Abstract
Little is known about the contribution of bacteria and fungi to decomposition of different carbon compounds in arctic soils, which are an important carbon store and possibly vulnerable to climate warming. Soil samples from a subarctic tundra heath were incubated with 13C-labeled glucose, acetic acid, glycine, starch, and vanillin, and the incorporation of 13C into different phospholipid fatty acids (PLFA; indicative of growth) and neutral lipid fatty acids (NLFA; indicative of fungal storage) was measured after 1 and 7 days. The use of 13C-labeled substrates allowed the addition of substrates at concentrations low enough not to affect the total amount of PLFA. The label of glucose and acetic acid was rapidly incorporated into the PLFA in a pattern largely corresponding to the fatty acid concentration profile, while glycine and especially starch were mainly taken up by bacteria and not fungi, showing that different groups of the microbial community were responsible for substrate utilization. The 13C-incorporation from the complex substrates (starch and vanillin) increased over time. There was significant allocation of 13C into the fungal NLFA, except for starch. For glucose, acetic acid, and glycine, the allocation decreased over time, indicating use of the storage products, whereas for vanillin incorporation into fungal NLFA increased during the incubation. In addition to providing information on functioning of the microbial communities in an arctic soil, our study showed that the combination of PLFA and NLFA analyses yields additional information on the dynamics of substrate degradation.
Bacteria and fungi comprise more than 90% of the soil microbial biomass and are the main agents for decomposition of organic matter in soil. Until recently it was thought that these two organism groups could be lumped together in this respect, and total microbial biomass or total activity (respiration) was often the only variable included in soil microbiology studies of decomposition and soil organic matter turnover (39). However, there is increasing evidence suggesting that whether decomposition is performed by bacteria or fungi, thereby channeling energy through the bacterial or the fungal food web, has profound effects on the ecosystem. Such effects can have direct influence on the higher trophic levels in the food web (30) or indirect effects on nutrient mineralization rates (14) and nutrient transfer (19, 20), and they can even determine the extent of carbon sequestration in the soil (37). The situation becomes even more complex when the impact of changes in climate, nitrogen availability, and litter input on the balance between bacteria and fungi is taken into account. The Arctic region has been identified as an area that will be especially vulnerable to these changes (3).
Little is known about the contribution of bacteria and fungi to the utilization of plant-derived carbon substrates in arctic soils. Differentiation of the bacterial and fungal contributions to decomposition has hitherto relied to a large extent on changes in bacterial and fungal biomasses, for example, by analysis of patterns of phospholipid fatty acids (PLFA) (40). PLFA are components of the cell membrane, and some of the PLFA extracted from the soil are characteristic for a certain microbial group in the environment. However, for changes in PLFA concentrations after the addition of substrates to be detected, substrates often have to be added at unrealistically large amounts. Even then only small changes in the PLFA concentrations will often be detected (35).
One way of overcoming these problems is to follow the incorporation of 13C label from added substrates into specific fatty acids (8, 17). This approach adds a new dimension—metabolic function—to the study of soil microbial communities without the need of cultivation. It also increases the sensitivity in tracing responses of organism groups to different substrates as the addition of substrates at low and more realistic concentrations with high specific 13C label will induce large changes in the 13C concentration of the PLFA without changing the total amount of PLFA.
Carbon-13 labeling has been used to follow uptake of recent photosynthates (11, 13, 27), pure substrates (10, 12, 32, 33, 41), and complex labeled plant material (28, 41, 43, 44) into PLFA although seldom in arctic soils. However, microorganisms incorporate carbon not only into phospholipids (indicating growth) but also into storage products, for example, when a nutrient other than carbon is limiting growth or under growth-restricting conditions. Thus, with excess carbon both bacteria and fungi will store carbon for later need, for example, as polyhydroxyalkanoate or glycogen (bacteria) and triacylglycerols (fungi). Thus, neutral lipid fatty acids (NLFA) of fungal origin can be used to indicate storage in fungi (4). Degraded PLFA, resulting in diacylglycerols, will also end up in the corresponding NLFA fraction, and NLFA has thus been suggested as an indicator of recently dead bacterial biomass (42). Therefore, the NLFA/PLFA ratio serves two purposes: for fungal lipids a higher NLFA/PLFA ratio would indicate allocation of lipids to energy storage while for bacterial lipids it would indicate turnover of this bacterial group. However, the latter will probably be of minor importance during short incubations. As far as we know, no studies on soil microorganisms have used incorporation of 13C from substrates to indicate both effects on growth (incorporation into PLFA) and storage (incorporation into NLFA).
We assessed the uptake of 13C-labeled substrates into lipid biomarkers of different microbial groups in a laboratory incubation experiment using soil from an arctic tundra heath. The selected substrates represented carbon sources present in soil. Glucose, acetic acid, and glycine are simple compounds common in plant root exudates, and glycine is also a nitrogen source. Starch is a very common polysaccharide in plant residues. Vanillin is a common product of lignin depolymerization (18) containing a phenol ring and is often used as a model substance to indicate lignin degradation. Starch and vanillin are therefore examples of more complex substrates and are supposedly more difficult to decompose. We followed the incorporation of the label into different PLFA and NLFA over time. We hypothesized that 13C from the simple compounds would be more rapidly incorporated into microbial PLFA than 13C from the more complex substrates (more rapid growth), and thus we expected 13C emanating from the complex substrates to increase in concentration in the PLFA and NLFA over time. We also hypothesized that bacteria would be better than fungi in utilizing simple compounds while the label from the more complex substrates would preferentially be incorporated into PLFA, indicating fungi (6, 29). We also expected 13C from the C-rich substrates to be incorporated into NLFA (fungal storage) to a larger extent than C from glycine, which also serves as a nitrogen source (4). However, with time the carbon in storage structures would decrease as it would be used for growth or maintenance energy.
MATERIALS AND METHODS
Soil samples.
The soil samples originated at a tundra heath in Abisko, northern Sweden (68°21′N, 18°49′E; 450 m above sea level). The vegetation of the heath was dominated by Cassiope tetragona (L.) D. Don. According to the climate records of the Abisko Scientific Research Station, the climate of Abisko is subarctic/alpine, with a mean annual air temperature of 0.07°C (1986 to 2006), precipitation of 308 mm, and a growing season of about 3 months.
Cored soil samples to 5-cm depth were taken from the six unmanipulated control plots of an experiment originally designed to simulate climatic warming and higher nutrient availability (24). The plots were distributed over an area of approximately 800 m2. The sampling took place on 24 August 2006. Each of the six soil samples was a composite of three separate soil cores, from which roots and stones were removed by hand. The well-drained 12- to 15-cm deep organic layer rests on top of rocky mineral soil and bedrock consisting of base-rich mica schists. The pH of the organic layer was 7.1 (36). The soil organic matter (SOM) content of the sampled uppermost 5-cm layer was approximately 89%, and the gravimetric water content was on average 248% (34). The total C pool at the site (upper 10 cm) was 2.79 ± 0.09 kg m−2, and the total N and P pools were 105 ± 5 g m−2 and 6.35 ± 0.26 g m−2, respectively (25). This gives a C/N ratio of 26.5 and a C/P ratio of approximately 440. The concentration of ammonium was 16 to 20 μg g−1 of SOM and that of microbial biomass C was 10 to 16 mg g−1 of SOM (25, 34).
Incubation with substrates.
For incubation with the various substrates, 10 0.4-g (fresh weight) subsamples of each soil sample were weighed into small minigrip bags. The five different substrates, universally 13C-labeled glucose (99%), glycine (99%), and starch (98%); ring-labeled vanillin (99%); and a 1:1 mixture of acetic acid labeled with 13C in each of the C atoms (Cambridge Isotope Laboratories, Andover, MA), were stirred into the soil in 100 μl of water in duplicates. The added amount was 0.5 mg of substrate g−1 of wet soil (≈2 mg of substrate g−1 of organic matter). This corresponds to 0.21 mg of 13C for glucose, 0.17 mg of 13C for glycine, 0.11 mg of 13C for acetic acid, 0.23 mg of 13C for starch, and 0.25 mg of 13C for vanillin per gram (fresh weight) of soil.
Half of the subsamples were incubated for 24 h, and the other half was incubated for 7 days at 15°C in dark. At the end of the incubation period, the soil was frozen and freeze dried. There were six replicates for each substrate and incubation time.
Analysis of fatty acids.
Fatty acids were extracted from the freeze-dried soil following a modified Bligh and Dyer method according to Frostegård et al. (22). The extracted lipids were fractionated on silicic acid columns (Bond Elut; Varian Inc., Palo Alto, CA) with chloroform (neutral lipids), with acetone (glycolipids), and with methanol (phospholipids). The NLFA and PLFA fractions were subjected to mild alkaline methanolysis, which transformed the fatty acids to free fatty acid methyl esters. Methyl nonadecanoate fatty acid (19:0) was used as an internal standard.
The fatty acid concentrations were determined by analyzing the free fatty acid methyl esters on a Hewlett-Packard 6890 gas chromatograph equipped with a 50-m HP5 capillary column (Hewlett-Packard) and with helium as the carrier gas. The gas chromatograph was interfaced with a Europa 20/20 isotope ratio mass spectrometer used for determination of the δ13C values (see reference 31 for further details on instrumentation).
Calculations for the isotope data were conducted following Boschker (7). The δ13C values of each lipid were corrected for the methyl group added during methanolysis (1). For each PLFA and NLFA, the absolute amount of 13C incorporated was calculated by relating the increase in the fraction 13C [F = R/(R + 1), where F is the fraction 13C and R is a 13C/12C ratio] after labeling to the fatty acid concentration and the amount of 13C in the added substrate.
The fatty acids are presented as the total number of carbon atoms, followed by a colon and then the number of double bonds. The prefixes a and i signify anteiso- and isobranching, respectively. The prefix cy indicates cyclopropyl fatty acids, while 10Me is a methyl group on the 10th carbon atom from the carboxyl end of the molecule. Terminally and mid-chain branched fatty acids were considered indicative of gram-positive bacteria (i15:0, a15:0, i16:0, 10Me16:0, i17:0, a17:0, 10Me17:0, and 10Me18:0), and cyclopropyl saturated (cy-17:0 and cy-19:0) and monounsaturated (16:1ω7 and 18:1ω7) fatty acids were considered indicative of gram-negative bacteria. The fatty acid 18:2ω6,9 was considered to represent fungi (21) in calculating the ratios of fungal-to-bacterial PLFA. The PLFA 18:1ω9 is often considered to be of fungal origin, but it can also be found in bacteria. The NLFA 18:1ω9 is of fungal origin and is often found to increase under excess of carbon (4). The results for 18:1ω9 were therefore used as additional evidence, together with 18:2ω6,9, for effects on fungi.
Data analysis.
The total fatty acid concentrations, total amounts of incorporated 13C in the fatty acids, and the various calculated ratios were tested for effects of substrate additions and incubation time using the linear mixed-model procedure of SPSS for Windows, version 14.0 (SPSS Inc., Chicago, IL). The model also included the interaction between substrate and incubation time and the field plot as a random factor.
Fatty acid concentration profiles and the 13C incorporation profiles (relative values) were analyzed by principal component analysis (PCA) using SIMCA-P software, version 11.5 (Umetrics, Umeå, Sweden). The data were unit variance scaled and centered prior to analysis. To assess whether the profiles differed among the substrates and incubation times, the extracted principal components (PCs) were analyzed by the linear mixed-model procedure as described above.
RESULTS
Substrate addition effects on fatty acid concentrations.
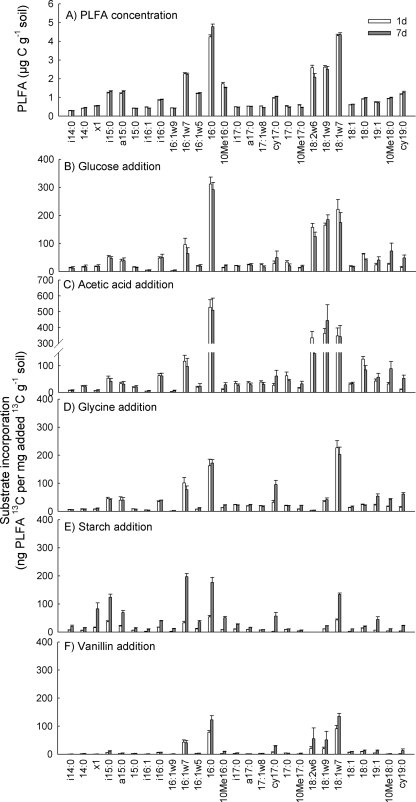
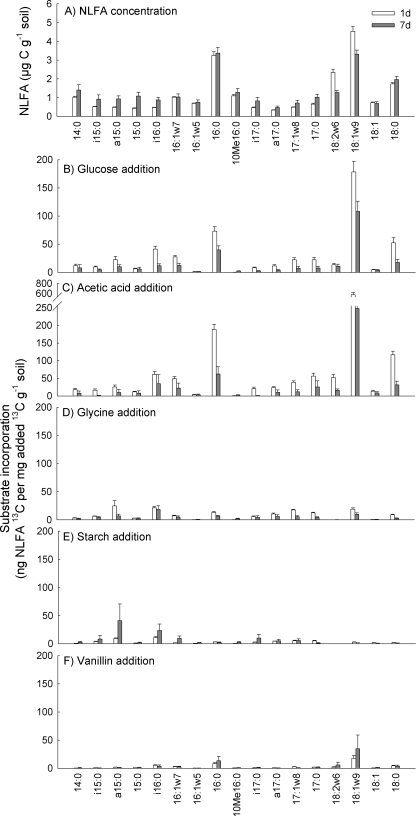
The total PLFA and NLFA concentrations and the NLFA-to-PLFA ratios were not significantly affected by substrate additions (P > 0.40) or incubation time (P > 0.29) (Table 1). Substrate additions did not alter the fatty acid profiles either, based on PCA of the individual fatty acid concentrations (data not shown). There were only minor effects of time on the PLFA (Fig. 1A) and NLFA patterns (see Fig. 3A) although the ratio of fungal-to-bacterial PLFA decreased with time (Table 1). Thus, the substrate additions were not large enough to significantly affect the PLFA and NLFA patterns.
TABLE 1.
Concentrations of PLFA and NLFA and ratios of NLFA to PLFA and fungi to bacteria in tundra heath soil following incubation with added glucose, acetic acid, glycine, starch, or vanillin for 1 or 7 days
| Fatty acid(s)a | Concn in soil for the indicated substrate and incubation period (μg of C g−1 of soil)b
|
Statistical significance (P)c | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Glucose addition
|
Acetic acid addition
|
Glycine addition
|
Starch addition
|
Vanillin addition
|
|||||||
| 1 day | 7 days | 1 day | 7 days | 1 day | 7 days | 1 day | 7 days | 1 day | 7 days | ||
| PLFATotal | 35.9 ± 1.7 | 32.5 ± 1.6 | 32.4 ± 0.9 | 31.2 ± 1.6 | 32.6 ± 1.4 | 32.5 ± 2.0 | 33.9 ± 1.4 | 34.8 ± 2.5 | 31.8 ± 1.1 | 34.2 ± 2.3 | NS |
| NLFATotal | 20.5 ± 1.7 | 20.0 ± 1.4 | 20.8 ± 1.9 | 22.3 ± 6.1 | 22.5 ± 2.6 | 21.5 ± 4.6 | 22.6 ± 1.9 | 32.9 ± 11.9 | 20.5 ± 1.9 | 23.6 ± 5.7 | NS |
| NLFA/PLFATotal | 0.60 ± 0.07 | 0.61 ± 0.01 | 0.64 ± 0.06 | 0.70 ± 0.17 | 0.71 ± 0.10 | 0.65 ± 0.11 | 0.70 ± 0.06 | 0.91 ± 0.34 | 0.64 ± 0.05 | 0.68 ± 0.15 | NS |
| NLFA/PLFA18:2ω6 | 0.80 ± 0.11 | 0.66 ± 0.11 | 0.90 ± 0.07 | 0.56 ± 0.07 | 1.10 ± 0.25 | 0.62 ± 0.02 | 0.88 ± 0.14 | 0.63 ± 0.08 | 0.88 ± 0.06 | 0.52 ± 0.06 | <0.05 |
| NLFA/PLFA18:1ω9 | 1.59 ± 0.19 | 1.49 ± 0.17 | 1.76 ± 0.14 | 1.28 ± 0.07 | 1.99 ± 0.43 | 1.19 ± 0.01 | 1.79 ± 0.33 | 1.43 ± 0.18 | 1.70 ± 0.10 | 1.22 ± 0.04 | NS |
| Fungi/bacteriaPLFA | 0.17 ± 0.02 | 0.13 ± 0.03 | 0.16 ± 0.02 | 0.13 ± 0.03 | 0.15 ± 0.02 | 0.11 ± 0.02 | 0.15 ± 0.02 | 0.13 ± 0.02 | 0.17 ± 0.02 | 0.14 ± 0.04 | <0.001 |
The subscript indicates the fatty acid(s) measured.
Values are means ± standard errors (n = 6).
Statistical significance for the effects of incubation time is shown. NS, P > 0.05. Substrate addition had no significant effects.
FIG. 1.
Original PLFA concentrations (A) and incorporation of 13C from added 13C-labeled glucose (B), acetic acid (C), glycine (D), starch (E), and vanillin (F) in PLFA in tundra heath soil following incubation for 1 or 7 days. The original concentration data shown in panel A are an average (plus standard error; n = 30) of samples amended with the various substrates, as the additions did not affect PLFA concentrations. The incorporation data in panels B to F are means plus standard errors (n = 6). d, days.
FIG. 3.
Original NLFA concentrations (A) and incorporation of 13C from added 13C-labeled glucose (B), acetic acid (C), glycine (D), starch (E), and vanillin (F) in NLFA in tundra heath soil following incubation for 1 or 7 days. The original concentration data in panel A are an average (plus standard error; n = 30) of samples amended with the various substrates, as the additions did not affect NLFA concentrations. The incorporation data in panels B to F are means plus standard errors (n = 6).
Incorporation of added label into PLFA.
Incorporation was calculated as 13C recovery in different lipid fractions proportioned to the 13C added in the substrate. The total incorporation of 13C in the sum of all PLFA per mg−1 of added label was significantly different between the substrates (Table 2). Acetic acid had highest recovery, followed by glucose. Incorporation of 13C label from glycine was on average 58% lower than that from acetic acid while utilization of starch and especially of vanillin was still considerably lower, on average 65% and 82% lower than that of acetic acid, respectively (Table 2).
TABLE 2.
13C incorporation into PLFA and NLFA in tundra heath soil following incubation with added glucose, acetic acid, glycine, starch, or vanillin for 1 or 7 days
| Fatty acid(s)a |
13C incorporation in soil for the indicated substrate and incubation period (μg 13C mg−1 of added 13C g−1 of soil)b
|
Statistical significancec | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Glucose addition
|
Acetic acid addition
|
Glycine addition
|
Starch addition
|
Vanillin addition
|
|||||||
| 1 day | 7 days | 1 day | 7 days | 1 day | 7 days | 1 day | 7 days | 1 day | 7 days | ||
| PLFATotal | 1.48 ± 0.13 | 1.45 ± 0.20 | 2.38 ± 0.24 | 2.30 ± 0.47 | 0.92 ± 0.11 | 1.09 ± 0.08 | 0.40 ± 0.06 | 1.24 ± 0.04 | 0.32 ± 0.03 | 0.54 ± 0.08 | S ***, T *, S×T * |
| NLFATotal | 0.51 ± 0.06 | 0.26 ± 0.06 | 1.27 ± 0.11 | 0.51 ± 0.20 | 0.15 ± 0.02 | 0.08 ± 0.02 | 0.06 ± 0.01 | 0.12 ± 0.05 | 0.05 ± 0.01 | 0.08 ± 0.04 | S ***, T ***, S×T *** |
| NLFA/PLFATotal | 0.36 ± 0.03 | 0.19 ± 0.05 | 0.54 ± 0.01 | 0.21 ± 0.04 | 0.17 ± 0.01 | 0.08 ± 0.02 | 0.19 ± 0.03 | 0.09 ± 0.04 | 0.17 ± 0.03 | 0.12 ± 0.06 | S ***, T ***, S×T *** |
| NLFA/PLFA18:2ω6 | 0.09 ± 0.01 | 0.08 ± 0.02 | 0.16 ± 0.01 | 0.02 ± 0.06 | 0.07 ± 0.07 | 0.09 ± 0.03 | —e | —e | 0.08 ± 0.01 | 0.07 ± 0.02 | NS |
| NLFA/PLFA18:1ω9 | 1.09 ± 0.16 | 0.58 ± 0.07 | 1.65 ± 0.23 | 0.55 ± 0.03 | 0.55 ± 0.04 | 0.23 ± 0.03 | 0.31 ± 0.06 | 0.05 ± 0.03 | 0.87 ± 0.08 | 0.53 ± 0.14 | S ***, T ***, S×T * |
| Fungi/bacteriaPLFA | 0.30 ± 0.05 | 0.24 ± 0.09 | 0.47 ± 0.06 | 0.21 ± 0.16 | 0.006 ± 0.001 | 0.006 ± 0.002 | 0d | 0.0001 ± 0.00007 | 0.16 ± 0.07 | 0.22 ± 0.16 | S ***, S×T * |
The subscript indicates the fatty acid(s) measured.
Values are means ± standard errors (n = 6).
Statistical significance for the effects of substrate addition (S), incubation time (T), and the interaction between the two (S×T) is shown. *, P < 0.05; ***, P < 0.001; NS, P > 0.05.
No 13C incorporation into fungi.
No 13C in the PLFA 18:2ω6,9.
For the added label, 13C recovery was estimated in 27 PLFA. The incorporation data as ng of 13C recovered in each PLFA per mg of added 13C per gram of soil showed that, in general, the largest amounts were incorporated into PLFA 16:0 and 18:1ω7, with clear differences between different PLFA (Fig. 1). Utilization of glucose and acetic acid, both easily available carbon sources, produced relatively similar incorporation patterns both after 1 and 7 days (Fig. 1B and C), and these patterns largely matched with the original PLFA concentration pattern (Fig. 1A). These samples were characterized by relatively high levels of incorporation into the fungal PLFA 18:2ω6,9 and 18:1ω9 and into the general PLFA 16:0 and 18:0, resulting in higher fungal/bacterial incorporation ratios than the ratios for the original PLFA profile (>0.2 and <0.2, respectively) (Tables 1 and 2).
Glycine, which was the only substrate containing nitrogen, differed from glucose and acetic acid as its 13C label was hardly recovered in any PLFA indicative of fungi, 18:2ω6,9 and 18:1ω9 (Fig. 1D), resulting in consistently lower fungal/bacterial incorporation ratios (<0.01) than glucose and acetic acid (>0.2) (Tables 1 and 2). The highest recovery of 13C from glycine was observed in 18:1ω7, with high levels also for i15:0, a15:0, i16:0, 16:1ω7c, and 16:0 (Fig. 1D). For the bacterial PLFA, the incorporation pattern was similar to the original PLFA concentration pattern (Fig. 1A).
After incubation for 1 day, 13C from starch had been slightly incorporated into microbial PLFA (Table 2 and Fig. 1E). However, there was a severalfold increase in the incorporation after incubation for 7 days, which was also shown in the total 13C incorporation (Tables 2 and 3). Carbon-13 from starch was mainly incorporated into microorganisms containing PLFA 16:1ω7, 18:1ω7, 16:0, i15:0, and a15:0 (Fig. 1E). The incorporation pattern of starch was also characterized by virtually no incorporation into the fungal PLFA 18:2ω6,9 and 18:1ω9 (Fig. 1E), resulting in a ratio of fungal-to-bacterial incorporation of <0.01, that is, similar to that found for glycine (Table 2). After 7 days of incubation a small amount of the label (3.7 ± 1.3 ng of PLFA 13C mg−1 of label 13C g−1 of soil) from starch was incorporated into a PLFA with a retention time between the times of 10Me17:0 and 18:2ω6,9. This PLFA was under the detection limit when soil was incubated with any other substrate and when incubation with starch had lasted only for 1 day.
TABLE 3.
Incorporation of 13C into PLFA and NLFA after 7 days relative to that after 1 day
| Substrate | Incorporation of 13C after 7 days (%)a
|
|
|---|---|---|
| PLFA | NLFA | |
| Glucose | 98 | 51 |
| Acetic acid | 98 | 40 |
| Glycine | 119 | 54 |
| Starch | 311 | 193 |
| Vanillin | 170 | 142 |
Percentage determined relative to the amount after 1 day.
Carbon-13 in vanillin was incorporated to the smallest degree of all substrates tested (Table 2). Incorporation of 13C was mainly limited to microbial groups containing PLFA 16:1ω7, 16:0, 18:2ω6,9, 18:1ω9, and 18:1ω7 (Fig. 1F). Thus, the incorporation pattern and the fungal-to-bacterial incorporation ratio were more similar to those of glucose and acetic acid than to those of glycine and especially starch (Table 2).
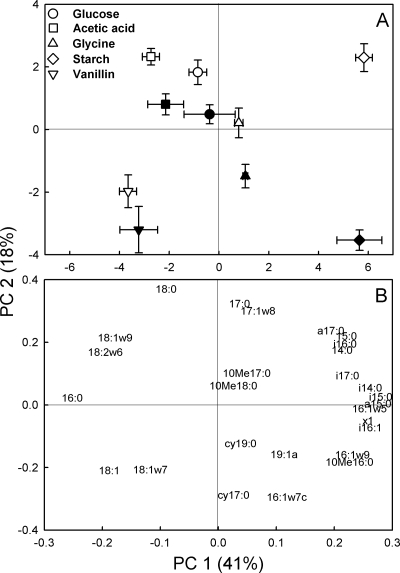
The relative incorporation of 13C (as a percentage of total incorporation) was subjected to PCA to visualize similarities and differences between the incorporation patterns (Fig. 2). The incorporation patterns significantly differed between the substrate additions (P < 0.001 for PC 1). The largest differences were between vanillin (Fig. 2, negative values on the PC 1 axis) and starch (positive values). The pattern for glycine addition was more similar to that of starch addition than the patterns for glucose and acetic acid additions, which were relatively similar to the pattern of vanillin addition.
FIG. 2.
PCA scores (A) and loadings (B) for relative 13C incorporation of added 13C-labeled glucose, acetic acid, glycine, starch, or vanillin in PLFA of tundra heath soil following incubation for 1 (white symbols) or 7 (black symbols) days. Explained variances for the PCs 1 and 2 are shown in parentheses. Values in panel A are means ± standard errors (n = 6).
The recovery of 13C was similar after 1 and 7 days for glucose, acetic acid, and glycine while it increased for vanillin and especially for starch (Table 3). However, there were similar changes in the incorporation patterns over time for all substrates, as indicated by a significant difference between the incubation times for PC 2 (P < 0.001) (Fig. 2). All substrates had more negative values along the PC 2 axis after 7 days than after 1 day. The main changes over time were thus similar for all substrates. These included an increasing recovery in the cyclopropyl PLFA cy-17:0 and cy-19:0 and in the PLFA with a methyl group, especially 10Me16:0, and a decreasing relative recovery with time in the PLFA 18:2ω6,9 and 18:0 except after the addition of vanillin (Fig. 1B and C and 2). The latter temporal change resulted in decreasing fungal/bacterial incorporation ratios with time for all substrates except vanillin (Table 2). The extent of the change over time significantly differed between the substrates (for the substrate-time interaction, P < 0.001 for PC 2) and was especially prominent for starch.
Incorporation of added label into NLFA.
Recovery of 13C was estimated in 17 NLFA (Fig. 3). The total 13C incorporation into NLFA was, similar to incorporation in the PLFA, highest with the addition of acetic acid, followed by glucose, glycine, starch, and vanillin in this order (Table 2). In contrast to the total 13C incorporation into PLFA, the recovery after 7 days was half of that after 1 day for glucose, acetic acid, and glycine, showing that the NLFA produced after 1 day was used during the later incubation (Table 3). As the recovery of 13C in the sum of NLFA for starch- and vanillin-amended samples increased in the course of the incubation, there was a highly significant substrate-time interaction (Tables 2 and 3).
The incorporation patterns of the samples amended with glucose and acetic acid were relatively similar to each other and were characterized by high levels of incorporation into the NLFA 18:1ω9, followed by 16:0 and 18:0, which indicates increased fungal storage of carbon into triacylglycerols (Fig. 3B and C). The level of incorporation of 13C label from glycine, starch, and vanillin into NLFA was low, as demonstrated by the NLFA-to-PLFA ratios that were half or less of the ratios for glucose and acetic acid (Table 2). However, vanillin-amended samples had virtually no 13C label in any NLFA other than NLFA indicative of fungi, including the general fatty acids 16:0 and 18:0. This implies incorporation into fungal storage products, as indicated for glucose and acetic acid additions (Fig. 3F).
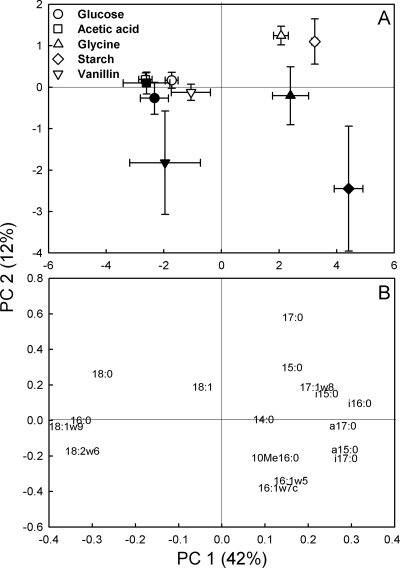
A PCA of the incorporation patterns of 13C from the different substrates (as a percentage of total incorporation) showed a significant substrate effect (P < 0.001) for PC 1; glucose, acetic acid, and vanillin had relatively similar patterns separating from those for glycine and starch (Fig. 4, PC 1 axis). The main difference was relatively higher recovery of 13C label in the NLFA indicative of fungi, 18:2ω6,9 and 18:1ω9, for the former three substrates. There was also a highly significant difference between the incubation times (P < 0.001) (Fig. 4, PC 2 axis), with the smallest differences for acetic acid and glucose and the largest for starch.
FIG. 4.
PCA scores (A) and loadings (B) for relative 13C incorporation of added 13C-labeled glucose, acetic acid, glycine, starch, or vanillin in NLFA of tundra heath soil following incubation for 1 (white symbols) or 7 (black symbols) days. Explained variances for the PCs 1 and 2 are shown in parentheses. Values in panel A are means ± standard errors (n = 6).
Although bacteria are not supposed to have a large amount of triacylglycerols, 13C label was also recovered in the NLFA indicative of bacteria, especially in the glycine- and starch-amended soils. For example, 13C from glycine was recovered in the NLFA a15:0, i16:0, and 17:1ω8 (Fig. 3D). For the starch-amended samples, mainly the NLFA a15:0 and i16:0 incorporated label, especially after 7 days (Fig. 3E).
DISCUSSION
In accordance with our first hypothesis, 13C from the simple compounds, glycine, glucose, and acetic acid, was more rapidly incorporated into PLFA during 1 day than the more complex substrates, starch and vanillin (Table 2). The recovery of 13C from the two latter substrates also increased over time, suggesting that all of the added substrate was not metabolized within 1 day (Table 3). However, comparison of the 13C incorporation from different substrates should be done with caution. First, we have no data on the extent of pool dilution, that is, the amount of the substrate present in soil solution. A larger pool dilution will result in an apparent lower recovery of the added label. It would be possible to estimate pool dilution for dissolved substances, such as glucose or glycine, but this would be difficult for starch because the continuous production of glucose from nonlabeled starch during the incubation period is of more importance than the actual starch content in soil. Second, the N content of the substrate is likely to affect not only the partitioning between growth and storage but also the extent to which substrate is used for making lipids or other building blocks of a cell. Last, the substrates differ in their metabolic function as precursors. Acetic acid is a precursor for lipid synthesis while glycine, for example, is a precursor for protein and purine synthesis. Thus, it is expected that the recovery of label from acetic acid in lipids is much larger than that from glycine (Table 2).
The 13C incorporation profiles for glucose- and acetic acid-amended soils and for glycine when only the bacterial PLFA were considered largely corresponded to the original PLFA concentration profile. A similar response was observed by Paterson et al. (32) to the addition of 13C-labeled glucose and fumaric acid within a mixture of sugars and amino and organic acids mimicking root exudates. They concluded that when the incorporation profile matches the abundance profile, substrate utilization drives the dominance of the microbial populations or that C uptake is noncompetitive and in proportion to population size and activity. Our results also indicate that glucose and acetic acid were used by both fungi and bacteria although the fungi/bacteria ratio was slightly higher for 13C incorporation than for the original PLFA pattern, especially after 1 day and for acetate (Tables 1 and 2), suggesting that fungi were initially more competitive than bacteria in taking up this substrate.
Glycine addition resulted in very low fungal/bacterial PLFA ratios of 13C incorporation (<0.01, compared to the initial values for fungal/bacterial ratios of PLFA concentrations of 0.11 to 0.15) (Tables 1 and 2). This is in agreement with the results of the 13C tracer experiment of Paterson et al. (32). That glycine uptake was dominated by bacteria and not by fungi may be related to the chemical properties of this amino acid as it was the only substrate used that contained nitrogen. An elevated nitrogen content in soil has earlier been shown to disfavor fungi compared to bacteria (13, 15, 16). Glycine was thus mainly taken up by bacteria, especially in the PLFA indicative of gram-negative bacteria (16:1ω7c, 18:1ω7, cy-17:0, and cy-19:0). Billings and Ziegler (5) also observed that N fertilizer addition to pine forest soil increases the activity of gram-negative bacteria and reduces that of gram-positive actinobacteria. However, results from a long-term field experiment conducted at the subarctic heath, from which the present soil samples were taken, are in contrast to this: NPK fertilizer application over 15 years led to a significant increase in the biomass of gram-positive relative to gram-negative bacteria (34). This change in the bacterial community composition may, however, be a result of fertilizer-induced alterations in the vegetation composition (34).
Amendment with starch and vanillin produced clearly different 13C incorporation patterns for PLFA, showing that our hypothesis that fungi are more important than bacteria in the degradation of complex substrates in these arctic soils could not be substantiated. Our results, therefore, contrast with the findings of Waldrop and Firestone (41), who reported that the same groups of microorganisms were responsible for degradation of added starch, xylose, and vanillin in temperate grassland and oak forest soils. This difference demonstrates that different organism groups can be responsible for degradation of common substrates in different soil habitats. In our study, starch was mainly utilized by bacteria containing monounsaturated PLFA 16:1ω7 and 18:1ω7, with simultaneous labeling of PLFA 16:0, i15:0, and a15:0. Thus, both gram-negative and gram-positive bacteria appeared to be involved in starch degradation. Starch was also taken up by a specialized group, as shown by the high level of 13C label in a PLFA not detectable in the original PLFA pattern. 13C recovery from starch was initially at a low level and increased with incubation time (Table 2). This is consistent with the fact that starch degradation begins outside the cell through the activity of induced extracellular enzymes. It is noteworthy that although the initial extracellular degradation of starch yields glucose, the 13C incorporation pattern for starch was different from that for glucose. The starch degraders may have a competitive advantage of uptake of the produced glucose due to their spatial position close to the exuded extracellular enzymes. An alternative explanation to the distinct patterns is that the concentration of the glucose produced from starch is lower than that of the actual glucose addition. High substrate concentrations have earlier been found to favor fungal growth (23).
Like starch, vanillin also had higher incorporation after 7 days than after 1 day (Table 3). However, in contrast to starch, vanillin was mainly taken up by fungi and bacteria with monounsaturated PLFA 16:1ω7 and 18:1ω7. This is in contrast to the results of Zak and Kling (46), who observed that arctic tundra soil fungi were unimportant in the degradation of vanillin relative to cellobiose and N-acetylglucosamine. Although the work of Zak and Kling (46) is, to the best of our knowledge, the only other experiment following 13C tracers into soil microbial lipids in arctic ecosystems, the poorly drained tundra soils of their study are very different from the dry heath of the present study. Fungi appear to be less important in poorly drained soils, which have anaerobic microsites, for example, due to flooding (9).
It is likely that the 13C signal is recycled within the community during incubation, which eventually leads to more even distribution among the PLFA with time. This phenomenon can be observed in that the 13C incorporation patterns became increasingly similar to the original PLFA pattern with time. During the first incubation day, recycling is probably of minor importance, even for those substrates that attained high levels of recovery within a day. Thus, short incubation times are recommended to enable detection of primary incorporation of label into different PLFA. The PLFA 10Me16:0, 10Me18:0, cy-17:0, and cy-19:0 contained a small amount of label after 1 day, with recovery increasing with time irrespective of substrate. Since this was also found for the substrates that did not increase recovery after the first day (glucose, acetic acid, and glycine), it may imply that these PLFA are indicative of organisms that preferentially fed on recycled material. For example, the 10Me-fatty acids are suggested to be indicative of actinomycetes (47), which are known to produce chitinases and are thereby potentially able to recycle fungal cell walls (38, 45).
A large amount of label from glucose and acetic acid was found in the NLFA related to fungi (18:2ω6,9 and especially 18:1ω9) as well as in the even-numbered saturated NLFA also common in fungi (Table 2 and Fig. 3). Increase in the amount of these NLFA in response to glucose addition has also been observed earlier (4). It appears that fungi preferentially store excess carbon in triglycerols as the NLFA 18:1ω9 (26). The large amount of label incorporated into neutral lipids also suggests that the substrate additions induced limitation of the fungi by a nutrient other than carbon, most likely nitrogen. The low level of incorporation of [13C]glycine into NLFA is a further indication of this. It is also possible that carbon will be stored under growth-restrictive conditions, explaining the initial increase in 13C in NLFA after vanillin addition. After addition of a substrate, it takes some time before growth commences in dormant organisms (lag period), and during this period there will be storage of carbon for later use. The recovery of the label in NLFA decreased over time for additions of glucose, acetic acid, and glycine, which suggests that the storage products were used in the course of the incubation.
Vanillin addition led to 13C incorporation especially into the NLFA 18:1ω9, indicating storage in fungi. However, although starch induced a similar excess of carbon to the microorganisms, the lack of substantial 13C incorporation from it into the NLFA 18:1ω9 is further evidence that bacteria is the main decomposer group of starch in this arctic soil. In the case of starch, the continued production of storage products was mainly due to increased incorporation into fatty acids assumed to be of bacterial origin, like i15:0, a15:0, i16:0, and i17:0. Bacteria do not normally accumulate triacylglycerols although it is not uncommon in bacteria belonging to the actinomycetes group (2). They may also be derived from turnover of bacteria as a neutral lipid, diacylglycerol, is produced when the phosphate group of a PLFA is removed by phosphatase enzyme activity (42). It is thus possible that the increase in the 13C NLFA fraction of a bacterial fatty acid partly indicates turnover (that is, death rate) during the incubation period, as suggested by White (42).
To conclude, the incorporation of 13C into PLFA and NLFA from different substrates added to organic tundra heath soil produced differential patterns, which showed that different groups of the soil microbial community were responsible for substrate utilization. While starch and to some extent glycine appeared to be used mainly by bacteria, fungi played a bigger role in the use of glucose, acetic acid, and especially vanillin although 13C was also found in bacterial PLFA. The use of 13C-labeled substrates enabled smaller amounts of the added substrate than conventional PLFA analyses since the substrate concentrations were high enough to work as tracers but low enough not to alter the original PLFA pattern. In accordance with our hypothesis, the recovery of 13C from the complex substrates increased over time. However, we did not find evidence that bacteria is better than fungi in utilizing simple compounds. We observed allocation to fungal storage lipids (NLFA) especially from the carbon-rich substrates, and eventually these lipids appeared to be used for growth. As distinct communities degraded different substrates, we recommend including several different substrates, possibly with and without nutrient additions, in assessing the functioning of soil microbial communities. Finally, our findings provide information on the functioning of the microbial communities in arctic soil, which can be utilized to estimate and predict consequences of microbial community changes on microbial activity under global change scenarios.
Acknowledgments
This work was supported by a Marie Curie Intra-European fellowship within the 6th European Community Framework Programme and by a grant from the Swedish Research Council to E.B.
We thank Anders Michelsen for the soil samples for this work and staff at the Abisko Scientific Research Station for their support. Per Bengtson gave valuable comments on the manuscript.
Footnotes
Published ahead of print on 10 April 2009.
REFERENCES
- 1.Abraham, W., C. Hesse, and O. Pelz. 1998. Ratios of carbon isotopes in microbial lipids as an indicator of substrate usage. Appl. Environ. Microbiol. 64:4202-4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alvarez, H. M., and A. Steinbüchel. 2002. Triacylglycerols in prokaryotic microorganisms. Appl. Microbiol. Biotechnol. 60:367-376. [DOI] [PubMed] [Google Scholar]
- 3.Arctic Council and International Arctic Science Committee. 2005. Impacts of a warming Arctic: Arctic climate impact assessment synthesis report. Cambridge University Press, Cambridge, United Kingdom.
- 4.Bååth, E. 2003. The use of neutral lipid fatty acids to indicate the physiological conditions of soil fungi. Microb. Ecol. 45:373-383. [DOI] [PubMed] [Google Scholar]
- 5.Billings, S. A., and S. E. Ziegler. 2008. Altered patterns of soil carbon substrate usage and heterotrophic respiration in a pine forest with elevated CO2 and N fertilization. Global Change Biol. 14:1025-1036. [Google Scholar]
- 6.Bittman, S., T. A. Forge, and C. G. Kowalenko. 2005. Responses of the bacterial and fungal biomass in a grassland soil to multiyear applications of dairy manure slurry and fertilizer. Soil Biol. Biochem. 37:613-623. [Google Scholar]
- 7.Boschker, H. T. S. 2004. Linking microbial community structure and functioning: stable isotope (13C) labeling in combination with PLFA analysis, p. 1673-1688. In G. A. Kowalchuk, F. J. de Bruijn, I. M. Head, A. D. Akkermans, and J. D. van Elsas (ed.), Molecular microbial ecology manual, vol. II, 2nd ed. Kluwer Academic Publishers, Dordrecht, The Netherlands. [Google Scholar]
- 8.Boschker, H. T. S., S. C. Nold, P. Wellsbury, D. Bos, W. de Graaf, R. Pel, R. J. Parkes, and T. E. Cappenberg. 1998. Direct linking of microbial populations to specific biogeochemical processes by 13C-labelling of biomarkers. Nature 392:801-805. [Google Scholar]
- 9.Bossio, D. A., and K. M. Scow. 1998. Impacts of carbon and flooding on soil microbial communities: phospholipid fatty acid profiles and substrate utilization patterns. Microb. Ecol. 35:265-278. [DOI] [PubMed] [Google Scholar]
- 10.Brant, J. B., E. W. Sulzman, and D. D. Myrold. 2006. Microbial community utilization of added carbon substrates in response to long-term carbon input manipulation. Soil Biol. Biochem. 38:2219-2232. [Google Scholar]
- 11.Butler, J. L., M. A. Williams, P. J. Bottomley, and D. D. Myrold. 2003. Microbial community dynamics associated with rhizosphere carbon flow. Appl. Environ. Microbiol. 69:6793-6800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeForest, J. L., D. R. Zak, K. S. Pregitzer, and A. J. Burton. 2004. Atmospheric nitrate deposition and the microbial degradation of cellobiose and vanillin in a northern hardwood forest. Soil Biol. Biochem. 36:965-971. [Google Scholar]
- 13.Denef, K., D. Roobroeck, M. C. W. Manimel Wadu, P. Lootens, and P. Boeckx. 2009. Microbial community composition and rhizodeposit-carbon assimilation in differently managed temperate grassland soils. Soil Biol. Biochem. 41:144-153. [Google Scholar]
- 14.De Ruiter, P. C., A.-M. Neutel, and J. C. Moore. 1994. Modeling food webs and nutrient cycling in agro-ecosystems. Trends Ecol. Evol. 9:378-383. [DOI] [PubMed] [Google Scholar]
- 15.De Vries, F. T., J. Bloem, N. van Eekeren, L. Brussaard, and E. Hoffland. 2007. Fungal biomass in pastures increases with age and reduced N inputs. Soil Biol. Biochem. 39:1620-1630. [Google Scholar]
- 16.De Vries, F. T., E. Hoffland, N. van Eekeren, L. Brussaard, and J. Bloem. 2006. Fungal/bacterial ratios in grasslands with contrasting nitrogen management. Soil Biol. Biochem. 38:2092-2101. [Google Scholar]
- 17.Evershed, R. P., Z. M. Crossman, I. D. Bull, H. Mottram, J. A. Dungait, P. J. Maxfield, and E. L. Brennand. 2006. 13C-labelling of lipids to investigate microbial communities in the environment. Curr. Opin. Biotech. 17:72-82. [DOI] [PubMed] [Google Scholar]
- 18.Flaig, W. 1964. Effects of micro-organisms in the transformation of lignin to humic substances. Geochim. Cosmochim. Acta 28:1523-1535. [Google Scholar]
- 19.Frey, S. D., E. T. Elliott, K. Paustian, and G. A. Peterson. 2000. Fungal translocation as a mechanism for soil nitrogen inputs to surface residue decomposition in a no-tillage agroecosystem. Soil Biol. Biochem. 32:689-698. [Google Scholar]
- 20.Frey, S. D., J. Six, and E. T. Elliott. 2003. Reciprocal transfer of carbon and nitrogen by decomposer fungi at the soil-litter interface. Soil Biol. Biochem. 35:1001-1004. [Google Scholar]
- 21.Frostegård, Å., and E. Bååth. 1996. The use of phospholipid fatty acid analysis to estimate bacterial and fungal biomass in soil. Biol. Fertil. Soils 22:59-65. [Google Scholar]
- 22.Frostegård, Å., A. Tunlid, and E. Bååth. 1991. Microbial biomass measured as total lipid phosphate in soils of different organic content. J. Microbiol. Methods 14:151-163. [Google Scholar]
- 23.Griffiths, B. S., K. Ritz, N. Ebblewhite, and G. Dobson. 1999. Soil microbial community structure: effects of substrate loading rates. Soil Biol. Biochem. 31:145-153. [Google Scholar]
- 24.Havström, M., T. V. Callaghan, and S. Jonasson. 1993. Differential growth responses of Cassiope tetragona, an arctic dwarf-shrub, to environmental perturbations among three contrasting high- and sub-arctic sites. Oikos 66:389-402. [Google Scholar]
- 25.Jonasson, S., A. Michelsen, I. K. Schmidt, and E. V. Nielsen. 1999. Responses in microbes and plants to changed temperature, nutrient, and light regimes in the arctic. Ecology 80:1828-1843. [Google Scholar]
- 26.Larsen, J., P. A. Olsson, and I. Jakobsen. 1998. The use of fatty acid signatures to study mycelial interactions between the arbuscular mycorrhizal fungus Glomus intraradicis and the saprotrophic fungus Fusarium culmorum in root-free soil. Mycol. Res. 102:1491-1496. [Google Scholar]
- 27.Leake, J. R., N. J. Ostle, J. I. Rangel-Castro, and D. Johnson. 2006. Carbon fluxes from plants through soil organisms determined by field 13CO2-labelling in an upland grassland. Appl. Soil Ecol. 33:152-175. [Google Scholar]
- 28.McMahon, S. K., M. A. Williams, P. J. Bottomley, and D. D. Myrold. 2005. Dynamics of microbial communities during decomposition of carbon-13 labeled ryegrass fractions in soil. Soil Sci. Soc. Am. J. 69:1238-1247. [Google Scholar]
- 29.Meidute, S., F. Demoling, and E. Bååth. 2008. Antagonistic and synergistic effects of fungal and bacterial growth in soil after adding different carbon and nitrogen sources. Soil Biol. Biochem. 40:2334-2343. [Google Scholar]
- 30.Moore, J. C., K. McCann, and P. C. de Ruiter. 2005. Modelling trophic pathways, nutrient cycling, and dynamic stability in soils. Pedobiologia 49:499-510. [Google Scholar]
- 31.Olsson, P. A., I. M. van Aarle, M. E. Gavito, P. Bengtson, and G. Bengtsson. 2005. 13C incorporation into signature fatty acids as an assay for carbon allocation in arbuscular mycorrhiza. Appl. Environ. Microbiol. 71:2592-2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paterson, E., T. Gebbing, C. Abel, A. Sim, and G. Telfer. 2007. Rhizodeposition shapes rhizosphere microbial community structure in organic soil. New Phytol. 173:600-610. [DOI] [PubMed] [Google Scholar]
- 33.Phillips, R. L., D. R. Zak, W. E. Holmes, and D. C. White. 2002. Microbial community composition and function beneath temperate trees exposed to elevated atmospheric carbon dioxide and ozone. Oecologia 131:236-244. [DOI] [PubMed] [Google Scholar]
- 34.Rinnan, R., A. Michelsen, E. Bååth, and S. Jonasson. 2007. Fifteen years of climate change manipulations alter soil microbial communities in a subarctic heath ecosystem. Global Change Biol. 13:28-39. [Google Scholar]
- 35.Rousk, J., and E. Bååth. 2007. Fungal and bacterial growth in soil with plant materials of different C/N ratios. FEMS Microbiol. Ecol. 62:258-267. [DOI] [PubMed] [Google Scholar]
- 36.Ruess, L., A. Michelsen, I. K. Schmidt, and S. Jonasson. 1999. Simulated climate change affecting microorganisms, nematode density and biodiversity in subarctic soils. Plant Soil. 212:63-73. [Google Scholar]
- 37.Six, J., S. D. Frey, R. K. Thiet, and K. M. Batten. 2006. Bacterial and fungal contributions to carbon sequestration in agroecosystems. Soil Sci. Soc. Am. J. 70:555-569. [Google Scholar]
- 38.Skujins, I., A. Pukite, and A. D. McLaren. 1970. Chitinase of Strepromycetes sp.: purification and properties. Enzymologia 39:353-370. [PubMed] [Google Scholar]
- 39.Stockdale, E. A., and P. C. Brookes. 2006. Detection and quantification of the soil microbial biomass—impacts on the management of agricultural soils. J. Agric. Sci. 144:285-302. [Google Scholar]
- 40.Tunlid, A., and D. C. White. 1992. Biochemical analysis of biomass, community structure, nutritional status and metabolic activity of microbial communities in soil, p. 229-262. In G. Stotzky, and J. M. Bollag (ed.), Soil biochemistry. Marcel Dekker, New York, NY.
- 41.Waldrop, M., and M. Firestone. 2004. Microbial community utilization of recalcitrant and simple carbon compounds: impact of oak-woodland plant communities. Oecologia 138:275-284. [DOI] [PubMed] [Google Scholar]
- 42.White, D. C. 1993. In situ measurement of microbial biomass, community structure and nutritional status. Philos. Trans. R. Soc. A 344:59-67. [Google Scholar]
- 43.Williams, M. A., D. D. Myrold, and P. J. Bottomley. 2006. Carbon flow from 13C-labeled straw and root residues into the phospholipid fatty acids of a soil microbial community under field conditions. Soil Biol. Biochem. 38:759-768. [Google Scholar]
- 44.Williams, M. A., D. D. Myrold, and P. J. Bottomley. 2007. Carbon flow from 13C-labeled clover and ryegrass residues into a residue-associated microbial community under field conditions. Soil Biol. Biochem. 39:819-822. [Google Scholar]
- 45.Williams, S. T., and C. S. Robinson. 1981. The role of streptomycetes in decomposition of chitin in acid soils. J. Gen. Microbiol. 127:55-63. [Google Scholar]
- 46.Zak, D. R., and G. W. Kling. 2006. Microbial community composition and function across an arctic tundra landscape. Ecology 87:1659-1670. [DOI] [PubMed] [Google Scholar]
- 47.Zelles, L. 1999. Fatty acid patterns of phospholipids and lipopolysaccharides in the characterisation of microbial communities in soil: a review. Biol. Fertil. Soils 29:111-129. [Google Scholar]