Abstract
Metabolic inactivation of leukotriene B4 (LTB4) is an innate mechanism to resolve tissue inflammation. We studied the nine Cyp4f genes in the mouse genome, measuring cutaneous transcript levels by real-time polymerase chain reaction, and LTB4 metabolism in mouse and human skin. Transcripts arising from Cyp4f13 and 4f16 ranked most abundant, Cyp4f14, 4f17, and 4f37 ranked least abundant, and Cyp4f18 and 4f39 ranked intermediate. Those from Cyp4f15 and Cyp4f40 were highly variable or too low to measure in some animals. Retinoic acid exposure induced microsomal LTB4 hydroxylation activities in mouse and human skin cells. Two NADPH-dependent LTB4 metabolites eluted identically with 20-OH and 20-COOH LTB4 reference standards. Collision induced dissociation of the precursor ion m/z 351 confirmed that LTB4 products from CYP4F3A and human epidermal keratinocytes are identical structurally to 20-OH LTB4. We conclude 20-hydroxylation is the major CYP-dependent LTB4 inactivation pathway in skin; this retinoid-inducible metabolic pathway has capacity to modulate tissue levels of pro-inflammatory lipids.
Introduction
Polymorphonuclear neutrophils infiltrating injured tissues are a principal source of leukotriene B4 (LTB4), synthesized from arachidonic acid via the 5-lipoxygenase pathway [1; 2]. Actions of this potent pro-inflammatory lipid are counterbalanced in part by cytochrome P450 enzymes (CYP4F gene products), which activate oxygen and catalyze successive oxidations at the LTB4 ω-terminus. This metabolism forms 20-OH LTB4 and, via an aldehyde intermediate, the biologically inactive 20-carboxy LTB4 metabolite [1]. CYP4F3A and CYP4F2 are the principal LTB4 hydroxylases in human neutrophils and hepatocytes, respectively [3; 4].
Mukhtar and colleagues characterized LTB4 hydroxylase enzyme activities in epidermis from humans, rats, and guinea pigs [5]. Recent studies of human epidermis suggest that CYP4F proteins in keratinocytes catalyze LTB4 hydroxylation and arise from one or more of five expressed CYP4F genes (CYP4F2, 4F3, 4F8, 4F11 and 4F12) [6; 7; 8]. Mechanisms regulating epidermal CYP4F expression and LTB4 hydroxylase activities are complex, involving both differentiation specific factors and retinoid X nuclear receptors in the context of pharmacological levels of retinoid acid exposure [6; 8]. CYP4F22 is the only CYP4F gene not yet well studied in human epidermis. CYP4F22 transcripts were detected in cultured human keratinocytes by reverse transcription polymerase chain reaction, and mutations in this gene lead to an epidermal barrier defect lamellar ichthyosis type 3 [9]. The mouse genome encodes nine Cyp4f genes, and not one of these has been studied in mouse epidermis or any other epithelia. The three most recently discovered Cyp4f genes (Cyp4f37, 4f39, and 4f40) have not been studied in any tissue. In this study, we aimed to characterize cutaneous expression of the nine mouse Cyp4f genes and to compare LTB4 hydroxylase activities in mouse and human skin cells.
Materials and methods
Animals, Tissues, and Cells
CD-1 strain mice (≈25 g) were from Charles Rivers (Wilmington, MA). Under isoflurane anesthesia, dorsal skin was shaved (≈9 cm2, Day -1) and treated topically (Day 0) with a 0.05% ethanolic solution of all-trans retinoic acid (≈5 μg/cm2; Sigma Aldrich Chemical Company, St Louis, MO) or an equal volume of vehicle (90 μl). Drug was re-applied every other day. Full-thickness skin was excised (dermis + epidermis, no subcutaneous tissue). Punch biopsies (6–8 mm) were taken for RNA and histological analyses, and then microsomes were prepared immediately from the remaining excised skin.
Human keratinocyte cultures were exactly as described [10] except for the culture media. We mixed equal volumes of Ca++-free Dulbecco’s Modified Eagle media (DMEM, #21068, Invitrogen, Carlsbad, CA) and Ham’s F12 (F12, #11765, Invitrogen). This DMEM:F12 was mixed with equal volumes of Medium 154-CF (Cascade, Portland, OR) and then supplemented with Cascade HKGS kit (#S-0001-K), gentamycin, sodium pyruvate (to 1 mM), and L-glutamine (to 2 mM). This tertiary mixture contained 0.075 mM Ca++ suitable for keratinocyte growth. When cultures achieved ≈75% confluence, cellular differentiation was initiated by adjusting medium Ca++ to 1.4 mM (Day 0).
Quantitative Real-time Polymerase Chain Reaction (qPCR)
Total RNA was extracted using a Mini Bead Beater (BioSpec Products, Bartlesville, OK) in conjunction with NucleoSpin RNA II kits (BD Bioscience Clontech; San Jose, CA). The RNA was subjected to a second DNase treatment (DNA-free kit, Ambion, Austin, TX) prior to reverse-transcription using a High-Capacity cDNA Reverse Transcription Kit (1 μg/20 μl reaction; Applied Biosystems, Foster City, CA). Template cDNA was diluted 1:10, and 4.5 μl were added per 10 μl of qPCR reaction.
Gene expression assays employed Taqman Assays-on-Demand qPCR kits and Universal PCR Master Mix containing AmpErase UNG (Applied Biosystems, Foster City, CA). Features of the proprietary assay kits and validation are described in detail [10]. We generated standard curves (2 pg/μl to 2 ag/μl) using purified, synthetic amplicons corresponding to genomic sequences amplified by each assay kit (Integrated DNA Technologies, Coralville, IA). Table 1 lists assay identifications, amplicon locations, and amplification efficiency values. Specificity was evaluated by adding water instead of reverse transcriptase and water instead of template cDNA.
Table 1.
Amplification efficiency values for the mouse gene qPCR assay kits.
| Transcripts Detected | Assay ID* | Efficiency** (%) | Reference Sequence (amplicon position) |
|---|---|---|---|
| CYP4F13 | Mm00504576_m1 | 1.97 (98%) | NM_130882 (363–442 bp) |
| CYP4F16 | Mm00775893_m1 | 1.90 (95%) | NM_024442 (186–335 bp) |
| CYP4F18 | Mm00499348_m1 | 2.00 (100%) | NM_024444 (280–430 bp) |
| CYP4F14 | Mm00491623_m1 | 1.97 (98%) | NM_022434 (351–450 bp) |
| CYP4F15 | Mm00506542_m1 | 1.97 (98%) | NM_134127 (1231–1320 bp) |
| CYP4F17 | m4f17-1398-45* | 2.05 (102%) | XM_139863 (451–550 bp) |
| CYP4F37 | m4f37-XM00-23* | 2.10 (105%) | XM_001001020 (175–270 bp) |
| CYP4F39 | m4f39-AK02-12* | 2.05 (102%) | AK028950 (21–120 bp) |
| CYP4F40 | m4f40-XM99-12* | 2.00 (100%) | XM_992966 (141–240 bp) |
| Cyclophilin A | Mm02342429_g1 | 1.97 (98%) | NM_008907 (200–320 bp) |
Assays were custom designed (asterisk) by Applied Biosystems or from the catalog inventory (no asterisk). Exact oligonucleotide sequences generating the specified amplicons are proprietary.
Efficiency is calculated as 10−1/slope, where the slope is calculated from a standard curve. A value of 2.0 equals 100% efficiency.
Leukotriene Metabolism
We prepared microsomes as described [11] from full-thickness mouse skin (5–7 mg protein/ml), fresh human epidermis (3–6 mg/ml), and cultured human keratinocytes (3–7 mg/ml). Microsomes were resuspended in 100 μl of buffer (0.1 M potassium phosphate, pH 7.4) and assayed without storage. Eighty μl were transferred to a reaction tube, reserving 20 μl for protein determinations. Leukotriene B4 (Biomol International, Plymouth Meeting, PA) was dried under nitrogen, reconstituted in 20 μl buffer, and mixed with microsomes (60 μM final concentration). Freshly prepared 50 mM NADPH (2 μl; 1 mM final concentration) initiated reactions, which proceeded at 37°C for 45 or 60 min while shaking. Supersomes from BD Biosciences (Franklin Lakes, NJ) contained recombinant CYP4F protein, NADPH cytochrome P450 reductase and cytochrome b5: CYP4F3A (BD Gentest #456273); CYP4F3B (#456274); CYP4F2 (#456272); and CYP4F12 (#456275). Ten pmol of recombinant P450/100 μl were assayed similarly but reacted only 15 min. For CYP4F3A, only 5 pmol were added.
Ice-cold ethanol (0.4 ml) containing 0.5 μg of prostaglandin B2 (Cayman Chemical Company, Ann Arbor, MI) stopped the reactions, and then proteins were allowed to precipitate during overnight storage at −30°C. Supernatant fractions recovered by centrifugation (1000×g, 10 min) were dried under nitrogen and resuspended in 10% methanol containing 0.1% glacial acetic acid (pH 4–5) prior to extraction using Bond Elut C18 (500 mg, #12102028, Varian Inc., Palo Alto, CA,) or Oasis HLB (30 mg, #WAT094225, Waters Corporation, Milford, MA) cartridges, according to the manufacturers’ instructions. Extracts were resolved by reversed-phase high performance liquid chromatography (HPLC) at 1 ml/min using a Dynamax Microsorb C18 column (5-μm, 4.6 × 250 mm; Varian Inc.). Initial isocratic solvent conditions (held for 3 min) were 75% solvent A (100:0.05; v:v; water:glacial acetic acid) and 25% solvent B (65:35:0.05; v:v:v; acetonitrile:methanol:glacial acetic acid). Solvent B was then increased linearly over 29 min to 100%, and held at 100% for 10 min. Retention times of products having maximal absorbance at 272-nm were compared with those for 20-OH and 20-COOH LTB4 reference compounds (Biomol). We estimated the mass of product formed from standard curves generated using LTB4 reference compound.
Liquid Chromatography/Mass Spectrometry (LC/MS)
Leukotriene metabolites resolved by reversed-phase HPLC were collected and analyzed by LC/MS to confirm analyte identity. We used two Phenomenex Luna ODS columns (5-μm, Torrence, CA) differing only in length (2.1 × 50 mm or 150 mm). Initial solvent conditions (0.2 ml/min) were: 80% solvent A (2.0 mM NH4Ac), 20% solvent B (35:65; v:v; methanol:acetonitrile). Solvent B was increased linearly from 20% to 100% in 8 min, and then held at 100% for 10 min. For MS, a ThermoFinnigan TSQ Quantum ultra mass spectrometer was operated in negative ion mode. The electrospray ionization source was fitted with a deactivated fused silica capillary (100-μm i.d.). Sheath gas and auxiliary gas were nitrogen. The ion transfer tube temperature was 30ºC. The spray voltage, tube lense voltage, pressure of sheath gas and auxiliary gas were optimized to achieve maximal response. Collision-induced dissociation (CID) was performed from 20 to 30 eV under 1.0 mTorr of argon. Selective reaction monitoring (SRM) was carried out to select a transition from a specific precursor ion to the most abundant characteristic fragment determined by CID experiments.
Results
Mouse skin Cyp4f gene expression
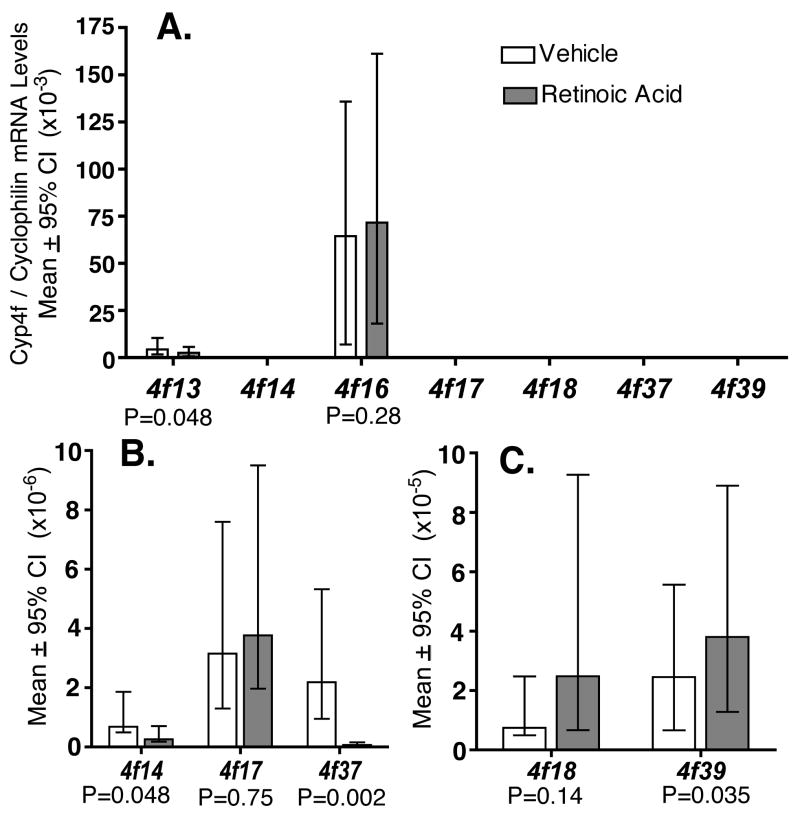
Quantitative real-time PCR assays measured transcripts in full-thickness mouse skin arising from nine Cyp4f genes--Cyp4f13, 4f14, 4f15, 4f16, 4f17, 4f18, 4f37, 4f39, and 4f40 (Fig. 1). Transcript abundance ranked highest for the Cyp4f13 and 4f16 genes, lowest for Cyp4f14, 4f17, and 4f37, and intermediate for Cyp4f18 and 4f39. Transcripts arising from Cyp4f15 were highly variable, ranging from intermediate to undetectable levels in individual mice. Transcripts arising from Cyp4f40 were present, but the levels were too low to measure reliably in most mice.
Figure 1.
Cyp4f expression levels in mouse skin exposed to vehicle (white bars) or pharmacological levels of all trans-retinoic acid (gray bars), for six days. Quantitative PCR measured CYP4F mRNA levels in full-thickness skin. Plots show normalized data [CYP4F/Cyclophilin A] as means ± 95% confidence intervals (CI), with probability (P) values for comparison of retinoic acid vs. vehicle. Standard deviations are not shown. Each group (n=7) contained nearly equal numbers of adult male and female mice. SPSS 15.0 statistical software (SPSS Inc, Chicago IL) generated descriptive statistics and P-values using the Kruskal-Wallis test for nonparametric data. Individual measurements at or below the lower limit of assay sensitivity were assigned the value zero; group data were not analyzed statistically if >20% of animals had zero values. This explains why Cyp4f15 and 4f40 data are not plotted. Panels B and C show results for subsets of the same data in Panel A, plotted on expanded Y-axes.
We measured changes in cutaneous Cyp4f gene expression in response to topical application of supraphysiological levels of all trans-retinoic acid. Histological examination of mouse skin confirmed the expected epidermal thickening due to retinoid-induced expansion of spinous and granular cell layers, as reported previously (data not shown) [12; 13]. After six days of retinoid exposure, cutaneous transcripts arising from Cyp4f13, 4f14, and 4f37 were down-regulated (P=0.048, 0.048, and 0.002, respectively, vs. vehicle), and transcripts arising from Cyp4f39 were up-regulated (P= 0.035). Changes in transcript abundance for the Cyp4f16, 4f17 and 4f18 genes were not statistically significant (P=0.28, 0.75, and 0.14, respectively).
Cutaneous LTB4 inactivation
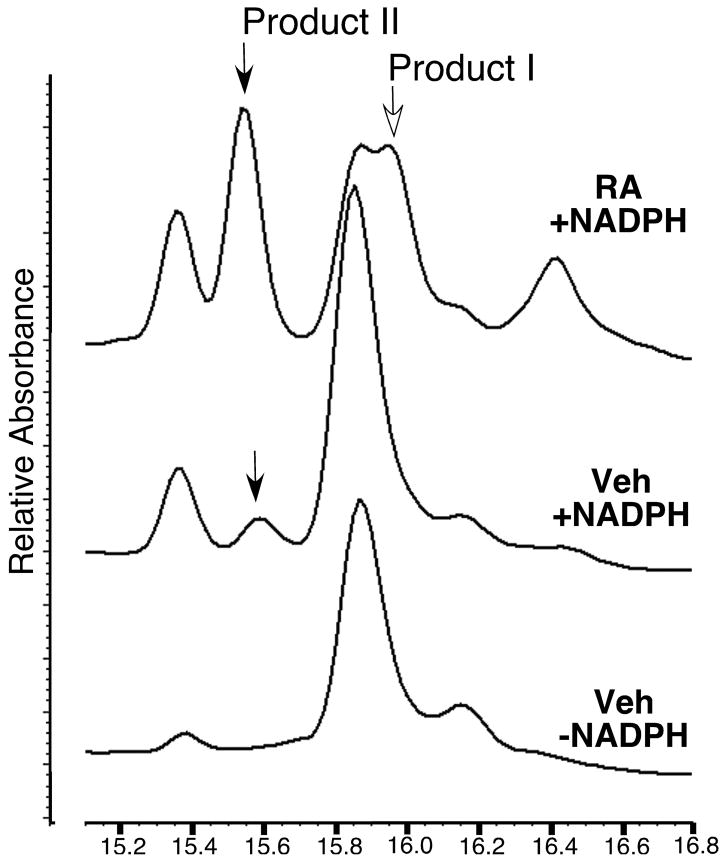
Exposure to supraphysiological levels of retinoic acid induced microsomal LTB4 hydroxylase activities in mouse skin, a result similar to that reported for human epidermal cells [6]. Two NADPH-dependent products (λmax 272-nm) were generated by retinoid treated mouse skin, and these eluted identically with the 20-OH LTB4 (Product I) and 20-COOH LTB4 (Product II) reference compounds, as resolved by reversed-phase HPLC (Fig. 2). Product I was not resolved from an unidentified NADPH-independent analyte (λmax 230-nm). Product II was resolved, and this was the only LTB4 metabolite detected in vehicle treated mouse skin. The rate of Product II formation was estimated at 0.08 and 0.27 (pmol/min/mg protein) for vehicle treated and retinoic acid treated mouse skin, respectively. Omitting NADPH from the reaction demonstrated specificity. In this case, none of the detectable analytes had an absorption spectrum consistent with a LTB4 derivative, regardless of the experimental treatment.
Figure 2.
Retinoic acid induces production of two NADPH-dependent products from LTB4 by adult mouse skin microsomes. Superimposed, reversed-phase HPLC chromatograms show truncated X-axes (15.2–16.8 min). Two NADPH-dependent products absorb light maximally at 272-nm and elute identically with 20-OH (open arrow, Product I, 16 min) and 20-COOH (closed arrows, Product II, 15.5 min) LTB4 reference compounds. The control chromatogram (-NADPH) is representative of results for vehicle and retinoid treated mouse skin. RA, all trans-retinoic acid treated skin. Veh, vehicle treated skin.
Mouse and human LTB4 hydroxylase activities
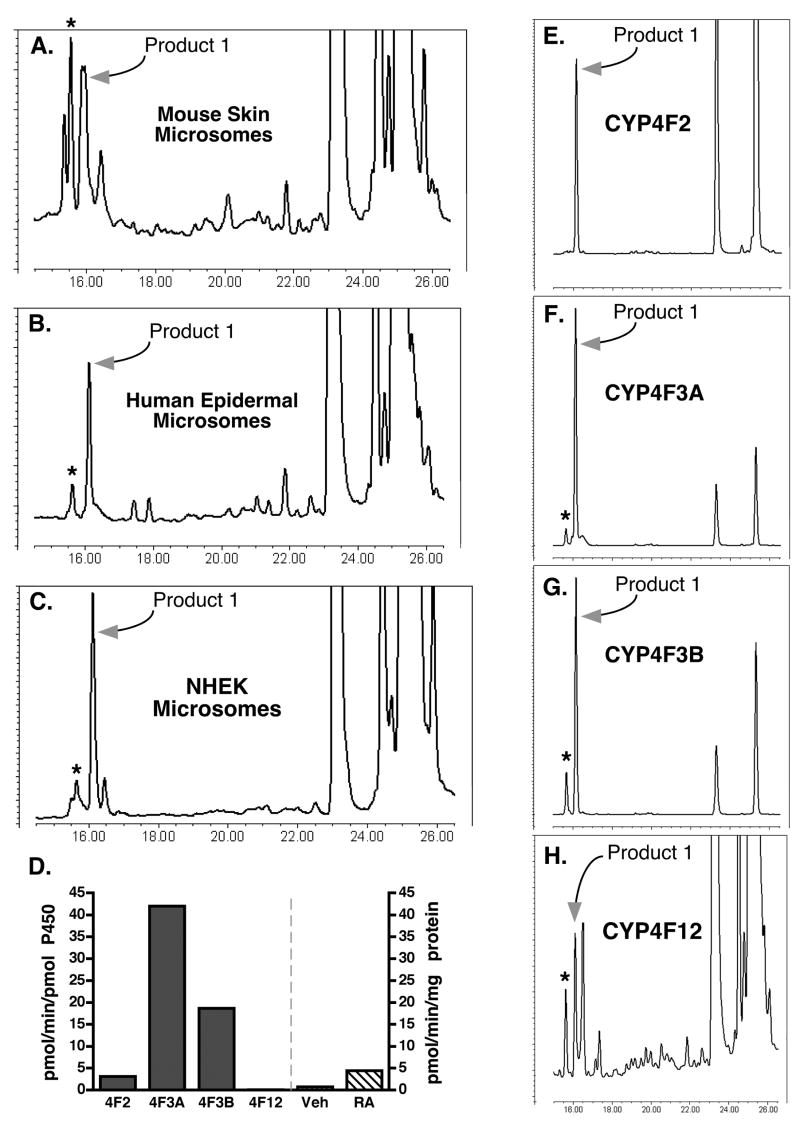
We compared retinoid-inducible LTB4 hydroxylase activities measured in full-thickness mouse skin with those in fresh human epidermal tissue and derived cell cultures (Fig. 3A–C). Normal human epidermal keratinocyte (NHEK) cultures that differentiated in vitro exhibited greater LTB4 hydroxylase activities than fresh skin tissues of either species, measured as the formation of Product I eluting with the 20-OH LTB4 reference compound. Product I from human epidermal cells resolved well in the reversed-phase system and was induced 6-fold in cultures that were differentiated in the presence of 0.1 μM all trans-retinoic acid (Fig. 3D) (0.74 vs. 4.4 pmol/min/mg for vehicle vs. retinoic acid treatments, respectively). Skin microsomes generated only two NADPH-dependent LTB4 products (λmax 272-nm), regardless of species of origin (mouse or human), and these eluted identically with 20-OH (Products I) and 20-COOH (Product II) LTB4 reference compounds when resolved by reversed-phase HPLC. If skin microsomes generated other hydroxylated products (e.g., ω-1, ω-2), the amounts were negligible by comparison as these can be resolved chromatographically from 20-OH and 20-COOH LTB4 [14].
Figure 3.
Mouse and human skin cells and recombinant human CYP4F proteins generate similar LTB4 metabolite profiles. Reversed-phase HPLC chromatograms show Product I (arrows) and Product II (asterisks) having absorption spectra (λmax 272-nm) and retention times indistinguishable from 20-OH and 20-COOH LTB4 reference compounds, respectively. A., Microsomes from full-thickness skin of adult mice treated topically every other day for 10 days with retinoic acid (from chromatogram in Fig. 2, top trace). B., Microsomes from freshly isolated human epidermis from neonatal foreskins. C., Microsomes from normal human epidermal keratinocyte (NHEK) cultures, derived from neonatal foreskins, differentiated 7-days in the presence of 0.1 μM all trans-retinoic acid (RA). D., Product I formation rates. Left Y-axis (pmol/min/pmol P450) is for recombinant human CYP4F2 (3.1), CYP4F3A (42), CYP4F3B (18.7), and CYP4F12 (0.06). Right Y-axis (pmol/min/mg protein) is for NHEK cultures differentiated in the presence of vehicle (0.74) or RA (4.4). E.-H., Recombinant CYP4F Supersomes. All Y-axes represent relative absorbance, except for Panel D.
20-Hydroxy LTB4 is the major epidermal product derived from LTB4
We compared Product I generated by microsomes from mouse [full-thickness] skin and from human epidermis with that generated by human CYP4F2, 4F3A, 4F3B, and 4F12 enzymes (Fig. 3E–H). All four recombinant proteins generated Product I from LTB4, which in all cases was indistinguishable from the 20-OH LTB4 reference compound assayed under similar conditions. CYP4F3A was the most efficient LTB4 20-hydroxylase (Fig. 3D). CYP4F3A and 4F3B generated a secondary product, which we presume is 20-COOH LTB4 derived from 20-OH LTB4. CYP4F12 was least efficient, generating several additional products.
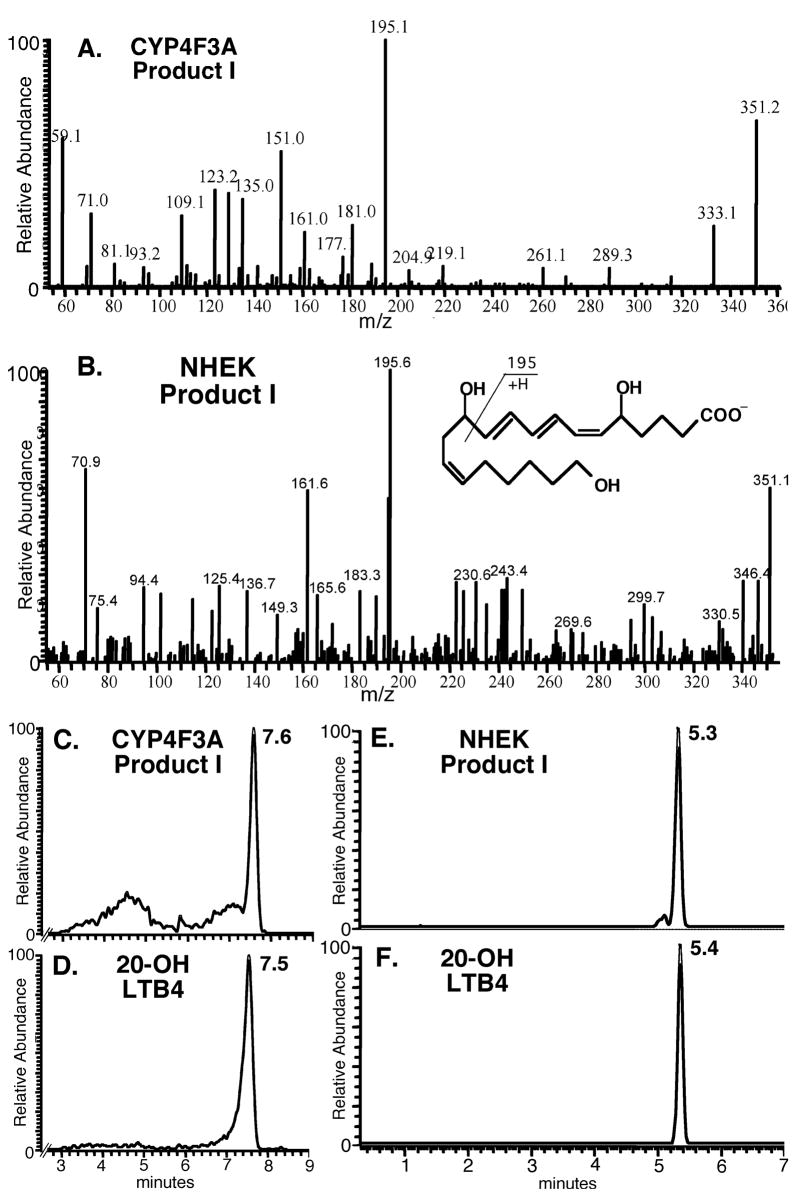
Since our results and those of others’ [15] agree that Product I is indistinguishable from 20-OH LTB4 by reversed-phase HPLC, we aimed next to characterize and compare the structures of Product I from epidermis and from recombinant CYP4F3A. For these structural studies we used NHEK cultures, which represent a single predominant cell type (unlike fresh tissue of either species). We manipulated the cultures to maximize enzyme expression and specific activities to levels normally associated with the outermost, differentiated epidermal layers. This material yielded sufficient LTB4 product to carry out CID LC/MS experiments on the precursor ion m/z 351.
Fragmentation patterns obtained for CYP4F3A and the epidermal keratinocyte samples are identical to those for the 20-OH LTB4 reference compound (Fig. 4A–B). The major fragment m/z 195 is characteristic for this compound, and transition from m/z 351 to 195 was selected in SRM experiments. Retention times for Product I from both CYP4F3A and epidermal keratinocyte samples are identical to that for the 20-OH LTB4 reference compound (Fig. 4C–F). As further evidence, we observed a single product peak when a mixture of 20-OH LTB4 standard and CYP4F3A Product I was subjected to LC/MS under the similar conditions (data not shown). Finally, pentafluorobenzyl ester derivatives of Product I were subjected to normal phase separation coupled with atmospheric pressure chemical ionization MS (data not shown). Under these conditions, Product I derived from CYP4F3A and that from epidermal keratinocytes are identical to the 20-OH LTB4 reference compound.
Figure 4.
Structures of Product I from CYP4F3A and differentiated human epidermal keratinocyte (NHEK) cultures are indistinguishable from that of the 20-OH LTB4 reference compound, determined by LC/MS. Keratinocytes were differentiated in vitro in the presence of retinoic acid for 10 days. A.-B., Product ion spectra of CYP4F3A Product I and epidermal Product I were the same as that for 20-OH LTB4 reference compound (Biomol). Inset shows predicted fragmentation of 20-OH LTB4 generating the dominant ion m/z 195. C.-F., LC retention times for CYP4F3 Product I and epidermal Product I were the same as that for 20-OH LTB4. Different retention times for 20-OH LTB4 correspond to use of two columns differing only in length.
Discussion
Neutrophils are one the earliest known sources of leukotriene hydroxylases, namely CYP4F gene products [2; 16; 17]. Our studies of CYP4F expression and LTB4 hydroxylation in human [6] and mouse skin cells provide essential information to begin understanding metabolic pathways controlling tissue levels of immune modulating lipids such as LTB4. The majority of mouse Cyp4f genes are expressed in skin at measurable levels, similar to recent results for CYP4F genes in human skin [6]. Christmas and colleagues reported recently that mouse CYP4F18 is orthologous to human CYP4F3A and that it is the principal LTB4 hydroxylase in mouse polymorphonuclear leukocytes [18]. Thus, the principal LTB4 hydroxylases in mouse neutrophils (CYP4F18) and human neutrophils (CYP4F3A) are also expressed in skin of the respective species. The Cyp4f37, 4f39, and 4f40 genes were presumed to be functional genes because they lacked identifiable defects in their genomic sequences [19]. We report these genes are transcribed in mouse skin but that in a subset of animals cutaneous Cyp4f40 expression was too low to measure reliably.
Our CID structure analyses agree with structural data reported previously for the reference compound 20-OH LTB4 [20]. Enzyme activity levels measured in fresh skin tissues were insufficient for LC/MS analyses, regardless of species. However, careful analyses employing reversed-phase HPLC suggest that 20-hydroxylation is the major NADPH-dependent LTB4 inactivation pathway in mouse and human skin and this pathway is retinoid-inducible in both species. The primary epidermal metabolite derived from LTB4 eluted identically to the 20-OH LTB4 reference compound when resolved on both reversed-phase (free acid) and normal phase (after pentafluorobenzyl derivatization) HPLC and detected by SRM. Compared with fresh skin tissues of either species, NHEK cultures had greater intrinsic LTB4 hydroxylation activities and greater induction of CYP4F enzymes by retinoic acid. This is attributable to the highly optimized in vitro differentiation system in which the majority of cells express the same differentiation state (vs. four differentiation states in epidermis in situ). Furthermore, drug delivery is highly efficiently in submerged cell cultures (vs. transcutaneous absorption through the epidermal water permeability barrier, including the lipophilic stratum corneum in situ). When assayed, NHEK cultures expressed predominately a late spinous-early granular phenotype [10]. We showed previously that CYP4F3 and CPYP4F2 are up-regulated in human keratinocytes during terminal differentiation, both in vivo and in vitro [6]. Mouse epidermal keratinocytes do not differentiate in vitro in the same manner as those from humans; hence, it is not possible to conduct comparable in vitro studies in both species.
Retinoids administered topically or systemically are important in the treatment of numerous inflammatory and hyperproliferative human skin diseases [21; 22; 23; 24]. Studies of NHEK cultures showed that retinoid X nuclear receptors, which can bind 9-cis and all-trans retinoic acid, mediate the induction of CYP4F enzymes by submicromolar levels of natural and synthetic retinoid ligands [6]. Retinoid X nuclear receptors can heterodimerize with retinoic acid receptors and several other nuclear receptors, and the partner(s) mediating retinoid-inducible CYP4F gene expression in human skin is unknown. Endogenous retinoids are important in normal skin structure and function [13], but we lack capacity to reliably measure and characterize endogenous (uninduced) CYP-dependent LTB4 metabolism due to the very low specific activities in situ. This is typical of many extrahepatic CYP proteins.
In this first report of cutaneous Cyp4f genes we provide compelling evidence that 20-hydroxylation is the main CYP-dependent LTB4 inactivation pathway; however, we cannot yet conclude which CYP4F enzyme(s) generate 20-OH and 20-COOH LTB4 in mouse skin. The mouse Cyp4f gene family is significantly expanded (vs. humans), rendering orthologous assignments tenuous [19]. Thus far only two recombinant mouse proteins have been characterized partially. CYP4F18, the main LTB4 hydroxylase in mouse polymorphonuclear leukocytes, generates mainly 19-OH LTB4, and to a lesser extent 18-OH LTB4 [18]. CYP4F14 catalyzes fatty acid hydroxylation (LTB4, 6-trans LTB4, hydroxy- and hydroperoxy- eicosatetraenoic acids, lipoxin A4, and prostaglandin A1). It showed >20-fold greater active towards 8-OH eicosatetraenoic acid (vs. LTB4) [4]. More work is needed to express and characterize individual proteins arising from the mouse Cyp4f genes and to generate antibodies interacting specifically with the individual gene products.
Leukotriene hydroxylation is perhaps the best-studied example of metabolic inactivation of pro-inflammatory lipids as an innate mechanism to resolve tissue inflammation. This pathway has been studied mainly in blood derived cells in which enzymatic pathways responsible for LTB4 synthesis and catabolism are highly expressed and first characterized. Our results support the novel concept that the epithelial component of skin (and possibly that in other extrahepatic tissues) also expresses CYP4F proteins and possesses innate capacity to inactivate pro-inflammatory lipids like LTB4 arising from neutrophils and other blood derived cells that infiltrate epithelia at sites of tissue injury.
Increased tissue LTB4 levels are considered an important indicator of diseased skin types and a critical mediator, at least in part, of the pathogenesis and pathophysiology of skin diseases. Measurements are reported in the ng/ml or nM concentration range, consistent with a Km value of <1 μM for recombinant human CYP4F3A towards LTB4 [15]. Examples include psoriasis (3–33 ng/g wet tissue [25] and 11 pg/mg protein in blister fluids [26]), eczema (27 ng/mg tissue protein [27] and 21 pg/mg protein in blister fluids, [26]), and atopic dermatitis (5 ng/g tissue [28] and 1 ng/ml (or 3 nM) in blister fluid [29]). In healthy atopic patients, LTB4 levels in skin chamber fluids ranged 1–12 ng/ml (or 4–36 nM) at 3–5 h after antigen challenge, 38% to 80% of which were ω-oxidation products [30]. Based on the sampling methods utilized, it is reasonable that LTB4 concentrations would approach the Km of a CYP4F LTB4 hydroxylase at localized microscopic foci of neutrophil-epithelial interactions within injured, inflamed, or lesional skin. Thus, existing biochemical data establish feasibility and support the novel concept that CYP4F-expressing epithelial cells have active roles in modulating tissue levels of pro-inflammatory lipids like LTB4 and participate in the cellular mechanisms resolving epithelial inflammation.
Skin keratinocytes provide a facile and physiologically relevant model to elucidate biochemical mechanisms resolving tissue inflammation and roles for CYP4F enzymes. Skin is a major target affected in adverse drug reactions, chronic inflammatory diseases and cancers in humans and also in mice having targeted gene disruptions resulting in spontaneous inflammation [31; 32]. In addition, other tissues such as lung and brain exhibit transient down- and then up-regulation of specific CYP4Fs, changes that track temporally with injury- or allergen-induced acute inflammation and subsequent resolution [33; 34; 35]. In response to traumatic brain injury, CYP4F expression was initially suppressed within 24 h and then induced over 2 week to levels exceeding control levels. Changes in the levels of specific CYP4F transcripts in specific rat brain regions correlated inversely with tissue LTB4 levels—an indicator of inflammation--and increased CYP4F protein levels localized to injury sites [35]. Acute inflammation is a critical defense mechanism, but if not resolved sequelae due to chronic inflammation can be more injurious than the initial insult. We propose that induction of CYP4F enzymes may be a general mechanism how diverse epithelia influence the course of inflammation-driven disease and recovery from environmentally induced cell damage [3; 6; 33; 35; 36; 37; 38; 39]. Induction of these enzymes, for example by retinoids, may be exploited clinically to promote healing of inflamed, injured epithelial tissues.
Acknowledgments
The authors appreciate contributions of Patricia Ladd. This work was supported in part by PHS grants AR47357 and AR45603 (D.S.K.), GM15431, ES13125, CA77839 and DK48831 (J.D.M), NS44174 (H.W.S.), ES00267 (cell culture, mass spectrometry shared resources); and resources in the Department of Veterans Affairs; Center for Human Genetics Research at Vanderbilt; Vanderbilt Ingram Cancer Center Tissue Acquisition & Pathology Shared Resource and the Cooperative Human Tissue Network.
Abbreviations
- CID
collision-induced dissociation
- HPLC
high performance liquid chromatography
- LC/MS
liquid chromatography/mass spectrometry
- LTB4
leukotriene B4
- NHEK
normal human epidermal keratinocyte
- qPCR
quantitative, real-time polymerase chain reaction
- SRM
selective reaction monitoring
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Murphy RC, Gijon MA. Biosynthesis and metabolism of leukotrienes. Biochem J. 2007;405:379–95. doi: 10.1042/BJ20070289. [DOI] [PubMed] [Google Scholar]
- 2.Peters-Golden M, Canetti C, Mancuso P, Coffey MJ. Leukotrienes: underappreciated mediators of innate immune responses. J Immunol. 2005;174:589–94. doi: 10.4049/jimmunol.174.2.589. [DOI] [PubMed] [Google Scholar]
- 3.Kalsotra A, Strobel HW. Cytochrome P450 4F subfamily: At the crossroads of eicosanoid and drug metabolism. Pharmacology and Therapeutics. 2006;112:589–611. doi: 10.1016/j.pharmthera.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 4.Kikuta Y, Kusunose E, Kusunose M. Prostaglandin and leukotriene omega-hydroxylases. Prostaglandins Other Lipid Mediat. 2002:68–69. 345–62. doi: 10.1016/s0090-6980(02)00039-4. [DOI] [PubMed] [Google Scholar]
- 5.Mukhtar H, Bik DP, Ruzicka T, Merk HF, Bickers DR. Cytochrome P-450-dependent omega-oxidation of leukotriene B4 in rodent and human epidermis. J Invest Dermatol. 1989;93:231–5. doi: 10.1111/1523-1747.ep12277578. [DOI] [PubMed] [Google Scholar]
- 6.Kalsotra A, Du L, Wang Y, Ladd PA, Kikuta Y, Duvic M, Boyd AS, Keeney DS, Strobel HW. Inflammation resolved by retinoid X receptor-mediated inactivation of leukotriene signaling pathways. FASEB J. 2008;22:538–47. doi: 10.1096/fj.07-9244com. [DOI] [PubMed] [Google Scholar]
- 7.Stark K, Schauer L, Sahlen GE, Ronquist G, Oliw EH. Expression of CYP4F12 in gastrointestinal and urogenital epithelia. Basic Clin Pharmacol Toxicol. 2004;94:177–83. doi: 10.1111/j.1742-7843.2004.pto940404.x. [DOI] [PubMed] [Google Scholar]
- 8.Stark K, Torma H, Oliw EH. Co-localization of COX-2, CYP4F8, and mPGES-1 in epidermis with prominent expression of CYP4F8 mRNA in psoriatic lesions. Prostaglandins Other Lipid Mediat. 2006;79:114–25. doi: 10.1016/j.prostaglandins.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 9.Lefevre C, Bouadjar B, Ferrand V, Tadini G, Megarbane A, Lathrop M, Prud’homme JF, Fischer J. Mutations in a new cytochrome P450 gene in lamellar ichthyosis type 3. Hum Mol Genet. 2006;15:767–76. doi: 10.1093/hmg/ddi491. [DOI] [PubMed] [Google Scholar]
- 10.Du L, Neis MM, Ladd PA, Lanza DL, Yost GS, Keeney DS. Effects of the differentiated keratinocyte phenotype on expression levels of CYP1–4 family genes in human skin cells. Toxicol Appl Pharmacol. 2006;213:135–44. doi: 10.1016/j.taap.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 11.Du L, Yermalitsky V, Hachey DL, Jagadeesh SG, Falck JR, Keeney DS. A biosynthetic pathway generating 12-hydroxy-5,8,14-eicosatrienoic acid from arachidonic acid is active in mouse skin microsomes. J Pharmacol Exp Ther. 2006;316:371–9. doi: 10.1124/jpet.105.093922. [DOI] [PubMed] [Google Scholar]
- 12.Didierjean L, Carraux P, Grand D, Sass JO, Nau H, Saurat JH. Topical retinaldehyde increases skin content of retinoic acid and exerts biologic activity in mouse skin. J Invest Dermatol. 1996;107:714–9. doi: 10.1111/1523-1747.ep12365603. [DOI] [PubMed] [Google Scholar]
- 13.Eichner R, Gendimenico GJ, Kahn M, Mallon JP, Capetola RJ, Mezick JA. Effects of long-term retinoic acid treatment on epidermal differentiation in vivo: specific modifications in the programme of terminal differentiation. Br J Dermatol. 1996;135:687–95. [PubMed] [Google Scholar]
- 14.Bylund J, Harder AG, Maier KG, Roman RJ, Harder DR. Leukotriene B4 omega-side chain hydroxylation by CYP4F5 and CYP4F6. Arch Biochem Biophys. 2003;412:34–41. doi: 10.1016/s0003-9861(03)00030-4. [DOI] [PubMed] [Google Scholar]
- 15.Christmas P, Jones JP, Patten CJ, Rock DA, Zheng Y, Cheng SM, Weber BM, Carlesso N, Scadden DT, Rettie AE, Soberman RJ. Alternative splicing determines the function of CYP4F3 by switching substrate specificity. J Biol Chem. 2001;276:38166–72. doi: 10.1074/jbc.M104818200. [DOI] [PubMed] [Google Scholar]
- 16.Kikuta Y, Mizomoto J, Strobel HW, Ohkawa H. Expression and physiological function of CYP4F subfamily in human eosinophils. Biochim Biophys Acta. 2007;1771:1439–45. doi: 10.1016/j.bbalip.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 17.Kikuta Y, Yamashita Y, Kashiwagi S, Tani K, Okada K, Nakata K. Expression and induction of CYP4F subfamily in human leukocytes and HL60 cells. Biochim Biophys Acta. 2004;1683:7–15. doi: 10.1016/j.bbalip.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 18.Christmas P, Tolentino K, Primo V, Zemski Berry K, Murphy RC, Chen M, Lee DM, Soberman RJ. CYP4F18 is the LTB4 omega -1/omega -2 hydroxylase in mouse polymorphonuclear leukocytes: Identification as the functional orthologue of human PMN CYP4F3A in the down-regulation of responses to LTB4. J Biol Chem. 2006;281:7189–96. doi: 10.1074/jbc.M513101200. [DOI] [PubMed] [Google Scholar]
- 19.Nelson DR, Zeldin DC, Hoffman SM, Maltais LJ, Wain HM, Nebert DW. Comparison of cytochrome P450 (CYP) genes from the mouse and human genomes, including nomenclature recommendations for genes, pseudogenes and alternative-splice variants. Pharmacogenetics. 2004;14:1–18. doi: 10.1097/00008571-200401000-00001. [DOI] [PubMed] [Google Scholar]
- 20.Wheelan P, Zirrolli A, Murphy RC. Negative ion electrospray tandem mass spectrometric structral characterization of leukotriene B4 (LTB4) and LTB4-derived metabolites. J Am Soc Mass Spectrom. 1996;7:129–139. doi: 10.1016/1044-0305(95)00629-X. [DOI] [PubMed] [Google Scholar]
- 21.Thacher SM, Vasudevan J, Chandraratna RA. Therapeutic applications for ligands of retinoid receptors. Curr Pharm Des. 2000;6:25–58. doi: 10.2174/1381612003401415. [DOI] [PubMed] [Google Scholar]
- 22.Winterfield L, Cather J, Cather J, Menter A. Changing paradigms in dermatology: nuclear hormone receptors. Clin Dermatol. 2003;21:447–54. doi: 10.1016/j.clindermatol.2003.08.011. [DOI] [PubMed] [Google Scholar]
- 23.Zouboulis CC. Retinoids--which dermatological indications will benefit in the near future? Skin Pharmacol Appl Skin Physiol. 2001;14:303–15. doi: 10.1159/000056361. [DOI] [PubMed] [Google Scholar]
- 24.Desai A, Kartono F, Del Rosso JQ. Systemic retinoid therapy: a status report on optimal use and safety of long-term therapy. Dermatol Clin. 2007;25:185–93. doi: 10.1016/j.det.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 25.Fogh K, Herlin T, Kragballe K. Eicosanoids in acute and chronic psoriatic lesions: leukotriene B4, but not 12-hydroxy-eicosatetraenoic acid, is present in biologically active amounts in acute guttate lesions. J Invest Dermatol. 1989;92:837–41. doi: 10.1111/1523-1747.ep12696858. [DOI] [PubMed] [Google Scholar]
- 26.Reilly DM, Parslew R, Sharpe GR, Powell S, Green MR. Inflammatory mediators in normal, sensitive and diseased skin types. Acta Derm Venereol. 2000;80:171–4. doi: 10.1080/000155500750042907. [DOI] [PubMed] [Google Scholar]
- 27.Hua Z, Fei H, Mingming X. Evaluation and interference of serum and skin lesion levels of leukotrienes in patients with eczema. Prostaglandins Leukot Essent Fatty Acids. 2006;75:51–5. doi: 10.1016/j.plefa.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 28.Fogh K, Herlin T, Kragballe K. Eicosanoids in skin of patients with atopic dermatitis: prostaglandin E2 and leukotriene B4 are present in biologically active concentrations. J Allergy Clin Immunol. 1989;83:450–5. doi: 10.1016/0091-6749(89)90132-2. [DOI] [PubMed] [Google Scholar]
- 29.Thorsen S, Fogh K, Broby-Johansen U, Sondergaard J. Leukotriene B4 in atopic dermatitis: increased skin levels and altered sensitivity of peripheral blood T-cells. Allergy. 1990;45:457–63. doi: 10.1111/j.1398-9995.1990.tb01097.x. [DOI] [PubMed] [Google Scholar]
- 30.Shalit M, Valone FH, Atkins PC, Ratnoff WD, Goetzl EJ, Zweiman B. Late appearance of phospholipid platelet-activating factor and leukotriene B4 in human skin after repeated antigen challenge. J Allergy Clin Immunol. 1989;83:691–6. doi: 10.1016/0091-6749(89)90084-5. [DOI] [PubMed] [Google Scholar]
- 31.Nathan C. Points of control in inflammation. Nature. 2002;420:846–52. doi: 10.1038/nature01320. [DOI] [PubMed] [Google Scholar]
- 32.Roujeau JC. Clinical heterogeneity of drug hypersensitivity. Toxicology. 2005;209:123–9. doi: 10.1016/j.tox.2004.12.022. [DOI] [PubMed] [Google Scholar]
- 33.Kalsotra A, Zhao J, Anakk S, Dash PK, Strobel HW. Brain trauma leads to enhanced lung inflammation and injury: evidence for role of P4504Fs in resolution. J Cereb Blood Flow Metab. 2007;27:963–74. doi: 10.1038/sj.jcbfm.9600396. [DOI] [PubMed] [Google Scholar]
- 34.Stoilov I, Krueger W, Mankowski D, Guernsey L, Kaur A, Glynn J, Thrall RS. The cytochromes P450 (CYP) response to allergic inflammation of the lung. Arch Biochem Biophys. 2006;456:30–8. doi: 10.1016/j.abb.2006.09.029. [DOI] [PubMed] [Google Scholar]
- 35.Wang Y, Zhao J, Kalsotra A, Turman CM, Grill RJ, Dash PK, Strobel HW. CYP4Fs expression in rat brain correlates with changes in LTB4 levels after traumatic brain injury. J Neurotrauma. 2008;25:1187–94. doi: 10.1089/neu.2008.0542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Curley CR, Monsuur AJ, Wapenaar MC, Rioux JD, Wijmenga C. A functional candidate screen for coeliac disease genes. Eur J Hum Genet. 2006;14:1215–22. doi: 10.1038/sj.ejhg.5201687. [DOI] [PubMed] [Google Scholar]
- 37.Kalsotra A, Anakk S, Brommer CL, Kikuta Y, Morgan ET, Strobel HW. Catalytic characterization and cytokine mediated regulation of cytochrome P450 4Fs in rat hepatocytes. Arch Biochem Biophys. 2007;461:104–12. doi: 10.1016/j.abb.2007.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kalsotra A, Cui X, Antonovic L, Robida AM, Morgan ET, Strobel HW. Inflammatory prompts produce isoform-specific changes in the expression of leukotriene B(4) omega-hydroxylases in rat liver and kidney. FEBS Lett. 2003;555:236–42. doi: 10.1016/s0014-5793(03)01240-7. [DOI] [PubMed] [Google Scholar]
- 39.Chaluvadi MR, Kinloch RA, Nyagode BA, Richardson TA, Raynor MJ, Sherman M, Antonovic L, Strobel HW, Dillehay DL, Morgan E. Regulation of Hepatic Cytochrome P450 Expression in Mice with Intestinal or Systemic Infections of Citrobacter rodentium. Drug Metab Dispos. 2008 Oct 29; doi: 10.1124/dmd.108.024240. ePub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]