SE5 mid-exponential-phase cells (OD of 0.418), late-exponential-phase cells (OD of 1.3), and stationary-phase cells (overnight culture). The numbers of invA mRNA molecules per CFU averaged 0.4, 1, and 0.004 copies/CFU in mid-exponential-, late-exponential-, and stationary-phase cells, respectively. It is important to note that nonhost environments are often stressful habitats for Salmonella and could seriously impair the biological activity of the Salmonella cells, potentially resulting in depressed transcription and hence a low copy number of mRNA molecules. Under such conditions, the efficiency of an assay based on the detection of mRNA as a sign of cell viability could be limited. Thus, a preenrichment step is essential to allow Salmonella present in the sample to increase in biomass and express de novo invA mRNA that will allow for its subsequent detection using this method. Accordingly, our results showed that invA mRNA was present in large enough quantities to be detected by the TaqMan assay, pointing to its reliability as a target for qRT-PCR detection.
invA mRNA detection in bagged spinach preenrichments previously spiked with live and dead Salmonella cells.
Twenty-four-hour sample preenrichments previously spiked with live and dead cells were analyzed for the presence of invA mRNA by qRT-PCR. The bagged spinach preenrichment, previously spiked with live Salmonella cells, was the only sample to result in a positive reaction by invA qRT-PCR (data not shown). These findings provided indisputable evidence that invA mRNA is a good target for detection of live Salmonella in this produce commodity.
Application of the qRT-PCR duplex assay to Salmonella detection in different produce commodities.
In order to assess the performance of the qRT-PCR detection assay developed for this study, four different produce commodities were artificially contaminated with five different S. enterica strains (four serovars) at levels of 106 and less than 10 CFU/25 g (Table 3). After 24 h, the preenrichments were used for detection of Salmonella using both the qRT-PCR assay and the BAM Salmonella culture method (18). Uninoculated samples from each commodity were used as negative controls. RNA was extracted from the preenrichments and used for invA qRT-PCR amplification in triplicate. All artificially contaminated samples were positive for Salmonella using both qRT-PCR and BAM methodologies (Table 3). Higher levels of inoculation were also positive for Salmonella by both methods (results not shown). Only lower inoculation levels are shown in order to highlight the power of this technique. Salmonella levels as low as 2 CFU/25 g were detected after preenrichment (24 ± 2 h). Absence of RT-PCR inhibitors was demonstrated by amplification of the RNA internal control since the RNA control would not have been amplified had there been PCR inhibitors present in the foods (Fig. 1B).
DISCUSSION
Numerous molecular assays have been developed for the detection of Salmonella in different matrices (8, 10, 23, 28, 32). Some of these are based on conventional PCR, and some others are based on qPCR technology, whether using SYBR green I or molecular probes (e.g., TaqMan). The main drawback of these methodologies is that they rely on the detection of DNA (8). DNA from dead cells can be detected, but FDA cannot take regulatory action unless the cells are shown to be viable. Thus, detection of DNA is not enough, and the FDA must obtain an isolate to take regulatory action. However, fulfillment of this requirement takes approximately 1 week and is somewhat laborious. This problem may be circumvented with the RNA detection technology described in this study (10, 16).
A novel qRT-PCR assay was developed that uses specific primers for the detection of invA mRNA (product of invA gene) of Salmonella spp. with TaqMan probes. This assay also includes an RNA internal control to detect potential PCR inhibitors that may be present in food samples. We also determined the number of invA mRNA molecules per CFU in different Salmonella strain SE5 growth stages and found that at least 1 copy of invA mRNA was present in every CFU in exponential growth. These levels of expression per CFU are similar to what was reported for S. enterica serovar Typhimurium strain ATCC 14028 (a maximum of 10 copies per CFU at exponential growth) (10). In our case, considering RNA losses that can occur during the RNA extraction procedure and DNase I treatment, we can estimate that the number of invA mRNA molecules per cell could be between 1 and 10 copies. Thus, a total population of approximately 108 CFU/ml would produce approximately 108 to 109 copies of invA mRNA/ml. The volume from each of the analyzed samples was 1 ml, and, after RNA extraction, the amount added to each reaction mixture was equivalent to 28 μl of the initial volume. invA mRNA amounts contained within this 28 μl were approximately equivalent to 106 copies, a value well above the detection limit of the qRT-PCR, which was 40 invA mRNA copies. Therefore, the selected specific RNA could be used as molecular targets for either identification and/or determination of metabolic activity of Salmonella cells in foods as well as environmental samples using the developed qRT-PCR method described herein.
It is important to note, however, that in non-host environments, Salmonella persists most likely in a starved and highly stressed state. As an example, we found that in stationary phase 4 out of 1,000 cells will actually contain invA mRNA, indicating that this mRNA in a non-host environment was negligible and most likely would go undetected using this methodology. However, the addition of a requisite preenrichment step in the culture medium substantially increases cell number and subsequent mRNA production accordingly. Thus, a preenrichment culture provides an essential preliminary step in the application of this assay to the reliable detection of Salmonella invA mRNA from the surface of various produce commodities.
It has become increasingly evident that there is a need for internal controls for PCR to rule out the presence of PCR inhibitors that can cause false-negative results from Salmonella-positive samples (19, 20). For this reason, we developed and included an exogenous RNA internal control. The inclusion of this internal control did not affect either the amplification or the detection limit of the qRT-PCR assay.
Numerous studies, performed to establish the use of RNA as proof of viability, employed heating (either autoclaving or boiling) of the cells (11). Some reports suggested that invA mRNA degrades rapidly in S. Typhimurium ATCC 14028 from 10 copies to less than 1 copy per CFU after 50 h of incubation in drinking and pond water (10). Also, Jacobsen and Hoben (21) showed that the spiking of soil manure with Salmonella cells followed by pasteurizing the samples at 65°C for 30 min resulted in a failure to detect invA mRNA by RT-PCR. However, invA DNA was still detectable, indicating that mRNA was being degraded rapidly. We performed a similar experiment where we spiked dead and live Salmonella cells into bagged spinach and tested the respective preenrichments for the presence of invA mRNA. Only the bagged spinach preenrichment, spiked with live cells, gave a positive signal by the qRT-PCR assay reported here. This clearly showed that the qRT-PCR assay, based on invA mRNA detection, has great potential to be used as a viability marker for Salmonella cells in food commodities.
This study reports a qRT-PCR TaqMan assay that allowed the fast and accurate detection of viable Salmonella cells in spinach, tomato, jalapeno peppers, and serrano peppers. The assay performed comparably to the traditional BAM Salmonella culture. It also is noteworthy that the method reported here performed optimally relative to several other PCR-based methods that have been recently shown to detect Salmonella using invA targets (5, 9, 25, 26, 28). DNA-based methods have the intrinsic limitation of detecting DNA from both dead and live cells, making the outcome suspect (5). Moreover, conventional PCR, while providing a target amplicon with relative ease and little expense, is incapable of revealing identity (i.e., primary sequence structure), thus failing to ensure the nature of the PCR product (25, 34). In addition, the use of a TaqMan assay had several advantages over the use of the SYBR green I assay, including greater sensitivity and greater sequence-specific hybridization that allowed for verification of PCR product identity (10, 21, 40). Our method also revealed substantial promise for detecting specific serovars of S. enterica—a claim unable to be guaranteed by previously reported invA-based methodologies (25, 27). Ready-to-use mixtures, such as the one used in this study, facilitate the performance of the assay while being both straightforward and reproducible.
It is noteworthy that, rather than performing replicates of several inoculations using the same strain, we opted to spike the same four produce matrices presented in Table 3 with two different Salmonella serovars, thereby offering a more powerful approach than simply repeating the experiment with the same strain multiple times. After all, the ultimate goal of the assay is to detect numerous disparate serovars, not simply S. Enteritidis alone, for example. Thus, increasing the biocomplexity of the test provided a more rigorous challenge to the capability of the method to detect Salmonella in general. Here, a move toward this end was taken by including a total of four different serovars comprising five distinct strains across the four produce types tested.
In conclusion, we have developed a method that has the potential to be used as an initial screening of the preenrichment cultures and as a preliminary proof of viability for Salmonella without precluding the need for the BAM method, which is necessary to yield a physical isolate for regulatory reasons. This assay will reduce the overall time and resources expended in the laboratory since only positive samples containing viable cells will be processed after the preenrichment step. Expenses associated with the assay were comparable to other previously reported methods at around $7 per sample, making the assay widely affordable for routine analysis. Despite this relative ease of performance and cost, it is important that future studies assess the use of this assay in additional food matrices. Also, in order for this method to be applied extensively, collaborative studies should be conducted to assess the interlaboratory reproducibility of this much-needed RNA-based assay.
Supplementary Material
Acknowledgments
We thank Jessica L. Jones for her assistance with the development of the qPCR assay. We also thank Deanne Deer for providing the DNA internal control used in this study.
N. González-Escalona acknowledges a Research Fellowship Program for the Center for Food Safety and Applied Nutrition administered by the Oak Ridge Associated Universities through a contract with the U.S. Food and Drug Administration.
Footnotes
Published ahead of print on 17 April 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Altekruse, S. F., M. L. Cohen, and D. L. Swerdlow. 1997. Emerging foodborne diseases. Emerg. Infect. Dis. 3:285-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrews, W. A., and T. S. Hammack. December 2007, posting date. Salmonella. In G. J. Jackson et al. (ed.), Bacteriological analytical manual online. Center for Food Safety and Applied Nutrition, U.S. Food and Drug Administration, Gaithersburg, MD. http://www.cfsan.fda.gov/∼ebam/bam-5.html.
- 3.Andrews, W. A., and T. S. Hammack. April 2003, posting date. Food sampling and preparation of sample homogenate. In G. J. Jackson et al. (ed.), Bacteriological analytical manual online. Center for Food Safety and Applied Nutrition, U.S. Food and Drug Administration, Gaithersburg, MD. http://www.cfsan.fda.gov/∼ebam/bam-1.html.
- 4.Blackstone, G. M., J. L. Nordstrom, and A. DePaola. 2005. A same day detection method for Salmonella species in shrimp using real time PCR, poster P-079. Abstr. 105th Gen. Meet. Am. Soc. Microbiol. American Society for Microbiology, Washington, DC.
- 5.Bohaychuk, V. M., G. E. Gensler, M. E. McFall, R. K. King, and D. G. Renter. 2007. A real-time PCR assay for the detection of Salmonella in a wide variety of food and food-animal matrices. J. Food Prot. 70:1080-1087. [DOI] [PubMed] [Google Scholar]
- 6.Boyd, E. F., J. Li, H. Ochman, and R. K. Selander. 1997. Comparative genetics of the inv-spa invasion gene complex of Salmonella enterica. J. Bacteriol. 179:1985-1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crump, J. A., S. P. Luby, and E. D. Mintz. 2004. The global burden of typhoid fever. Bull. W. H. O. 82:346-353. [PMC free article] [PubMed] [Google Scholar]
- 8.Drahovska, H., J. Turna, E. Piknova, T. Kuchta, I. Szitasova, A. Skarkova, and M. Sasik. 2001. Detection of Salmonella by polymerase chain reaction targeted to fimC gene. Biologia 56:611-616. [Google Scholar]
- 9.Feder, I., J. C. Nietfeld, J. Galland, T. Yeary, J. M. Sargeant, R. Oberst, and M. L. Tamplin. 2001. Comparison of cultivation and PCR-hybridization for detection of Salmonella in porcine fecal and water samples. J. Clin. Microbiol. 39:2477-2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fey, A., S. Eichler, S. Flavier, R. Christen, M. G. Hofle, and C. A. Guzman. 2004. Establishment of a real-time PCR-based approach for accurate quantification of bacterial RNA targets in water, using Salmonella as a model organism. Appl. Environ. Microbiol. 70:3618-3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fykse, E. M., G. Skogan, W. Davies, J. S. Olsen, and J. M. Blatny. 2007. Detection of Vibrio cholerae by real-time nucleic acid sequence-based amplification. Appl. Environ. Microbiol. 73:1457-1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galan, J. E., C. Ginocchio, and P. Costeas. 1992. Molecular and functional characterization of the Salmonella invasion gene invA: homology of InvA to members of a new protein family. J. Bacteriol. 174:4338-4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galan, J. E., J. Pace, and M. J. Hayman. 1992. Involvement of the epidermal growth factor receptor in the invasion of cultured mammalian cells by Salmonella Typhimurium. Nature 357:588-589. [DOI] [PubMed] [Google Scholar]
- 14.Galan, J. E., and R. Curtiss III. 1991. Distribution of the invA, -B, -C, and -D genes of Salmonella Typhimurium among other Salmonella serovars: invA mutants of Salmonella Typhi are deficient for entry into mammalian cells. Infect. Immun. 59:2901-2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gonzalez-Escalona, N., J. Martinez-Urtaza, J. Romero, R. T. Espejo, L. A. Jaykus, and A. Depaola. 2008. Determination of molecular phylogenetics of Vibrio parahaemolyticus strains by multilocus sequence typing. J. Bacteriol. 190:2831-2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gonzalez-Escalona, N., A. Fey, M. G. Hofle, R. T. Espejo, and A. Guzman. 2006. Quantitative reverse transcription polymerase chain reaction analysis of Vibrio cholerae cells entering the viable but non-culturable state and starvation in response to cold shock. Environ. Microbiol. 8:658-666. [DOI] [PubMed] [Google Scholar]
- 17.Hammack, T. S., R. M. Amaguana, M. L. Johnson, and W. H. Andrews. 2003. Effectiveness of universal pre-enrichment broth for recovery of Salmonella from selected dairy foods. J. Assoc. Off. Anal. Chem. Int. 86:714-718. [PubMed] [Google Scholar]
- 18.Hammack, T. S., I. E. Valentin-Bon, A. P. Jacobson, and W. H. Andrews. 2004. Relative effectiveness of the Bacteriological Analytical Manual method for the recovery of Salmonella from whole cantaloupes and cantaloupe rinses with selected preenrichment media and rapid methods. J. Food. Prot. 67:870-877. [DOI] [PubMed] [Google Scholar]
- 19.Hartman, L. J., S. R. Coyne, and D. A. Norwood. 2005. Development of a novel internal positive control for TaqMan based assays. Mol. Cell. Probes 19:51-59. [DOI] [PubMed] [Google Scholar]
- 20.Hoorfar, J., N. Cook, B. Malorny, M. Wagner, D. De Medici, A. Abdulmawjood, and P. Fach. 2004. Diagnostic PCR: making internal amplification control mandatory. Lett. Appl. Microbiol. 38:79-80. [DOI] [PubMed] [Google Scholar]
- 21.Jacobsen, C. S., and W. E. Holben. 2007. Quantification of mRNA in Salmonella sp. seeded soil and chicken manure using magnetic capture hybridization RT-PCR. J. Microbiol. Methods 69:315-321. [DOI] [PubMed] [Google Scholar]
- 22.Jones, T. F., and D. E. Gerber. 2001. Perceived etiology of foodborne illness among public health personnel. Emerg. Infect. Dis. 7:904-905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krascsenicsova, K., L. Piknova, E. Kaclikova, and T. Kuchta. 2008. Detection of Salmonella enterica in food using two-step enrichment and real-time polymerase chain reaction. Lett. Appl. Microbiol. 46:483-487. [DOI] [PubMed] [Google Scholar]
- 24.Lundberg, U., U. Vinatzer, D. Berdnik, A. von Gabain, and M. Baccarini. 1999. Growth phase-regulated induction of Salmonella-induced macrophage apoptosis correlates with transient expression of SPI-1 genes. J. Bacteriol. 181:3433-3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Malorny, B., J. Hoorfar, C. Bunge, and R. Helmuth. 2003. Multicenter validation of the analytical accuracy of Salmonella PCR: towards an international standard. Appl. Environ. Microbiol. 69:290-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malorny, B., C. Bunge, and R. Helmuth. 2007. A real-time PCR for the detection of Salmonella Enteritidis in poultry meat and consumption eggs. J. Microbiol. Methods 70:245-251. [DOI] [PubMed] [Google Scholar]
- 27.Malorny, B., J. Hoorfar, M. Hugas, A. Heuvelink, P. Fach, L. Ellerbroek, C. Bunge, C. Dorn, and R. Helmuth. 2003. Inter-laboratory diagnostic accuracy of a Salmonella specific PCR-based method. Int. J. Food Microbiol. 89:241-249. [DOI] [PubMed] [Google Scholar]
- 28.Malorny, B., E. Paccassoni, P. Fach, C. Bunge, A. Martin, and R. Helmuth. 2004. Diagnostic real-time PCR for detection of Salmonella in food. Appl. Environ. Microbiol. 70:7046-7052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nde, C. W., M. K. Fakhr, C. Doetkott, and C. M. Logue. 2008. An evaluation of conventional culture, invA PCR, and the real-time PCR iQ-Check kit as detection tools for Salmonella in naturally contaminated premarket and retail turkey. J. Food Prot. 71:386-391. [DOI] [PubMed] [Google Scholar]
- 31.Novinscak, A., C. Surette, and M. Filion. 2007. Quantification of Salmonella spp. in composted biosolids using a TaqMan qPCR assay. J. Microbiol. Methods 70:119-126. [DOI] [PubMed] [Google Scholar]
- 32.Oikonomou, I., K. Halatsi, and A. Kyriacou. 2008. Selective PCR: a novel internal amplification control strategy for enhanced sensitivity in Salmonella diagnosis. Lett. Appl. Microbiol. 46:456-461. [DOI] [PubMed] [Google Scholar]
- 33.Pfaffl, M. W., and M. Hageleit. 2001. Validities of mRNA quantification using recombinant RNA and recombinant DNA external calibration curves in real-time RT-PCR. Biotechnol. Lett. 23:275-282. [Google Scholar]
- 34.Rahn, K., S. A. De Grandis, R. C. Clarke, S. A. McEwen, J. E. Galan, C. Ginocchio, R. Curtiss III, and C. L. Gyles. 1992. Amplification of an invA gene sequence of Salmonella Typhimurium by polymerase chain reaction as a specific method of detection of Salmonella. Mol. Cell. Probes 6:271-279. [DOI] [PubMed] [Google Scholar]
- 35.Rodriguez-Lazaro, D., M. Hernandez, T. Esteve, J. Hoorfar, and M. Pla. 2003. A rapid and direct real time PCR-based method for identification of Salmonella spp. J. Microbiol. Methods 54:381-390. [DOI] [PubMed] [Google Scholar]
- 36.Takayama, K., and S. A. Kjelleberg. 2000. The role of RNA stability during bacterial stress responses and starvation. Environ. Microbiol. 2:355-365. [DOI] [PubMed] [Google Scholar]
- 37.Tirado, C., and K. Schmidt. 2001. WHO surveillance programme for control of foodborne infections and intoxications: preliminary results and trends across greater Europe. J. Infect. 43:80-84. [DOI] [PubMed] [Google Scholar]
- 38.U.S. Bureau of the Census. 1996. Statistical abstract of the United States. U.S. Government Printing Office, Washington, DC.
- 39.Valasek, M. A., and J. J. Repa. 2005. The power of real-time PCR. Adv. Physiol. Educ. 29:151-159. [DOI] [PubMed] [Google Scholar]
- 40.Wittwer, C. T., M. G. Herrmann, A. A. Moss, and R. P. Rasmussen. 1997. Continuous fluorescence monitoring of rapid cycle DNA amplification. BioTechniques 22:130-138. [DOI] [PubMed] [Google Scholar]
- 41.Wolffs, P. F., K. Glencross, R. Thibaudeau, and M. W. Griffiths. 2006. Direct quantitation and detection of Salmonellae in biological samples without enrichment, using two-step filtration and real-time PCR. Appl. Environ. Microbiol. 72:3896-3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.