Abstract
Gut microbiota carry out key functions in health and participate in the pathogenesis of a growing number of diseases. The aim of this study was to develop a custom microarray that is able to identify hundreds of intestinal bacterial species. We used the Entrez nucleotide database to compile a data set of bacterial 16S rRNA gene sequences isolated from human intestinal and fecal samples. Identified sequences were clustered into separate phylospecies groups. Representative sequences from each phylospecies were used to develop a microbiota microarray based on the Affymetrix GeneChip platform. The designed microbiota array contains probes to 775 different bacterial phylospecies. In our validation experiments, the array correctly identified genomic DNA from all 15 bacterial species used. Microbiota array has a detection sensitivity of at least 1 pg of genomic DNA and can detect bacteria present at a 0.00025% level of overall sample. Using the developed microarray, fecal samples from two healthy children and two healthy adults were analyzed for bacterial presence. Between 227 and 232 species were detected in fecal samples from children, whereas 191 to 208 species were found in adult stools. The majority of identified phylospecies belonged to the classes Clostridia and Bacteroidetes. The microarray revealed putative differences between the gut microbiota of healthy children and adults: fecal samples from adults had more Clostridia and less Bacteroidetes and Proteobacteria than those from children. A number of other putative differences were found at the genus level.
In the healthy adult, there are 1011 to 1014 bacteria colonizing the intestine. This outnumbers the total tissue cells in the body by at least an order of magnitude. The composition and activity of this complex microbial system (called microbiota or microflora) have a major influence on health and disease (9). Commensal microbiota contribute to the trophic functions of the gut (producing fermentation products and vitamins that can be used by intestinal epithelial cells), stimulate the immune function of the gastrointestinal tract, transform or excrete toxic substances, protect the host against invasion by pathogenic species, and modulate gut motility (28, 36). At the same time, recent research incriminates a dysfunctional cross-talk between the host and the microbiota in the pathogenesis of a growing number of disorders, such as irritable bowel syndrome, inflammatory bowel disease, allergic diseases, and gastrointestinal cancer (28).
While the intestine in a newborn contains no microbes, immediately after birth the intestine of the infant is colonized by enterobacteria and enterococci. Gradual changes in microbiota composition occur during childhood, with a general reduction in the number of aerobes and facultative anaerobes and an increase in the populations of obligate anaerobic species (27). It is considered that by 2 years of age the microbiota resembles that of an adult, which is dominated in health and disease by species from only four phyla, Firmicutes (predominantly Clostridia; 50 to 70% total bacterial numbers), Bacteroidetes (10 to 30%), Proteobacteria (up to 10%), and Actinobacteria (up to 5%), with 90% believed to be obligate anaerobes (4, 10, 11, 22).
Traditionally, microorganisms were detected in intestinal samples and feces by microscopic, biochemical, or physiological methods, or by culturing on selective nutrient media. However, since most intestinal microbiota species are obligate anaerobes, their isolation and culturing are difficult (21, 38, 42). In recent years, new methods based on the use of microarray technology have been utilized for the characterization of complex microbial communities (18, 32, 41, 43, 47). Microarrays represent an excellent choice for the high-throughput analysis of bacterial populations, because many different probes can be placed on one slide or synthesized on one chip, and samples thus can be tested for the presence of many different species simultaneously. Environmental and clinical samples can be interrogated directly, circumventing any need for culturing, and thus nonculturable species can be reliably detected.
Several types of microarrays have been used to date to characterize the composition of microbial communities (47). Community genome arrays are constructed using whole genomic DNA (gDNA) isolated from pure culture strains (46). Functional gene arrays contain genes encoding key enzymes that are involved in various biochemical processes, and they are useful for monitoring physiological changes in microbial communities (14, 45). Phylogenetic oligonucleotide arrays contain probes derived from rRNA sequence information and are ideally suited for the analysis of microbial community composition structure and variance. Different types of phylogenetic arrays have been designed for these purposes (26, 30, 31).
A number of projects performed in the last several years focused on sampling the diversity of human microbiota by the cloning and subsequent sequencing of the 16S rRNA genes isolated from gastrointestinal and fecal samples (5, 10, 13, 23, 38). In this project, we have designed, developed, and validated a custom microbiota microarray containing 16S rRNA genes probes to 775 different microbial phylospecies of human intestinal bacteria. We also have tested the applicability of this array to profiling the microbiota populations in fecal samples isolated from two adult and two child volunteers.
MATERIALS AND METHODS
Compilation of 16S rRNA gene sequences identified from human intestinal and fecal samples.
To compile a list of 16S rRNA gene sequences identified from intestinal microbiota, a search of the Entrez nucleotide database (http://www.ncbi.nlm.nih.gov/) was carried out in October 2006. Sequence retrieval was limited to sequences of 1,200 bp or longer. The search yielded a total of 15,735 microbial sequences representing microbiota from stools or intestinal bacteria obtained from mucosal biopsies.
All sequence analysis steps were carried out in collaboration with Qiong Wang and Jim Cole from the Ribosomal Database Project (RDP; Michigan State University) (8). All 16S rRNA gene sequences were mapped to the RDP database, aligned by RDP Aligner, and classified into Bergey's taxonomy by RDP Classifier (40). Using the alignment, distance matrices among sequences were calculated for the complete data set using Jukes Cantor correction. DOTUR (37) was employed to cluster all sequences into separate phylogroups (10). Based on previous studies (1, 10, 13), a cutoff value of 0.020 was utilized to define group boundaries (corresponding to 98% nucleotide similarity within each group). This clustering produced a total of 852 groups. During the microarray design process, unique nucleotide probes were to be sought for each individual target sequence supplied; therefore, one representative nucleotide sequence was chosen for each phylospecies. A consensus nucleotide sequence for each group was produced, and the actual 16S rRNA gene sequence within each group that was closest (in terms of nucleotide similarity) to the consensus was chosen as the representative 16S rRNA gene sequence of that phylospecies. The lowest common lineage node among all sequences within each group was used as the group designation.
The 852 representative sequences were extracted and aligned. The aligned data set was truncated to include 16S rRNA nucleotides only between nucleotide positions 28 and 1491 (Escherichia coli 16S rRNA gene positions). This was necessary to ensure that no microarray probes were designed to the regions outside 28 to 1491, since only nucleotides in that region can be amplified by universal 16S rRNA gene PCR primers. The truncated set was used as an input for the design algorithm.
Design of microbiota microarray.
The custom microbiota microarray was designed based on the Affymetrix GeneChip platform (array format 100-2186; 11-μm feature size) (2, 3). “Prokaryotic antisense” was chosen as the target type. Potential probes were pruned with all of the submitted 16S rRNA gene sequences, with the reverse complement of the submitted 16S rRNA gene sequences, and with a standard human genome library. The probe length was 25 nucleotides. Each species was represented on the array by at least one probe set. The minimum number of different oligonucleotide probes per probe set was 5, and the maximum was 11. As controls, standard human, mouse, and rat control sets were included on the GeneChip together with E. coli and prokaryotic spike set controls.
Probe sets to 775 phylospecies were present in the final design (Table 1). Note that for the same species, the algorithm can produce several probe sets with different specificities if the unique probe set is not optimal. The examination of the complete set of probes showed that the distribution of probe %GC was similar to that for Affymetrix E. coli and Bacillus subtilis arrays.
TABLE 1.
Phylogenetic distribution of bacterial phylospecies from human intestine that can be interrogated by microbiota array
| Class | No. of phylospecies |
|---|---|
| Cyanobacteria | 1 |
| Alphaproteobacteria | 9 |
| Betaproteobacteria | 17 |
| Gammaproteobacteria | 11 |
| Deltaproteobacteria | 4 |
| Epsilonproteobacteria | 6 |
| Clostridia | 527 |
| Mollicutes | 12 |
| Bacilli | 24 |
| Actinobacteria | 29 |
| Spirochaetes | 4 |
| Bacteroidetes | 126 |
| Fusobacteria | 3 |
| Verrucomicrobiae | 1 |
| Lentisphaerae | 1 |
| Total | 775 |
Cultivation of bacterial species and isolation of gDNA.
Sixteen bacterial species were chosen for validation experiments (Table 2) from the list of known phylospecies among 775, so that they (i) are available from ATCC, (ii) can be cultured, (iii) represent different bacterial classes, and (iv) represent bacteria with different average GC contents. All bacterial samples were purchased from the ATCC and stored at −80°C. For DNA preparation, frozen stocks were cultured in either 15-ml centrifuge tubes or T-25 tissue culture flasks containing medium recommended by the ATCC for that species. Anaerobic cultures were sealed inside a GasPak bag together with a gas generator packet (BD GasPak EZ) and placed together with aerobic cultures in a shaking water bath at 37°C, and all cultures were incubated for 1 to 2 days. Once a sufficient cell density was reached, the cultures were collected and then frozen at −80°C.
TABLE 2.
Bacteria used to validate the microbiota array
| ATCC no. | Bacterial species | Class | Gram stain | %GC contenta |
|---|---|---|---|---|
| 27539 | Bifidobacterium catenulatum | Actinobacteria | + | 60 |
| 15707 | Bifidobacterium longum | Actinobacteria | + | 60 |
| 25559 | Eggerthella lenta | Actinobacteria | + | 64 |
| 27274 | Enterococcus faecalis | Bacilli | + | 37 |
| 4356 | Lactobacillus acidophilus | Bacilli | + | 35 |
| 8483 | Bacteroides ovatus | Bacteroidetes | − | 43 |
| 8492 | Bacteroides uniformis | Bacteroidetes | − | 43 |
| 9689 | Clostridium difficile | Clostridia | + | 29 |
| 638 | Clostridium paraputrificumb | Clostridia | + | 30 |
| 9714 | Clostridium sordellii | Clostridia | + | 30 |
| 19403 | Clostridium sphenoides | Clostridia | + | 30 |
| 8486 | Eubacterium limosum | Clostridia | + | 47 |
| 27210 | Ruminococcus albus | Clostridia | + | 42 |
| 51649 | Holdemania filiformis | Mollicutes | + | 38 |
| 25586 | Fusobacterium nucleatum | Fusobacteria | − | 27 |
| 25922 | Escherichia coli | Gammaproteobacteria | − | 51 |
The %GC was estimated for nonsequenced species based on other studies or phylogenetic homology.
ATCC 638 was recently reclassified as Clostridium bifermentans in the NCBI and RDP databases.
A ZR fungal/bacterial DNA kit (Zymo Research) was used for all DNA extractions. Total DNA was isolated successfully from all species; however, due to problems with the cultivation of Ruminococcus albus, only small quantities of gDNA were prepared, limiting its use in validation experiments.
PCR amplification of 16S rRNA genes.
Two primers that are frequently used in microbial diversity studies to amplify a near-full-length 16S rRNA gene sequence and considered conserved for Bacteria were utilized for the PCR amplification of 16S rRNA genes: Bact-27F (AGRGTTTGATCMTGGCTCAG) and Univ-1492R (GGYTACCTTGTTACGACTT). PCR was carried out in a 50-μl volume using either PrimeStar HS DNA polymerase (Takara) or Taq 2× Mastermix (New England Biolabs). Amplified DNA was purified with a QIAquick PCR purification kit (Qiagen).
Isolation of gDNA from fecal samples.
Fecal samples were obtained from two healthy children and two healthy adults. Samples were frozen immediately after passage and were kept at −80°C. Total gDNA was isolated from fecal samples using a QIAamp DNA stool mini kit (Qiagen). Isolated gDNA was passed through a Zymo-spin IV-HRC filter (Zymo Research) to remove any remaining inhibitors. For microarray experiments, three separate PCRs, each containing 500 ng of gDNA, were carried out for each fecal DNA sample. The resultant mixtures were pooled. Amplified DNA was purified with a QIAquick PCR purification kit (Qiagen).
Microarray experiments and analysis.
gDNA and PCR-amplified DNA were fragmented with DNase I (37°C, 10 min) to provide the bulk of DNA fragments with sizes of 100 to 500 bp. Control experiments identified the optimal DNase I concentration as 0.075 U per 1 μg of gDNA and 0.04 U per 1 μg of PCR-amplified gDNA. Fragmented DNA was end labeled with biotin. Hybridization, washing, staining, and array scanning were performed according to the standard Affymetrix protocol for prokaryotic array format 100. GCOS (Affymetrix) was used to process the raw microarray data (the parameters used were α1 = 0.03, α2 = 0.05, and τ = 0.015, corresponding to a 97% statistical confidence of probe set detection). To obtain a consensus presence call for a probe set on replicate arrays, a particular probe set had to be called present (α ≤ 0.03) on one array and be present or marginal (0.03 < α ≤ 0.05) on another array; otherwise, the phylospecies was considered not to be reliably detected. The signal values were normalized only for the experiments that used DNA from fecal samples, because in all other tests only a small fraction of probe sets showed signal values above the level of background noise. CARMAweb was used to normalize signal intensities (35). The MAS5 algorithm was used for Perfect Match correction, the Loess algorithm was used for array normalization, and the median Polish algorithm was used to obtain expression summaries (i.e., signal values) for each probe set on a chip (7). Signal values were averaged for replicate arrays as geometric means. To estimate the total hybridization signal for larger taxonomical groups, signal values for all phylospecies in that group that were detected on a particular array were summed. If a phylospecies was not called present by the GCOS algorithm, the signal values of the corresponding probe set(s) were ignored.
Measurements of signal ratios.
Six microbiota arrays were used to measure differences in probe set signal intensities, corresponding to different amounts of bacterial DNA added to each array. Equal amounts of gDNA (1 μg) from Bifidobacterium longum, Bacteroides uniformis, Lactobacillus acidophilus, and Clostridium sphenoides were mixed together and subsequently fragmented with DNase I. The amount of each species gDNA added to arrays was 12.5 ng (array FC1), 25 ng (array FC2), 50 ng (array FC3), 100 ng (array FC4), 150 ng (array FC5), or 200 ng (array FC6). To account for array-to-array variability in global hybridization affinity, 100 ng of gDNA from Holdemania filiformis was added to each array (fragmented separately). To ensure that each hybridization sample contained the same amount of fragmented bacterial DNA, fragmented gDNA from E. coli was used to adjust the total gDNA content of each sample to 1 μg. During the analysis of each array, the signal values obtained for the H. filiformis probe set were adjusted to the same baseline value, and all other signals were multiplied by this normalization factor. Array FC1 was used as a baseline experiment, and all signal ratios were calculated by dividing signal values from other arrays by the corresponding values from array FC1.
qPCR.
Quantitative real-time PCR (qPCR) was carried out on an ABI Prism 7000 sequence detection system using platinum SYBR green qPCR supermix (Invitrogen). To selectively amplify 16S rRNA genes from chosen bacterial groups, forward primer 27F was combined with a group-specific primer identified in ARB PLEASE! (http://www.arb-home.de/probelib.html) or ProbeBase (25). Selected primers were checked for acceptable group coverage and correct specificity in RDP (8). Primer sequences are listed online (http://www.wright.edu/∼oleg.paliy/Papers/MF_array/MF_array.html). The amplification of all bacterial 16S rRNA genes with conserved primers (27F-Eub338R) was used as the sample control. The total reaction volume was 25 μl. Each reaction mixture contained 0.4 μM of each primer, 1× SYBR green supermix, and fecal gDNA. For each primer pair, separate qPCR reactions were carried out with template concentrations of 500 pg and 4 ng. All reactions were performed in duplicate. The cycling parameters were 95°C for 15 s, 55°C for 30 s, and 72°C for 30 s. The specificity of each amplification was assessed by a melting curve analysis of the amplified sample, and the correct amplicon size was confirmed by agarose gel electrophoresis. The efficiency of amplification for the control primer pair was estimated from the standard curve generated from a series of fourfold template dilutions; the efficiency of amplification for each test primer pair was calculated for each individual sample tested. The amplification efficiency for different primer pairs was relatively similar but not identical; these differences have been taken into account during relative abundance calculations. Mathematical formulas used to calculate relative gDNA abundances and measurement errors are provided online (http://www.wright.edu/∼oleg.paliy/Papers/MF_array/MF_array.html).
RESULTS
Microarray development.
The goal of this project was to develop a custom community microarray specifically designed to interrogate quantitatively the presence and relative abundance of bacteria in human intestinal and fecal samples. In order to enable the custom microarray to detect as many different intestinal bacterial species as possible, a broad search of the Entrez nucleotide database was performed. We identified more than 15,000 different bacterial 16S rRNA gene sequence entries with human intestinal origins. Upon manual examination, we found that this data set was not nonredundant; there were multiple sequence entries for the same bacterial species, and we found several cases where the same 16S rRNA gene sequence was present in the database with different sequence identity numbers. This redundancy necessitated the use of the clustering approach to distribute all of the sequences into separate phylospecies (also called species-level operational taxonomic units) based on a procedure described previously for the analysis of microbial diversity (5, 10, 13). Using a 98% sequence similarity cutoff to limit allowable sequence variability within individual phylospecies (1, 10, 13), a total of 852 distinct phylospecies were defined. The majority of bacteria belonged to the class Clostridia in the phylum Firmicutes (10). The use of this clustering procedure ensured that the distribution and the overall number of phylospecies are relatively insensitive to a continuous increase in the number of 16S rRNA gene sequences obtained from human intestinal flora, because we expect that many new sequences would cluster into existing phylospecies groups (10).
The Affymetrix GeneChip platform was chosen for the development of a custom microbiota microarray. As the specificity of the GeneChips can be as low as a single-nucleotide difference between oligonucleotides, species-specific probes could be designed for each species of microbiota. Even though only 16S rRNA gene sequences were available for the design of oligoprobes, the focus of this custom microarray on only those microbial species identified in human intestinal microbiota improved the chances of finding unique oligoprobes to each species. Indeed, we have been able to design probes for 775 different phylospecies (Table 1). Between 5 and 11 probes were designed for each phylospecies, with 84% of probe sets containing 11 probes. The designed microarray was successfully produced in collaboration with Affymetrix, Inc. (Santa Clara, CA).
Correct identification of pure bacterial cultures.
To test the ability of the microarray to correctly identify the presence of bacteria in samples, we have hybridized 250 ng of gDNA isolated from individual bacterial species listed in Table 2 to separate arrays. Chosen species span a number of different classes and had a wide range of genome %GC. Among 16 bacterial species used, 15 were reliably detected (with at least 97% confidence) by the microbiota array. The array correctly discriminated among closely related species within the same genus (Table 2). Because the standard hybridization mixture contains herring sperm DNA (carrier) and B2 oligomers (hybridization standard), a negative control array was run with hybridization mixture lacking any bacterial DNA. As expected, the scanned microarray was largely blank, with only several probe sets showing signal values marginally above the level of background noise. DNA isolated from cell pellets believed to be Eggerthella lenta did not produce any significant signal. When we used the total gDNA isolated from that bacterial culture to sequence the 16S rRNA gene, the 16S rRNA gene was identified as belonging to the genus Propionibacterium in the phylum Actinobacteria. Some of these bacteria are known to live on human skin, thus they might have contaminated and eventually overtaken the culture during culture handling and growth (12). Our microbiota array did not contain any probes for Propionibacterium; the sequencing thus corroborated the results of the array hybridization to that gDNA sample. Among the other 15 bacterial samples, cross-hybridization to other probe sets was low; on average, each individual array called present 4.6 extra probe sets (representing 0.4% of all probe sets on the array); the signal intensity for these cross-hybridizing probes was usually at least an order of magnitude less than that for the probe set correctly matching the loaded bacterial gDNA. To test if such cross-hybridization can be reduced by the use of multiple replicates, we hybridized an additional array to total gDNA (250 ng) from Bacteroides uniformis, because the first array run for this sample identified the highest number of additional sequences (13 probe sets) as being present in the sample. The second array had a total of four nonmatching probe sets called present. When the results of the two replicate arrays were analyzed, only one probe set was called present in both replicates in addition to the one corresponding to B. uniformis; that other probe set corresponded to another Bacteroides phylospecies. This indicated that the use of multiple arrays per sample significantly reduces the nonspecific detection of bacterial phylospecies.
PCR amplification of 16S rRNA genes leads to increased sensitivity of detection.
The amplification of 16S ribosomal gDNA in a PCR previously has led to the increased sensitivity of the community microarrays (18, 44). To specifically amplify near-full-length bacterial 16S rRNA genes, we used two phylogenetically conserved 16S primers, as described in Materials and Methods. Successful 16S rRNA gene amplification was confirmed for all validation species by agarose gel electrophoresis (data not shown). Because no gel purification was carried out, the amplified DNA samples contained a mixture of total gDNA (used as a template for PCR amplification) and amplified 16S rRNA genes and were designated 16S+gDNA. Such 16S+gDNA samples (250 ng starting gDNA, 10 cycles of PCR amplification) were hybridized to microbiota array separately for each validation species. Fifteen bacterial species were detected correctly on the array; 16S+gDNA from Propionibacterium spp. produced the correct negative result (as described above). In analyzing the hybridization results between two choices of hybridization sample (gDNA and 16S+gDNA), the use of 16S amplification (10 cycles) led to an increase in the probe signal for matching probe sets (5.5-fold on average) and thus would improve the sensitivity of detection. The increased sensitivity of detection led to a higher level of the cross-hybridization of 16S+gDNA samples: an average of 9.1 extra probe sets (0.9% of all probe sets) were called present in these experiments. However, the use of a second replicate array for the 16S+gDNA sample from Clostridium sphenoides led to a decrease in the number of cross-hybridizing probe sets from 21 to 1 when results from both arrays were combined.
Correct identification of bacterial DNA in complex mixtures.
Because the intestinal and fecal samples are expected to contain hundreds of different species of bacteria, we wanted to test if the designed microarray can correctly detect the presence of individual bacterial species in complex mixtures. To that end, we mixed 100 ng of gDNA from each of the 15 species (Ruminococcus was not used due to the lack of gDNA) and hybridized this mixture to two microbiota arrays. For two other arrays, 16S rRNA gene-specific PCR amplification was carried out on the combined gDNA mixture prior to hybridization. We detected reliably (97% confidence) all species on each array, with the exception of Propionibacterium spp. (for which there were no probes on the array). Clostridium sordellii was not detected on one array but was detected on three others. Good concordance in signal intensity values was observed between replicate arrays (Spearman rank correlation of 0.95 to 0.99). The use of replicates led to a more than twofold decrease in cross-hybridization. The PCR amplification of gDNA resulted in an increased strength of hybridization signal and in a lower variability of signal intensity among probe sets matching our bacterial species.
To determine the sensitivity limit of the microbiota array for different types of samples, several other mixtures of bacterial gDNA were hybridized to the arrays. The detection limit was estimated to be at least 4 ng for total unamplified gDNA and at least 1 ng for 16S+gDNA when gDNA was subjected to 10 cycles of 16S rRNA gene-specific PCR amplification. The detection limit can be decreased significantly by increasing the number of PCR amplification cycles; with 30 cycles of PCR amplification, we reliably detected 1 pg (10−12 g) of starting gDNA material. If necessary, the PCR amplification can be extended up to 35 to 40 cycles total, which would allow us to use this microarray as a clinical detection tool (41, 43).
Sensitivity of detection of spiked-in bacterial DNA in human gDNA.
It is believed that different microbiota species are present in the human gastrointestinal tract in widely different numbers spanning several orders of magnitude. Therefore, one important characteristic of any potential high-throughput detection tool would be its ability to detect small amounts of particular bacterial species in an otherwise large sample. In our next set of validation experiments, we spiked gDNA from several bacterial species into a sample of gDNA of nonbacterial origin. As nonbacterial DNA, total gDNA isolated from the human cell line HeLa-2 was used. We were able to detect 4 ng of total unamplified gDNA for three out of the four gDNAs tested (0.1% of the total sample applied). When 10 cycles of 16S rRNA gene-specific PCR amplification were used on the total sample, we detected 1 ng of bacterial gDNA (0.025% total sample). This limit was reduced to a mere 10 pg by increasing the number of PCR cycles to 30. This represents 0.00025% of the initial gDNA sample, which compares well with other designed community microarrays (29, 32) (note that human gDNA is not expected to be amplified with bacterial universal primers; therefore, the final relative abundance of bacterial gDNA in the 16S rRNA gene-amplified spike-in samples is higher than that of the original ratio). If we assume the highest relative species abundance in a complex bacterial community to be 1% of total DNA, the ability of the microarray to detect bacterial presence is in a 4,000-fold range.
Detection of differences in gDNA amounts among samples.
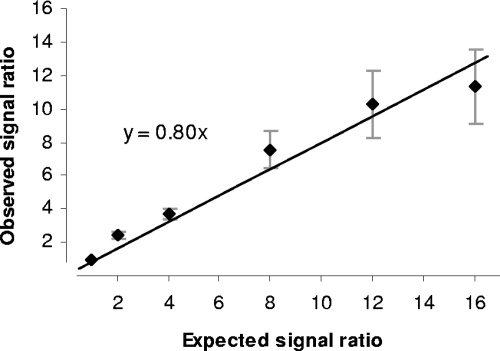
The expected future uses of the designed microarray include not only a qualitative detection of different bacterial species in intestinal and fecal samples but also a quantitative comparison of the abundance of a particular species of intestinal bacteria among different samples. To examine if microbiota array can detect differences in bacterial gDNA amounts among different samples, a series of six hybridization experiments were carried out. Each sample contained gDNA from the same mix of bacterial species, but different amounts of such gDNA mix were added to each array. The loads were calculated so as to provide a range of abundance differences of 2-, 4-, 8-, 12-, and 16-fold. Each sample was hybridized individually to a separate array, and observed signal ratios among samples were compared to expected values (Fig. 1). Overall, a good concordance between expected and obtained signal ratio values was established. A close to linear relationship was observed (R2 = 0.95 for linear fit), with a line slope coefficient of 0.8. The variation in signal ratios increased with an increase in expected ratio values, as evident from the size of error bars in Fig. 1. The observed signal ratios also seemed to deviate more from the expected values for higher (n-fold) differences, which might indicate a potential saturation of the hybridization mixture with DNA fragments for array experiments containing larger amounts of each species' DNA.
FIG. 1.
Linearity of signal ratio measurements. Expected signal ratios among different microarrays are plotted on the x axis, and observed signal ratios are plotted on the y axis. Signal value ratios for probe sets corresponding to four bacterial species hybridized to the arrays were averaged (arithmetic means). Error bars correspond to standard deviations of signal ratios among different probe sets. The trend line represents the best fit of the linear model y = ax + b to the average data. Parameter b was forced to 0.
Identification of bacterial phylospecies in gDNA from human fecal samples.
To examine the ability of the microbiota array to quantitatively profile the microbiota composition, we obtained fresh fecal samples from four volunteers: two adults and two children 12 and 15 years of age. gDNA isolated from each sample was subjected to 16S rRNA gene-specific PCR amplification, and the amplified samples were hybridized to the arrays. Three individual PCR amplifications were pooled together to assemble each hybridization sample. The pooling of multiple PCRs was critical in reducing the variability of microarray data resulting from previously described PCR bias (data not shown) (34). Two microbiota arrays were used per stool sample. An excellent replicate reproducibility of microarrays was achieved (Spearman rank correlation of 0.97 to 0.99 for signal values).
Between 227 and 232 species were called present in fecal samples from the children, while we detected 191 and 208 species in the adult stools. Table 3 lists the composition of each fecal sample at the bacterial class level. In general, similar microbiota composition profiles were observed for all four samples and were dominated by Clostridia and Bacteroidetes, which together accounted for 88 to 98% of the total hybridization signal. Fecal samples from adult volunteers had relatively higher proportions of Clostridia (86 to 89% in adults, 77 to 79% in children), whereas Bacteroidetes were more abundant in fecal samples from children (9% in adults, 12 to 13% in kids). Child fecal samples had much higher numbers of Proteobacteria (7 to 12% in children, 0 to 2% in adults); at the same time, Verrucomicrobiae were more abundant in adults (because of the low number of fecal samples available, tests of statistical significance of detected differences were not performed).
TABLE 3.
Detection of bacterial classes in fecal samples from children and adults
| Class | Child 1
|
Child 2
|
Adult 1
|
Adult 2
|
||||
|---|---|---|---|---|---|---|---|---|
| No.a | %b | No. | % | No. | % | No. | % | |
| Cyanobacteria | 0 | 0 | 0 | 1 | <0.1 | |||
| Alphaproteobacteria | 0 | 0 | 0 | 0 | ||||
| Betaproteobacteria | 4 | 4.2 | 0 | 1 | <0.1 | 1 | 1.3 | |
| Gammaproteobacteria | 2 | 2.4 | 6 | 11.3 | 0 | 2 | 0.2 | |
| Deltaproteobacteria | 1 | 0.5 | 1 | 0.5 | 0 | 1 | 0.5 | |
| Epsilonproteobacteria | 0 | 0 | 0 | 0 | ||||
| Clostridia | 179 | 78.8 | 186 | 76.5 | 164 | 88.6 | 172 | 85.5 |
| Mollicutes | 2 | <0.1 | 3 | 0.1 | 4 | 0.6 | 2 | 0.1 |
| Bacilli | 7 | 1.4 | 2 | 0.1 | 2 | 0.1 | 3 | 0.4 |
| Actinobacteria | 6 | 0.1 | 0 | 1 | <0.1 | 0 | ||
| Spirochaetes | 0 | 0 | 0 | 1 | <0.1 | |||
| Bacteroidetes | 25 | 12.5 | 33 | 11.5 | 18 | 9.3 | 21 | 9.3 |
| Fusobacteria | 0 | 0 | 0 | 0 | ||||
| Verrucomicrobiae | 1 | 0.1 | 0 | 1 | 1.4 | 1 | 2.6 | |
| Lentisphaerae | 0 | 1 | <0.1 | 0 | 1 | <0.1 | ||
Number of phylospecies detected for each bacterial class.
Combined percent contribution of phylospecies in each bacterial class to the total hybridization signal measured by microbiota microarray.
Though we observed a good consistency of bacterial composition among samples at the class and order levels, significant variability was found at the phylospecies levels. Signal for 227 to 232 species was detected in fecal samples from children; however, only 143 of these were detected in both (62%). For adult stools, 109 species were present in both samples (55%). In general, variability between adult samples was higher (the Spearman rank correlation of hybridization signal values between child samples was 0.65, and that between adult samples was 0.57), which might be attributed to the differences between adult volunteers in gender, age, or ethnic origin (the children differed only in age). However, the diversity of the microbiota was similar among all volunteers at the genus level: 39 and 42 different genera were detected in the child fecal samples, whereas 34 and 40 different genera were present in adult stools.
The distribution of signal among species and genera displayed a hyperbolic relationship, where relatively few members accounted for the majority of observed signal. For all four fecal samples, the 10 genera with the highest total signal values for all present species in these genera accounted for 84 to 88% of the overall signal. The most abundant phylospecies (Faecalibacterium prausnitzii in children, Papillibacter spp. in adults) was present at 4 to 6% of the total hybridization signal, whereas the lowest signal for a detected phylospecies was at a 0.001 to 0.004% level.
At the genus level, the microbiota composition in children was dominated by Faecalibacterium (26% total signal), Ruminococcus (17% total signal), and Bacteroides (9% total signal). Fecal samples in adults were dominated by Papillibacter (25% total signal), Ruminococcus (16% total signal), and Faecalibacterium (10% total signal). In children's stools, Papillibacter was present only at 6% of the total abundance level on average. Roseburia was another abundant genus, present at a 4 to 8% overall signal level in all samples. Note, however, that due to the variability in hybridization affinities among probe sets, and because bacteria have different numbers of 16S rRNA gene copies per genome, comparisons of the total signal among different species and groups within the same sample should be used with caution and only as an approximation of relative species abundance. Other notable differences among samples included the presence of Haemophilus spp. in children's samples and the detection of Verrucomicrobium spp. (class Verrucomicrobiae) and Phascolarctobacterium spp. (class Clostridia) in stool samples from adults. Interestingly, one of the children had a strikingly high signal for members of Gammaproteobacteria, including Klebsiella and E. coli. Gammaproteobacteria accounted for 11% of the total signal in this fecal sample. This sample was taken from a 15-year-old child volunteer consuming a western diet; no medical history was available for this volunteer that would allow us to explain the observed phenomenon.
We did not detect a significant presence of Bifidobacterium or Lactobacillus, two genera often used in probiotic formulations, in any of the fecal samples. Similar findings were reported in other microbiota community studies (31, 38). The analysis of the matching of the universal primers used in this study to amplify full-length 16S rRNA genes to the RDP sequence database (8) indicated that out of the 151 Bifidobacteriales 16S rRNA gene sequences, only 98 matched the 27F primer used (the reverse primer matched 90% of full-length Bifidobacteriales sequences). During further analysis, a new 27F forward primer was developed containing degenerate nucleotides in four different positions (AGRGTTYGATYMTGGCTCAG); this degenerate primer matched 132 Bifidobacteriales sequences. Using this new forward primer in combination with the original reverse primer, new PCR amplifications were carried out on the fecal gDNA isolated from one of the child volunteers, and the mixture was hybridized to the microarray. Seven different phylospecies of bifidobacteria were detected in this new amplified sample; however, the hybridization signal was not high: Bifidobacteriales constituted only 1.2% of the overall signal on the array. This finding is consistent with the study of Palmer et al. (31), who also detected very low numbers of bifidobacteria in both babies and adults. No improvement in the detection of lactobacilli was achieved with the new primers, indicating that either these species were not present in the samples or that their relative abundance was below the detection threshold (0.001% of the overall sample).
Confirmation of microarray results with qPCR.
We used qPCR to confirm the findings of our microarray analysis of microbiota in human fecal samples. Five bacterial groups that showed differences in the presence or relative abundance between adult and child samples as measured by microarray were chosen for validation (Table 4). gDNA isolated from stool samples of child number 2 and adult number 1 was amplified with group-specific primer combinations. The amplification of total bacterial gDNA with universal primers was used as a measure of the total gDNA amount in each reaction. Very good concordance between microarray and qPCR results was observed (Table 4). The only significant difference was a relatively low abundance of Haemophilus in the child sample. Nevertheless, Haemophilus gDNA was detected consistently in all qPCR tests of the child's sample, whereas gDNA from this bacterial group was never detected in the adult sample. The low relative abundance of Haemophilus in our qPCR tests of child samples might be explained by the lower annealing efficiency of the Haemophilus-specific reverse primer (due to a low primer melting temperature). Overall, qPCR tests validate the good quantitative performance of the designed microarray on the clinical fecal samples.
TABLE 4.
Validation of microarray results by qPCR
| Bacterial groupa | Child 2
|
Adult 1
|
||
|---|---|---|---|---|
| Arrayb (%) | qPCRc (%) | Array (%) | qPCR (%) | |
| Faecalibacterium | 24.5 | 21.06 ± 2.91 | 6.8 | 10.73 ± 1.44 |
| Enterobacteriales | 9.6 | 11.28 ± 1.03 | ND | 0.17 ± 0.06 |
| Haemophilus | 1.7 | 0.11 ± 0.02 | ND | ND |
| Verrucomicrobium | ND | ND | 1.4 | 2.31 ± 0.93 |
| Phascolarctobacterium | ND | ND | 2.9 | 2.05 ± 0.12 |
The chosen primers cover most of the 16S rRNA gene sequences in the RDP database belonging to each group.
Combined percent contribution of phylospecies in each bacterial group to the total hybridization signal measured by microbiota microarray. ND, not detected.
Relative abundance of gDNA of each bacterial group compared to the total bacterial gDNA as measured by qPCR. Data are shown as arithmetic means ± standard deviations.
DISCUSSION
This study describes the design and validation of a custom high-throughput microbiota microarray. The developed array contains thousands of 16S rRNA gene oligoprobes, allowing the simultaneous interrogation of the presence and relative abundance level of 775 different bacterial phylospecies identified in the human intestine. Validation experiments produced excellent results: all 15 bacterial species used in these tests were correctly detected in pure cultures and in a complex mixture containing gDNA from all species, whereas Propionibacterium gDNA correctly produced no significant signal on the array. The choice of the Affymetrix GeneChip platform for the development of the gut microbiota array combined with specific features of our design allowed us to produce a microarray with selectivity, sensitivity, and linearity of measurements exceeding those of other recently developed community microarrays (16, 32, 41). The detection limit of the microarray was at least 1 pg when gDNA was enriched for 16S rRNA genes by PCR amplification. The microarray was able to detect bacterial DNA present at 0.00025% of the total gDNA sample, which represents an estimated 4,000-fold dynamic range of detection. Cross-hybridization among different probe sets was low and was almost completely reduced by the use of multiple replicate arrays per sample. The array was able to consistently detect quantitative differences in species abundance among gDNA samples, which will allow us to use this tool to quantitatively assess changes in microbiota composition among intestinal and fecal samples.
Several challenges exist in the use of microarray technology to interrogate the phylogenetic structure of complex bacterial communities. Due to the large number of different species usually found in such bacterial populations, the individual species often are present at a low level of relative abundance, which makes their reliable detection difficult. Similarly to previous reports (18, 44), our validation experiments indicate that the use of the 16S rRNA gene-specific PCR amplification of gDNA dramatically increases the sensitivity of detection (4 ng for unamplified gDNA versus 1 pg for gDNA subjected to 30 cycles of PCR amplification). However, the PCR amplification of 16S rRNA genes also is known to introduce biases in the relative abundances of different DNA species (17, 34, 39). Several approaches can be taken to reduce the variability of product-to-template ratios among different species within the same PCR mixture. These include lowering the number of PCR amplification cycles whenever possible, using multiple PCRs for each template sample, and shortening the ramp time between denaturation and annealing steps in the PCR cycle (6, 20, 33). Finally, rigorous statistical tests should be used to estimate the robustness of observed signal values and (n-fold) differences among samples.
Similar biases also can manifest during the hybridization of gDNA mixture to the microarray probes, with large variances in the GC content of the interrogated sequences (and corresponding oligoprobes) accounting for the majority of hybridization bias. To counter this potential issue, the design of microarray probes was restricted to the consideration of oligomers with a limited range of allowable %GC (see the probe GC distribution figure at http://www.wright.edu/∼oleg.paliy/Papers/MF_array/MF_array.html).
A third challenge exists in the evaluation of bacterial abundance based on the observed hybridization signal. The use of the microbiota array enables us to generate signal values for each interrogated 16S rRNA gene sequence. This quantitative data can be used to extrapolate the relative number of cells of each species within each sample. However, because bacteria have different numbers of 16S rRNA gene copies per genome, which can range from 1 to at least 15 (1, 19), the hybridization signal for each species is reflective of the species abundance and also is a function of the number of 16S rRNA genes that the species possesses. To account for such genome differences, we are developing an algorithm that would adjust the hybridization signal for each species by the number of 16S rRNA genes that species is known (or in many cases, is predicted) to possess.
All of the above-mentioned biases are specific to each individual sequence/species and are cancelled out in comparisons of two or more microarrays (we assume here that for a given DNA sequence, the same bias generally is observed independently of the other molecules in the mixture). Therefore, the designed microarray can quantitatively analyze microbiota composition among different intestinal and stool samples without any additional adjustments required.
Our pilot microarray experiments on fecal samples isolated from adult and child volunteers indicate that the microbiota composition is dominated by Bacteroidetes and Clostridia, with much higher overall hybridization signals displayed by the latter group. This is consistent with numerous previous studies that used a variety of microbiological and molecular biology techniques (4, 10, 11, 22, 38). However, we also detected differences in the relative abundance of several bacterial groups between adults and older children. Traditionally, it has been thought that by 2 years of age the microbiota in children resembles that of an adult (27); however, the microbiota of children beyond a few years of age have not been studied (15, 31). This study is the first to suggest that older children have microbiota profiles that are different from those of adults. Because only two volunteers were available to us from each subject group, no firm conclusions can be made at this point, since the observed differences might be explained simply by random fluctuations in the relative abundance of various bacterial groups or by other sociogeographical factors. Further studies employing much higher numbers of subjects are needed to provide results of statistical significance.
Among our initial findings, we observed that older children (12 to 15 years old) harbor a relatively higher proportion of Bacteroidetes and fewer Firmicutes than adults. An interesting study by Ley et al. indicated that obese individuals harbor more Firmicutes and fewer Bacteroidetes than lean individuals (24). Furthermore, a gradual increase in Bacteroidetes was correlated with the loss of body weight over time. None of the volunteer subjects enrolled in our pilot study were obese, but the children were smaller in size and body weight, by virtue of age, than adults. Therefore, the ratio between these two groups of bacteria may be reflective of body weight as well as the age of subjects.
Currently, there are increasing questions about the human intestinal microbiome, as we recognize its relevance to many diseases that cannot adequately be understood or managed with today's advances in technology and medicine. The microbiota microarray developed in this project has the potential to be used for further quantitative studies of human microbiota and its dynamics in intestinal disorders such as irritable bowel syndrome, inflammatory bowel disease, and colon cancer. The developed microarray also can be used in the clinical setting as a rapid screening tool for measuring the bacterial presence in human intestine. The potential for the accurate screening of microbiota in patients suffering from a variety of intestinal disorders can provide useful information that may direct the choice of the most appropriate therapy.
Acknowledgments
We thank Benjamin Withman, Laura Rigsbee, and Richard Agans for manuscript proofreading, the members of the WSU Center for Genomics Research for access to the facility, and the Michael Leffak group for valuable comments and a gift of human gDNA. The analysis of the 16S rRNA gene data set was carried out with the help of Qiong Wang and Jim Cole (Michigan State University).
This work was supported by the National Institutes of Health (O.P.), by the Wright State University Boonshoft School of Medicine (O.P. and S.M.), and by the Kampf fund (S.M.).
Footnotes
Published ahead of print on 10 April 2009.
REFERENCES
- 1.Acinas, S. G., L. A. Marcelino, V. Klepac-Ceraj, and M. F. Polz. 2004. Divergence and redundancy of 16S rRNA sequences in genomes with multiple rrn operons. J. Bacteriol. 186:2629-2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Affymetrix. 2001. GeneChip arrays for gene expression analysis. Technical note. Affymetrix, Santa Clara, CA.
- 3.Affymetrix. 2001. GeneChip arrays provide optimal sensitivity and specificity for microarray expression analysis. Technical note. Affymetrix, Santa Clara, CA.
- 4.Bibiloni, R., M. Mangold, K. L. Madsen, R. N. Fedorak, and G. W. Tannock. 2006. The bacteriology of biopsies differs between newly diagnosed, untreated, Crohn's disease and ulcerative colitis patients. J. Med. Microbiol. 55:1141-1149. [DOI] [PubMed] [Google Scholar]
- 5.Bik, E. M., P. B. Eckburg, S. R. Gill, K. E. Nelson, E. A. Purdom, F. Francois, G. Perez-Perez, M. J. Blaser, and D. A. Relman. 2006. Molecular analysis of the bacterial microbiota in the human stomach. Proc. Natl. Acad. Sci. USA 103:732-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chandler, D. P., J. K. Fredrickson, and F. J. Brockman. 1997. Effect of PCR template concentration on the composition and distribution of total community 16S rRNA genes clone libraries. Mol. Ecol. 6:475-482. [DOI] [PubMed] [Google Scholar]
- 7.Choe, S. E., M. Boutros, A. M. Michelson, G. M. Church, and M. S. Halfon. 2005. Preferred analysis methods for Affymetrix GeneChips revealed by a wholly defined control dataset. Genome Biol. 6:R16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cole, J. R., B. Chai, R. J. Farris, Q. Wang, S. A. Kulam, D. M. McGarrell, G. M. Garrity, and J. M. Tiedje. 2005. The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 33:D294-D296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ducluzeau, R. 1988. Role of experimental microbial ecology in gastroenterology, p. 7-26. In E. Bergogne-Berezin (ed.), Microbial ecology and intestinal infections. Springer-Verlag, Berlin, Germany.
- 10.Eckburg, P. B., E. M. Bik, C. N. Bernstein, E. Purdom, L. Dethlefsen, M. Sargent, S. R. Gill, K. E. Nelson, and D. A. Relman. 2005. Diversity of the human intestinal microbial flora. Science 308:1635-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frank, D. N., A. L. St. Amand, R. A. Feldman, E. C. Boedeker, N. Harpaz, and N. R. Pace. 2007. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. USA 104:13780-13785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao, Z., C. H. Tseng, Z. Pei, and M. J. Blaser. 2007. Molecular analysis of human forearm superficial skin bacterial biota. Proc. Natl. Acad. Sci. USA 104:2927-2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gill, S. R., M. Pop, R. T. Deboy, P. B. Eckburg, P. J. Turnbaugh, B. S. Samuel, J. I. Gordon, D. A. Relman, C. M. Fraser-Liggett, and K. E. Nelson. 2006. Metagenomic analysis of the human distal gut microbiome. Science 312:1355-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He, Z., T. J. Gentry, C. W. Schadt, L. Wu, J. Liebich, S. C. Chong, Z. Huang, W. Wu, B. Gu, P. Jardine, C. Criddle, and J. Zhou. 2007. GeoChip: a comprehensive microarray for investigating biogeochemical, ecological and environmental processes. ISME J. 1:67-77. [DOI] [PubMed] [Google Scholar]
- 15.Hopkins, M. J., R. Sharp, and G. T. Macfarlane. 2001. Age and disease related changes in intestinal bacterial populations assessed by cell culture, 16S rRNA abundance, and community cellular fatty acid profiles. Gut 48:198-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huyghe, A., P. Francois, Y. Charbonnier, M. Tangomo-Bento, E. J. Bonetti, B. J. Paster, I. Bolivar, D. Baratti-Mayer, D. Pittet, and J. Schrenzel. 2008. Novel microarray design strategy to study complex bacterial communities. Appl. Environ. Microbiol. 74:1876-1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanagawa, T. 2003. Bias and artifacts in multitemplate polymerase chain reactions (PCR). J. Biosci. Bioeng. 96:317-323. [DOI] [PubMed] [Google Scholar]
- 18.Kim, P. I., B. D. Erickson, and C. E. Cerniglia. 2005. A membrane-array method to detect specific human intestinal bacteria in fecal samples using reverse transcriptase-PCR and chemiluminescence. J. Microbiol. Biotechnol. 15:310-320. [Google Scholar]
- 19.Klappenbach, J. A., P. R. Saxman, J. R. Cole, and T. M. Schmidt. 2001. rrndb: the Ribosomal RNA Operon Copy Number Database. Nucleic Acids Res. 29:181-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kurata, S., T. Kanagawa, Y. Magariyama, K. Takatsu, K. Yamada, T. Yokomaku, and Y. Kamagata. 2004. Reevaluation and reduction of a PCR bias caused by reannealing of templates. Appl. Environ. Microbiol. 70:7545-7549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Langendijk, P. S., F. Schut, G. J. Jansen, G. C. Raangs, G. R. Kamphuis, M. H. Wilkinson, and G. W. Welling. 1995. Quantitative fluorescence in situ hybridization of Bifidobacterium spp. with genus-specific 16S rRNA-targeted probes and its application in fecal samples. Appl. Environ. Microbiol. 61:3069-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lay, C., L. Rigottier-Gois, K. Holmstrom, M. Rajilic, E. E. Vaughan, W. M. de Vos, M. D. Collins, R. Thiel, P. Namsolleck, M. Blaut, and J. Dore. 2005. Colonic microbiota signatures across five northern European countries. Appl. Environ. Microbiol. 71:4153-4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ley, R. E., F. Backhed, P. Turnbaugh, C. A. Lozupone, R. D. Knight, and J. I. Gordon. 2005. Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. USA 102:11070-11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ley, R. E., P. J. Turnbaugh, S. Klein, and J. I. Gordon. 2006. Microbial ecology: human gut microbes associated with obesity. Nature 444:1022-1023. [DOI] [PubMed] [Google Scholar]
- 25.Loy, A., M. Horn, and M. Wagner. 2003. probeBase: an online resource for rRNA-targeted oligonucleotide probes. Nucleic Acids Res. 31:514-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loy, A., A. Lehner, N. Lee, J. Adamczyk, H. Meier, J. Ernst, K. H. Schleifer, and M. Wagner. 2002. Oligonucleotide microarray for 16S rRNA gene-based detection of all recognized lineages of sulfate-reducing prokaryotes in the environment. Appl. Environ. Microbiol. 68:5064-5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mackie, R. I., A. Sghir, and H. R. Gaskins. 1999. Developmental microbial ecology of the neonatal gastrointestinal tract. Am. J. Clin. Nutr. 69:1035S-1045S. [DOI] [PubMed] [Google Scholar]
- 28.Mai, V., and J. G. Morris, Jr. 2004. Colonic bacterial flora: changing understandings in the molecular age. J. Nutr. 134:459-464. [DOI] [PubMed] [Google Scholar]
- 29.Maynard, C., F. Berthiaume, K. Lemarchand, J. Harel, P. Payment, P. Bayardelle, L. Masson, and R. Brousseau. 2005. Waterborne pathogen detection by use of oligonucleotide-based microarrays. Appl. Environ. Microbiol. 71:8548-8557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mitterer, G., M. Huber, E. Leidinger, C. Kirisits, W. Lubitz, M. W. Mueller, and W. M. Schmidt. 2004. Microarray-based identification of bacteria in clinical samples by solid-phase PCR amplification of 23S ribosomal DNA sequences. J. Clin. Microbiol. 42:1048-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Palmer, C., E. M. Bik, D. B. Digiulio, D. A. Relman, and P. O. Brown. 2007. Development of the human infant intestinal microbiota. PLoS Biol. 5:e177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Palmer, C., E. M. Bik, M. B. Eisen, P. B. Eckburg, T. R. Sana, P. K. Wolber, D. A. Relman, and P. O. Brown. 2006. Rapid quantitative profiling of complex microbial populations. Nucleic Acids Res. 34:e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peplies, J., F. O. Glockner, and R. Amann. 2003. Optimization strategies for DNA microarray-based detection of bacteria with 16S rRNA-targeting oligonucleotide probes. Appl. Environ. Microbiol. 69:1397-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Polz, M. F., and C. M. Cavanaugh. 1998. Bias in template-to-product ratios in multitemplate PCR. Appl. Environ. Microbiol. 64:3724-3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rainer, J., F. Sanchez-Cabo, G. Stocker, A. Sturn, and Z. Trajanoski. 2006. CARMAweb: comprehensive R- and bioconductor-based web service for microarray data analysis. Nucleic Acids Res. 34:W498-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sartor, R. B. 2008. Microbial influences in inflammatory bowel diseases. Gastroenterology 134:577-594. [DOI] [PubMed] [Google Scholar]
- 37.Schloss, P. D., and J. Handelsman. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suau, A., R. Bonnet, M. Sutren, J. J. Godon, G. R. Gibson, M. D. Collins, and J. Dore. 1999. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl. Environ. Microbiol. 65:4799-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wagner, A., N. Blackstone, P. Cartwright, M. Dick, B. Misof, P. Snow, G. P. Wagner, J. Bartels, M. Murtha, and J. Pendleton. 1994. Surveys of gene families using polymerase chain-reaction—PCR selection and PCR drift. Syst. Biol. 43:250-261. [Google Scholar]
- 40.Wang, Q., G. M. Garrity, J. M. Tiedje, and J. R. Cole. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261-5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang, R. F., M. L. Beggs, B. D. Erickson, and C. E. Cerniglia. 2004. DNA microarray analysis of predominant human intestinal bacteria in fecal samples. Mol. Cell. Probes 18:223-234. [DOI] [PubMed] [Google Scholar]
- 42.Wilson, K. H., and R. B. Blitchington. 1996. Human colonic biota studied by ribosomal DNA sequence analysis. Appl. Environ. Microbiol. 62:2273-2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilson, K. H., W. J. Wilson, J. L. Radosevich, T. Z. DeSantis, V. S. Viswanathan, T. A. Kuczmarski, and G. L. Andersen. 2002. High-density microarray of small-subunit ribosomal DNA probes. Appl. Environ. Microbiol. 68:2535-2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wilson, W. J., C. L. Strout, T. Z. DeSantis, J. L. Stilwell, A. V. Carrano, and G. L. Andersen. 2002. Sequence-specific identification of 18 pathogenic microorganisms using microarray technology. Mol. Cell. Probes 16:119-127. [DOI] [PubMed] [Google Scholar]
- 45.Wu, L., D. K. Thompson, G. Li, R. A. Hurt, J. M. Tiedje, and J. Zhou. 2001. Development and evaluation of functional gene arrays for detection of selected genes in the environment. Appl. Environ. Microbiol. 67:5780-5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu, L., D. K. Thompson, X. Liu, M. W. Fields, C. E. Bagwell, J. M. Tiedje, and J. Zhou. 2004. Development and evaluation of microarray-based whole-genome hybridization for detection of microorganisms within the context of environmental applications. Environ. Sci. Technol. 38:6775-6782. [DOI] [PubMed] [Google Scholar]
- 47.Zhou, J. 2003. Microarrays for bacterial detection and microbial community analysis. Curr. Opin. Microbiol. 6:288-294. [DOI] [PubMed] [Google Scholar]