Abstract
We examined the bacterial communities of epilithic biofilms in 17 streams which represented a gradient ranging from relatively pristine streams to streams highly impacted by acid mine drainage (AMD). A combination of automated ribosomal intergenic spacer analysis with multivariate analysis and ordination provided a sensitive, high-throughput method to monitor the impact of AMD on stream bacterial communities. Significant differences in community structure were detected among neutral to alkaline (pH 6.7 to 8.3), acidic (pH 3.9 to 5.7), and very acidic (pH 2.8 to 3.5) streams. DNA sequence analysis revealed that the acidic streams were generally dominated by bacteria related to the iron-oxidizing genus Gallionella, while the organisms in very acidic streams were less diverse and included a high proportion of acidophilic eukaryotes, including taxa related to the algal genera Navicula and Klebsormidium. Despite the presence of high concentrations of dissolved metals (e.g., Al and Zn) and deposits of iron hydroxide in some of the streams studied, pH was the most important determinant of the observed differences in bacterial community variability. These findings confirm that any restoration activities in such systems must focus on dealing with pH as the first priority.
The acid mine drainage (AMD) produced by active and abandoned mines, as well as acidity generated from weathering of naturally exposed pyrite (more generally termed acid rock drainage), can have a significant influence on the ecology of the receiving waters; it can inhibit the growth of organisms across a broad range of trophic levels, including diatoms (15), protozoans (36), aquatic invertebrates (11), and piscivorous birds (24) in freshwater lakes, rivers, and streams. This may occur as a direct consequence of a lower pH, especially in catchments that have a low capacity to buffer these highly acidic solutions, or it may be indirectly due to increased concentrations of dissolved metals, such as iron, aluminum, copper, zinc, cadmium, arsenic, and mercury, as well as metal precipitates, which can cloak the stream beds of the receiving waters (26).
Despite the widespread impact of AMD on aquatic ecosystems, the effects on freshwater bacterial communities are poorly understood. Several studies, conducted in a broad range of geographic regions, indicate the existence of a characteristic bacterial community in AMD-affected environments (10). The bacteria associated with AMD-contaminated environments include acidophilic organisms, such as Acidobacterium capsulatum, Acidithiobacillus ferrooxidans, Acidithiobacillus thiooxidans, Acidiphilium spp., Acidocella spp., and Acidisphaera spp., all of which can tolerate high concentrations of protons in the surrounding water (4). The low pH of AMD-affected waters inhibits the growth of many organisms, but it may promote the growth of certain bacteria, including Acidithiobacillus ferroxidans (14), Leptospirillum ferrooxidans (17), and Ferrimicrobium acidiphilium, which oxidize Fe(II) (stabilized under acidic conditions) to Fe(III), liberating energy for growth.
The aim of this study was to determine if characteristic bacterial communities occur in streams differentially impacted by AMD. Automated ribosomal intergenic spacer analysis (ARISA) was used as a high-throughput, low-cost molecular method to assess differences in the predominant bacterial community structures both in and between streams. Redundancy analysis was then used to identify environmental drivers that might best explain the observed bacterial community heterogeneity. In addition, the compositions of bacterial communities were determined by sequence analysis of cloned 16S rRNA gene fragments, and quantitative PCR (qPCR) was used to determine the relative abundance of bacterial and eukaryotic organisms at each site. We hypothesize that whole bacterial communities are sensitive descriptors of ecological conditions that are capable of differentiating between stream sites impacted in various ways by AMD.
MATERIALS AND METHODS
Experimental outline.
The principal study area was the west coast of the South Island of New Zealand. Coal mining has occurred in this region for over 130 years, and the region contains a large number of active and abandoned mines. Seventeen streams in a number of catchments were chosen to obtain broad geographic coverage representing streams that are highly impacted by AMD (and possibly acid rock drainage from naturally exposed pyrite), as well as less impacted sites and unimpacted reference streams (categories are shown in Table 1). The streams and sites are referred to by abbreviations (for the full names and geographic locations of the stream sites, see Table S1 in the supplemental material).
TABLE 1.
Streams categorized according to the relative impact of mining activity on each stream site
| Category | Site characteristicsa | Site(s) |
|---|---|---|
| I | Pristine, neutral pH (pH 6.7 to 7.4) | LA, MU, TW, BR |
| II | Pristine, naturally acidic (pH 3.9 to pH 5.7) | DR, RU |
| III | Moderately impacted by abandoned mines | BF, WM, DU |
| IV | Highly impacted by active and abandoned mines | GA, WE, SWA, OB, PO, PT |
| V | Highly impacted by active and abandoned mines, undergoing restorative treatment | NG |
| VI | Highly impacted by abandoned mines, but with carbonate geology (pH 8.3) | NMC |
The pH is the pH of water samples collected during April and May 2008.
Sampling procedure.
Streams were sampled during April and May 2008. At each site, physicochemical measurements were recorded within a 10-m reach. Stream temperature was recorded for 30 days prior to sampling using HOBOware Lite data loggers (Onset Computer Corporation, Massachusetts). The pH and specific conductivity of stream water were recorded in the field using an Accumet AP85 pH-conductivity meter (Fisher Scientific Ltd., Pittsburgh, PA). The wetted width was measured at three locations along the reach, and the depth was measured at up to five locations across each width. Water velocity was measured with a Flow-Mate 2000 flow meter (Marsh McBirney Ltd., Maryland), and discharge was calculated using the method of Gore (16). On three different occasions during the month, water samples (100 ml) were collected for nutrient analysis. Samples were filtered in the field (Whatman GF/F filters; pore size, 0.7 μm) and frozen until they were analyzed. Concentrations of dissolved reactive phosphorus, nitrate/nitrite nitrogen, and ammoniacal nitrogen were measured using standard colorimetric protocols (1). For analysis of dissolved metals, water samples (250 ml) were collected on a single sampling occasion at the same time as biofilm samples (see below) and filtered (pore size, 0.45 μm). The concentrations of Al and Zn in the stream water were then determined by inductively coupled plasma mass spectrometry at Hills Laboratories (Hamilton, New Zealand).
At many of our study sites, metal hydroxides had precipitated onto the stream bed. At most of the sites affected by AMD there were orange metal hydroxides, which were likely ferric hydroxides given the low pH (pH <4.1) of most of these sites. As there was no visible sand or silt deposition at any of the study sites, the mass of inorganic matter on individual cobbles was used as a proxy for the mass of metal hydroxide deposited, using the method Niyogi et al. (29). At each site, five randomly selected cobbles that were similar sizes were placed in plastic bags. In the laboratory, the precipitate was scrubbed from each cobble and rinsed, and the dry mass and inorganic mass were calculated using standard protocols (1). We then estimated the amount of inorganic mass (ash mass) per cm2 using the three dimensions of the cobbles (5).
Microbial community analysis was performed for samples of epilithic biofilm. Five cobbles that were similar sizes (maximum length, ∼20 cm) were randomly collected within the 10-m reach from each stream. The biofilm biomass was removed from each cobble by abrasion using a sterile sponge (Speci-Sponge; VWR International). Each sponge was then placed into an individual sterile Whirl-Pak bag with ∼15 ml of stream water to ensure complete immersion, sealed, and stored in the dark at 4°C until it was analyzed. Biofilm biomass was separated from the sponges using a laboratory stomacher (Stomacher 400; Seward, Norfolk, United Kingdom), and the sample material was transferred into centrifuge tubes before it was pelleted by centrifugation (8,000 × g, 20 min). DNA was then extracted from pelleted biofilm samples within 1 week after sample collection using a modified method of Miller et al. (21, 27).
Relative abundance of bacterial and eukaryotic DNA.
To assess changes in the proportions of bacterial and eukaryotic organisms, triplicate qPCRs targeting 16S rRNA and 18S rRNA genes, respectively, were performed for each biofilm sample.
To quantify eukaryotic 18S rRNA genes, a TaqMan fast reagent starter kit (Applied Biosystems Ltd., Melbourne, Australia) containing two eukaryote-specific unlabeled PCR primers and a FAM dye-labeled 18S TaqMan minor groove binder probe was used according to the manufacturer's instructions. Bacterial 16S rRNA genes were quantified using the method of Nadkarni et al. (28), and a dilution series of Escherichia coli K-12 strain MG1655 DNA (GenBank accession no. U00096) was used to generate a standard curve for the qPCR. DNA was amplified using an ABI Prism 7900HT qPCR machine (Applied Biosystems Ltd), and were data analyzed using SDS software (version 2.3; Applied Biosystems Ltd.).
ARISA of biofilm bacterial DNA.
The diversity of bacterial communities was assessed by performing ARISA with each biofilm sample (all five biofilm samples extracted from each of the 17 streams studied) using the method of Lear and Lewis (23). Sample data were then manipulated as described by Lear et al. (21) such that each DNA sample provided 800 variables, which represented the length (in bp) of the 16S-23S intergenic spacer region of constituent bacteria.
To visualize patterns in bacterial community structure based on the ARISA data, nonmetric multidimensional scaling (MDS) was done using the Bray-Curtis matrix. The relationship between data sets was plotted using MDS, and the statistical significance of differences between ARISA data sets was analyzed using permutational multivariate analysis of variance (PERMANOVA) (25). The relationship between the ARISA data and measured environmental variables was investigated using distance-based multivariate multiple regression on the basis of the Bray-Curtis measure using the DISTLM routine (25), constructed using forward selection. All tests were performed using type III sums of squares (as any missing data points caused the data to be unbalanced) and 9,999 permutations with the reduced model (3). Statistical analyses were completed using the Primer 6 (version 6.1.11) computer program (Primer-E Ltd., Plymouth, United Kingdom) with the PERMANOVA+ add-on package (2).
Sequence analysis of cloned bacterial 16S rRNA genes.
Clone libraries of bacterial 16S rRNA genes were constructed using DNA extracted from three streams (BR, DU, and OB), which represented communities from pristine streams with a neutral pH (pH 7.1) (BR), acidic streams that were moderately impacted by AMD (pH 4.5) (DU), and very acidic streams that were highly impacted by AMD (pH 2.8) (OB). To identify the dominant groups of bacteria present, a conserved region of the bacterial 16S rRNA gene was amplified by PCR with the universal bacterial primers PB36 (5′-AGR GTT TGA TCM TGG CTC AG-3′) (34) and PB38 (5′-GKT ACC TTG TTA CGA CTT-3′) (34). PCRs were performed using Promega GoTaq Green DNA polymerase (Invitro Technologies Ltd., New Zealand) as follows: (i) 95°C for 5 min; (ii) 35 cycles of 95°C for 45 s, 55°C for 40 s, and 72°C for 90 s; and then (iii) 72°C for 10 min. PCR products were purified using a Zymo DNA Clean and Concentrator-5 kit (Ngaio Diagnostics Ltd., New Zealand) and cloned using a Promega pGEM-T Easy vector system (Invitro Technologies Ltd., New Zealand) and E. coli One Shot TOP10 competent cells (Invitrogen, New Zealand) according to the manufacturer's instructions. Transformants were screened for an insert using PCR, and products were purified as previously described. For each DNA library, the nucleotide sequences of 96 clones were obtained using a contract sequencing facility (Macrogen Inc., Seoul, Korea). Sequences were analyzed using the NCBI BLAST database to identify the most closely matching sequences. A failure to accurately identify an organism to the genus level was defined as a 16S rRNA gene sequence with less than 97% similarity to any sequence currently deposited in the NCBI GenBank database, as suggested by Drancourt et al. (9).
Nucleotide sequence accession numbers.
The 264 sequences obtained in this study have been deposited in the GenBank database (www.ncbi.nlm.nih.gov) under accession no. FJ203732 to FJ203994.
RESULTS
Physicochemical data.
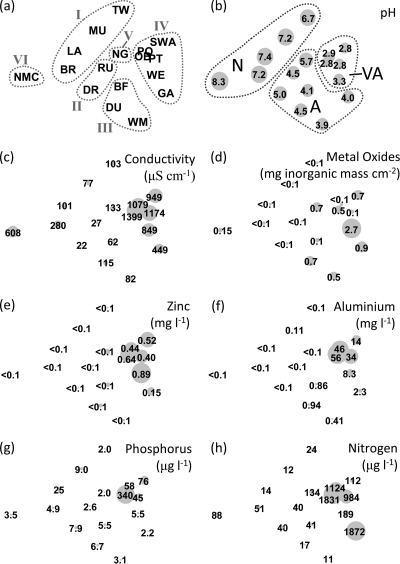
The pH of the stream water varied from pH 2.8 to 8.3 for the 17 streams (for details, see Table S2 in the supplemental material). The conductivity also varied widely for the streams, ranging from 22 to 1,399 μS cm−1. The mass of inorganic matter (ash mass) deposited on cobbles was generally low, except in stream WE (2.73 mg cm−2), in which the mass was more than three times greater than the mass in any other stream. The levels of dissolved aluminum and zinc were highest (>5 mg liter−1 and >0.2 mg liter−1, respectively) in streams OB, PT, SWA, PO, and WE. The concentration of dissolved reactive phosphorus was highest in stream OB (340 μg liter−1), which also contained high concentrations of dissolved inorganic nitrogen (>1 mg liter−1), as did streams PO and GA. Streams with the lowest pH (pH <4) generally exhibited greater conductivity and contained higher concentrations of Zn, Al, N, and P.
Relative abundance of eukaryotic and bacterial DNA.
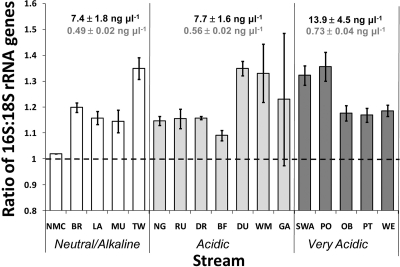
The concentrations of bacterial DNA in biofilm material were greater than the concentrations of eukaryotic DNA in the same samples for all streams (Fig. 1), as measured using qPCR for bacterial 16S rRNA genes and eukaryotic 18S rRNA genes. The average concentrations were 9.8 ng μl−1 and 0.6 ng μl−1 for bacterial and eukaryotic DNA in extracts, respectively. In general, the concentrations of both bacterial and eukaryotic DNA increased with acidity.
FIG. 1.
Ratio of the concentration of 16S rRNA genes to the concentration of 18S rRNA genes, comparing of the cycle threshold values for qPCR performed with biofilm DNA extract from each stream. The error bars indicate standard errors for five replicates per stream site. The dashed line indicates a 1:1 ratio. The values above the bars are average concentrations of bacterial 16S rRNA-primed DNA (black) and average concentrations of 18S rRNA-primed DNA (gray).
ARISA of biofilm DNA.
Multivariate analysis of ARISA data (Fig. 2) showed that there was strong clustering of data from the five samples collected from each stream. A PERMANOVA revealed significant differences in ARISA data among streams (P < 0.0001) but not within streams (P = 0.556). All subsequent statistical analyses were conducted using bacterial community data that were averages for the five rocks sampled in each stream (Fig. 3). The pH and conductivity accounted for 13% and 9% of the total variation in the ARISA data (Table 2) and were the only significant variables (P < 0.05) among the variables measured. Indeed, the ARISA data separated strongly according to pH in MDS plots (Fig. 3b), and data for more acidic streams were located on the right in plots. The data were separated into three clusters, representing streams that refer to neutral/alkaline (pH 6.7 to 8.3), acidic (pH 3.9 to 5.7), and very acidic (pH 2.8 to 3.5). Significant differences in bacterial community structure were detected for each of the combinations of these stream categories (P ≤ 0.0001 for all combinations, as determined by PERMANOVA), and the greatest difference in bacterial community structure was the difference between neutral/alkaline and very acid streams (the average similarity was 3.6, compared to an average similarity of 9.0 for the acid and very acid streams, where the PRIMER similarity values ranged from 0 to 100 [perfect similarity]). SIMPER analysis revealed that 20% of the difference in ARISA data between samples obtained from neutral/alkaline and very acidic streams was due to only seven ARISA peaks. These peaks (at 382, 421, 436, 501, 526, 649, and 788 bp [data not shown]) comprised 34% of the area of the ARISA peaks for samples taken from very acidic streams but only 9% and 2% of the amplified DNA fragments detected for the acidic and neutral/alkaline streams, respectively. The lowest variability in bacterial community profiles occurred with the very acidic streams, which formed the most compact cluster on the MDS plot (PRIMER multivariate dispersion values were 1.34, 1.10, and 0.66 for neutral/alkaline, acidic, and very acidic streams, respectively, where larger values indicate greater dispersion of the data). Significant differences were also detected between sample data for pristine streams (categories I and II), moderately impacted streams (category III), and highly impacted streams (categories IV, V, and VI) (P ≤ 0.0001 for all combinations, as determined by PERMANOVA). The greatest difference in bacterial community structure was the difference between samples from pristine and highly impacted streams (the average similarity was 5.1, compared to 7.2 for a comparison of moderately and highly impacted streams).
FIG. 2.
Differences in bacterial community profiles for biofilm samples from five rocks for 17 different streams. The plot is a nonmetric MDS plot of ARISA traces derived from a Bray-Curtis distance matrix of samples. 2D Stress = 0.25.
FIG. 3.
Differences in bacterial community profiles for rocks in different streams. Each plot was constructed from identical data using nonmetric MDS of ARISA traces derived from a Bray-Curtis distance matrix of samples. The data points are average traces for five bacterial community profiles for each site. 2D Stress = 0.18. The sizes of the gray circles reflect the relative values for the monitored water quality parameters recorded for each site. For plots (a and b), the data are grouped according to site category (a) (categories are described in Table 1) or pH (b) (N, neutral to alkaline [pH >6.7]. A, acidic [pH 3.9 to 5.7]; VA, very acidic [pH <3.5]).
TABLE 2.
Relationship between bacterial community data (averages for the five rock samples taken from each stream) based on the Bray-Curtis distance measure and recorded environmental variables analyzed with a forward selection procedure using a DISTLM routinea
| Variable | R2 (cumulative) | SS (tr) | Pseudo-F | Pb |
|---|---|---|---|---|
| pH of stream water | 0.13 | 5,506 | 1.44 | 0.0001 |
| Conductivity of stream water | 0.23 | 2,954 | 0.74 | 0.0027 |
| Aluminumc | 0.29 | 8,349 | 2.30 | 0.2534 |
| Stream discharge | 0.35 | 7,799 | 2.13 | 0.5935 |
| Dissolved inorganic nitrogenc | 0.40 | 5,089 | 1.32 | 0.5808 |
| Water temp | 0.46 | 7,220 | 1.95 | 0.4982 |
| Inorganic mass | 0.50 | 8,009 | 2.19 | 0.6698 |
| Zincc | 0.56 | 4,432 | 1.14 | 0.5142 |
| Dissolved reactive phosphorusc | 0.60 | 5,288 | 1.38 | 0.7652 |
Variables were fitted sequentially, and the tests whose results are shown are conditional tests for including each successive variable, assuming that the variables which precede it in the table were already included in the model. The variables stream depth and stream width were excluded from the model as a correlation matrix revealed multicollinearity with measurements of stream discharge (r > 0.90). The concentrations of aluminum and zinc were weakly correlated with the conductivity of the stream water (0.80 > r < 0.90).
P values were obtained using 9,999 permutations of residuals with a reduced model.
Concentration in filtered stream water.
Bacterial community composition.
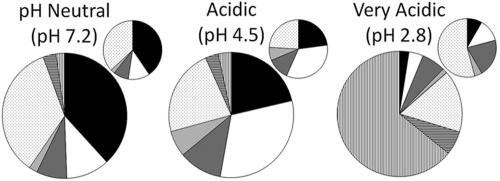
Three clone libraries of 16S rRNA genes were constructed from DNA extracted from stream sites BR, DU, and OB. As MDS plots revealed that bacterial community data clustered according to the pH of the stream water (Fig. 3b), these clone libraries were used to provide an example of the bacterial communities present in the neutral/alkaline, acidic, and very acidic streams examined in this study. All streams contained a large proportion of organisms with novel sequence data (<97% sequence identity to the closest known genus) that could not accurately be attributed to known bacterial genera (69%, 73%, and 40% of the clones in the neutral, acidic, and very acidic streams, respectively).
Analysis of 16S rRNA gene clone libraries revealed that both neutral and acidic streams contained a high proportion of alphaproteobacteria (38% and 21% of the clones for the neutral\alkaline and acidic streams, respectively) (Fig. 4; see Table S3 in the supplemental material). A large proportion of clones obtained from the acidic stream were related to the betaproteobacteria (29%); ∼70% of these organisms were most closely related to the iron-oxidizing bacterium Gallionella ferruginea (accession no. L07897), and phylogenetic analysis suggested that diverse organisms related to the genus Gallionella were present in the stream (data not shown). Organisms related to G. ferruginea were not detected in the neutral/alkaline stream. Sequence analysis of DNA cloned from the acidic stream revealed that presumed acidophilic bacteria related to genera such as Acidocella, Acidiphilium, and Acidobacterium (8, 31, 32, 37) were prevalent and comprised 7% of the total population of clones. Analysis of clones obtained from the very acidic stream revealed large representation by a few eukaryotic species (64% of clones), which were not deliberately targeted using the universal bacterial primers. These clones were related primarily to chloroplast DNA sequences of Klebsormidium, Navicula, and Euglena.
FIG. 4.
Compositions of bacterial communities in the biofilms of a stream with a neutral pH (BR), an acidic stream (DU), and a very acidic stream (OB). Groups were identified by sequence analysis of 16S rRNA genes and are indicated as follows: black sections, alphaproteobacteria; white sections, betaproteobacteria; dark gray sections, gammaproteobacteria; light gray sections, deltaproteobacteria; dotted sections, other bacteria identified to the phylum level; sections with horizontal stripes, other bacteria not identified to the phylum level; sections with vertical stripes, eukaryotes. The smaller graphs show the same data but include only bacteria identified to the phylum level. The clone sequence accession numbers are FJ203732 to FJ203994.
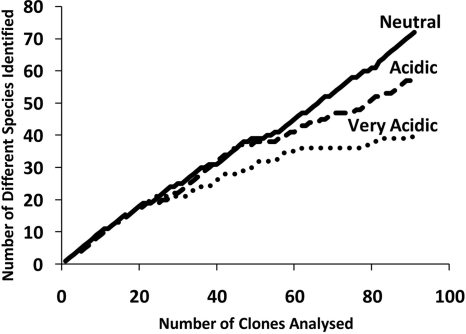
Species accumulation curves based on the abundance of 16S rRNA gene fragments did not reach an asymptote, indicating that the full extent of bacterial diversity was not described by the analysis of ∼96 clones for each stream site (Fig. 5) (similar trends were also observed for the ARISA data [data not shown]). Nevertheless, the data suggest that bacterial diversity decreased with increasing acidity, which was supported by Shannon-Weiner diversity indices of 3.5, 3.3, and 2.7 for the neutral/alkaline, acidic, and very acidic streams, respectively.
FIG. 5.
Species accumulation curves for 16S rRNA gene sequences derived from clone libraries of DNA from biofilms in a stream with a neutral pH (BR), an acidic stream (DU), and a very acidic stream (OB), amplified using universal bacterial primers PB36 and PB38. A new species was considered to be present if the amplified DNA sequence shared <99% similarity with any other sequence analyzed for the same clone library.
DISCUSSION
While most studies conducted to date have considered only the site-specific responses of bacterial communities exposed to AMD, we identified differences in bacterial community structure among a number of streams and catchments impacted differently by AMD. In support of our original hypothesis, descriptors of the structure of whole bacterial communities (obtained using ARISA) were sufficiently sensitive to discriminate between streams exposed to different degrees of impact by AMD.
If the biogeography of complex environmental communities is largely shaped by only a small number of key regulatory factors (e.g., pH and temperature), then predictive models might enable useful estimates of bacterial community structure to be generated from measurements of only a small number of environmental variables, as is routinely done to predict the structure of freshwater macroinvertebrate communities (e.g., RIVPACS [38]). In the present study, differences in bacterial community structure between sites were shown to be largely driven by pH and conductivity. No significant effect was detected for any other recorded parameter commonly associated with the impact of mine drainage (such as the concentration of zinc or the mass of precipitated metal hydroxides). In support of this, the bacterial communities in pristine streams with naturally low pHs (pH < 5.5) were more similar to the bacterial communities in streams impacted by AMD than to the bacterial communities in other pristine sites with near-neutral stream water (pH >6.5). The presumptive effect of pH was further emphasized by the inclusion of data for site NMC, which drains a catchment influenced by the abandoned Strongman Mine north of Greymouth and has elevated levels of conductivity, nitrogen, and metal hydroxide deposition. Despite the location of stream NMC in a catchment affected by mining activities, the pH of the water in this stream is naturally high (∼pH 8), because it is influenced by underlying limestone. It is therefore interesting that the bacterial community structure in stream NMC was most similar to that in pristine streams with a comparable, near-neutral pH, providing additional evidence of the very important role of pH in shaping the bacterial community structure in such streams. Furthermore, the bacterial community structure at site NG was more similar to that in the pristine streams than to that in any of the other highly impacted streams (with the exception of site NMC), despite elevated concentrations of some metals, such as zinc. Interestingly, this site is downstream of Mangatini Stream, which has recently been amended with lime (since April 2008) in an effort to alleviate the effects of acidic drainage on the stream. This may indicate that there was partial recovery of the bacterial community following pH adjustment.
Since multivariate analysis of the ARISA data revealed significant differences in bacterial community structure associated with pH, clone libraries of 16S rRNA were constructed to provide examples of the dominant organisms present in neutral, acidic, and very acidic streams. The majority of bacteria identified in this study have not been cultured yet or accurately identified to the genus level. Nevertheless, differences in bacterial community composition between streams were apparent, and organisms detected in the acidic and very acidic streams have been found in abundance in other sites affected by AMD. For example, DNA sequences most closely related to sequences of the iron-oxidizing genus Gallionella, as well as sequences of the acidophilic bacterial genera Acidocella, Acidiphilium, and Acidobacteria, were abundant in a clone library of the acidic stream. Such organisms have previously been detected in abundance at a large number of affected sites (7, 18, 20) and may be a useful indicator of the impact of AMD. For the very acidic stream, DNA sequence analysis revealed that the filamentous alga Klebsormidium and the diatom Navicula were abundant. Again, the competitive advantage of these genera at sites affected by AMD is well documented (6). Klebsormidium can be a good indicator of iron concentrations in water (35), while diatoms, such as Navicula, are known to remain active in very acidic (pH <3.0) environments (19). Increases in algal biomass and primary production have previously been observed following stream acidification (pH <5.0) and may be attributed to reduced grazing pressure by macroinvertebrates and the low-pH preference of some algal species (such as Klebsormidium sp., which may be found in abundance in highly acidic freshwater environments [33]). The increased abundance of bacterial DNA in the more acidic streams may be directly related to the increased abundance of algae such as Klebsormidium sp., which provide a large surface area for bacterial attachment while also exuding labile dissolved organic matter, stimulating bacterial growth (30). However, without further sequence analysis, it remains possible that the increased abundance of “bacterial DNA” detected by qPCR was in fact due to increased numbers of algal chloroplasts, which contain genes closely related to bacterial 16S rRNA genes and were found in high abundance in a clone library of the very acidic stream.
The results of this study suggest that analysis of bacterial biofilm community structure by ARISA may provide a sensitive measure of the extent of ecosystem degradation and be a useful tool for monitoring the success of ecological restoration in streams impacted by AMD. Importantly, the successful restoration of any degraded ecosystem requires knowledge of the major factors contributing to the deterioration of that environment. While a number of recent studies highlight the importance of concentrations of metals such as Fe, Zn, and Cu for bacterial community structure in natural environmental systems (12), the findings of the present study support the concept that pH is a universal and primary factor influencing the biogeography of microbial communities, as recently suggested by Fierer et al. (13). In conclusion, we found that the combination of ARISA and multivariate analysis is a sensitive, high-throughput method for monitoring the impact of human activity on freshwater streams and for identifying the key drivers of ecosystem change at such sites.
Supplementary Material
Acknowledgments
This work was funded by the Foundation for Research, Science and Technology, New Zealand (grants UA0X306 and CRLX0401).
We thank Kristine Boxen for her kind assistance with the ARISA of bacterial DNA.
Footnotes
Published ahead of print on 10 April 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.American Public Health Association. 1998. Standard method for the examination of water and wastewater, 20th ed. American Public Health Association, Washington, DC.
- 2.Anderson, M. J., R. N. Gorley, and K. R. Clarke. 2008. PERMANOVA+ for PRIMER: guide to software and statistical methods. PRIMER-E Ltd., Plymouth, United Kingdom.
- 3.Anderson, M. J., and T. J. Willis. 2003. Canonical analysis of principal coordinates: a useful method of constrained ordination for ecology. Ecology 84:511-525. [Google Scholar]
- 4.Baker, B. J., and J. F. Banfield. 2003. Microbial communities in acid mine drainage. FEMS Microbiol. Ecol. 44:139-152. [DOI] [PubMed] [Google Scholar]
- 5.Biggs, B. J. F., and C. Kilroy. 2000. Stream periphyton manual. New Zealand Ministry for the Environment/National Institute of Water and Atmospheric Research, Christchurch.
- 6.Bray, J., P. A. Broady, D. K. Niyogi, and J. S. Harding. 2008. Periphyton communities in New Zealand streams impacted by acid mine drainage. Mar. Freshw. Res. 59:1084-1091. [Google Scholar]
- 7.Bruneel, O., R. Duran, C. Casiot, F. Elbaz-Poulichet, and J.-C. Personne. 2006. Diversity of microorganisms in Fe-As-rich acid mine drainage waters of Carnoules, France. Appl. Environ. Microbiol. 72:551-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cleaver, A. A., N. P. Burton, and P. R. Norris. 2007. A novel Acidimicrobium species in continuous cultures of moderately thermophilic, mineral-sulfide-oxidizing acidophiles. Appl. Environ. Microbiol. 73:4294-4299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drancourt, M., C. Bollet, A. Carlioz, R. Martelin, J.-P. Gayral, and D. Raoult. 2000. 16S ribosomal DNA sequence analysis of a large collection of environmental and clinical unidentifiable bacterial isolates. J. Clin. Microbiol. 38:3623-3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Druschel, G. K., B. J. Baker, T. M. Gihring, and J. F. Banfield. 2004. Acid mine drainage biogeochemistry at Iron Mountain, California. Geochem. Trans. 5:13-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dsa, J. V., K. S. Johnson, D. Lopez, C. Kanuckel, and J. Tumlinson. 2008. Residual toxicity of acid mine drainage-contaminated sediment to stream macroinvertebrates: relative contribution of acidity vs. metals. Water Air Soil Pollut. 194:185-195. [Google Scholar]
- 12.Duarte, S., C. Pascal, A. Alves, A. Correia, and F. Cassio. 2008. Copper and zinc mixtures induce shifts in microbial communities and reduce litter decomposition in streams. Freshw. Biol. 53:91-101. [Google Scholar]
- 13.Fierer, N., J. L. Morse, S. T. Berthrong, E. S. Bernhardt, and R. B. Jackson. 2007. Environmental controls on the landscape-scale biogeography of stream bacterial communities. Ecology 88:2162-2173. [DOI] [PubMed] [Google Scholar]
- 14.Fowler, T. A., P. R. Holmes, and F. K. Crundwell. 1999. Mechanism of pyrite dissolution in the presence of Thiobacillus ferroxidans. Appl. Environ. Microbiol. 65:2987-2993.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerhardt, A., L. J. de Bisthoven, K. Guhr, A. M. V. M. Soares, and M. J. Pereira. 2008. Phytoassessment of acid mine drainage: Lemna gibba bioassay and diatom community structure. Ecotoxicology 17:47-58. [DOI] [PubMed] [Google Scholar]
- 16.Gore, J. 1996. Discharge measurements and streamflow analysis, p. 53-74. In F. R. Hauer and G. A. Lamberti (ed.), Methods in stream ecology. Academic Press, New York, NY.
- 17.Johnson, D. B., N. Okibe, and K. B. Hallberg. 2004. Differentiation and identification of iron-oxidising acidophilic bacteria using cultivation techniques and amplified ribosomal DNA restriction enzyme analysis. J. Microbiol. Methods 60:299-313. [DOI] [PubMed] [Google Scholar]
- 18.Johnson, D. B., S. Rolfe, K. B. Hallberg, and E. R. Iversen. 2001. Isolation and phylogenetic characterization of acidophilic microorganisms indigenous to acidic drainage waters at an abandoned Norwegian copper mine. Environ. Microbiol. 3:630-637. [DOI] [PubMed] [Google Scholar]
- 19.Kelly, M. 1988. Mining and the freshwater environment. Elsevier Applied Science, London, United Kingdom.
- 20.Kisimoto, N., Y. Kosako, and T. Tano. 1991. Acidobacterium capsulatum gen. nov., sp. nov.: an acidophilic chemoorganotrophic bacterium containing menaquinone from an acidic environment. Curr. Microbiol. 22:1-7. [Google Scholar]
- 21.Lear, G., M. J. Anderson, J. G. Smith, K. Boxen, and G. Lewis. 2008. Spatial and temporal heterogeneity of the bacterial communities within stream epilithic biofilms. FEMS Microbiol. Ecol. 65:463-473. [DOI] [PubMed] [Google Scholar]
- 22.Reference deleted.
- 23.Lear, G., and G. D. Lewis. 2009. Impact of catchment land use on bacterial communities within stream biofilms. Ecol. Indicators 9:848-855. [Google Scholar]
- 24.Levings, C. D., K. L. Barry, J. A. Grout, G. E. Piercey, A. D. Marsden, A. P. Coombs, and B. Mossop. 2004. Effects of acid mine drainage on the estuarine food web, Britannia Beach, Howe Sound, British Columbia, Canada. Hydrobiologia 525:185-202. [Google Scholar]
- 25.McArdle, B. H., and M. J. Anderson. 2001. Fitting multivariate models to community data: a comment on distance-based redundancy analysis. Ecology 82:290-297. [Google Scholar]
- 26.McKnight, D. M., and G. L. Feder. 1984. The ecological effect of acid conditions and precipitation of hydrous metal oxides in a Rocky Mountain stream. Hydrobiologia 119:129-138. [Google Scholar]
- 27.Miller, D. N., J. E. Bryant, E. L. Madsen, and W. C. Ghiorse. 1999. Evaluation and optimization of DNA extraction and purification procedures for soil and sediment samples. Appl. Environ. Microbiol. 65:4715-4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nadkarni, M. A., F. E. Martin, N. A. Jacques, and N. Hunter. 2002. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology 148:257-266. [DOI] [PubMed] [Google Scholar]
- 29.Niyogi, D. K., J. W. Lewis, and D. McKnight. 2001. Litter breakdown in mountain streams affected by mine drainage: biotic mediation of abiotic controls. Ecol. Appl. 11:506-516. [Google Scholar]
- 30.Niyogi, D. K., W. M. Lewis, and J. W. Lewis. 2003. Direct and indirect effects of mine drainage on bacterial processes in mountain streams. J. N. Am. Benthol. Soc. 22:276-291. [Google Scholar]
- 31.Pankrator, T. A., Y. M. Serkebaeva, I. S. Kulichevskaya, W. Liesack, and S. N. Dedysh. 2008. Substrate-induced growth and isolation of Acidobacteria from acid Sphagnum peat. ISME J. 2:551-560. [DOI] [PubMed] [Google Scholar]
- 32.Roling, W. F. M., S. Ortega-Lucach, S. R. Larter, and I. M. Head. 2006. Acidophilic microbial communities associated with a natural, biodegraded hydrocarbon seepage. J. Appl. Microbiol. 101:290-299. [DOI] [PubMed] [Google Scholar]
- 33.Sabater, S., T. Buchaca, J. Cambra, J. Catalan, H. Guash, N. Ivorra, I. Munoz, E. Navarro, M. Real, and A. Romani. 2003. Structure and function of benthic algal communities in an extremely acidic river. J. Phycol. 39:481-489. [Google Scholar]
- 34.Saul, D. J., A. Rodrigo, R. Reeves, L. Williams, K. Borges, H. Morgan, and P. Bergquist. 1993. Phylogeny of twenty Thermus isolates constructed from 16S rRNA gene sequence data. Int. J. Syst. Bacteriol. 43:754-760. [DOI] [PubMed] [Google Scholar]
- 35.Stevens, A. E., B. C. McCarthy, and M. L. Vis. 2001. Metal content of Klebsormidium-dominated (Chlorophyta) algal mats from acid mine drainage waters in southeastern Ohio. J. Torrey Bot. Soc. 128:226-233. [Google Scholar]
- 36.Tremaine, S. C., and A. L. Mills. 1991. Impact of water column acidification on protozoan bacterivory at the lake sediment-water interface. Appl. Environ. Microbiol. 57:775-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wakao, N., and S. Yamanaka. 2002. Enhanced growth of Acidocella facilis and related acidophilic bacteria at high concentrations of aluminium. Microbes Environ. 17:98-104. [Google Scholar]
- 38.Wright, J. F., M. T. Furse, and P. D. Armitage. 1993. RIVPACS—a technique for evaluating the biological quality of rivers in the U.K. Eur. Water Pollut. Control 3:15-25. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.