Abstract
The vacuole has crucial roles in stress resistance and adaptation of the fungal cell. Furthermore, in Candida albicans it has been observed to undergo dramatic expansion during the initiation of hyphal growth, to produce highly “vacuolated” subapical compartments. We hypothesized that these functions may be crucial for survival within the host and tissue-invasive hyphal growth. We also considered the role of the late endosome or prevacuole compartment (PVC), a distinct organelle involved in vacuolar and endocytic trafficking. We identified two Rab GTPases, encoded by VPS21 and YPT72, required for trafficking through the PVC and vacuole biogenesis, respectively. Deletion of VPS21 or YPT72 led to mild sensitivities to some cellular stresses. However, deletion of both genes resulted in a synthetic phenotype with severe sensitivity to cellular stress and impaired growth. Both the vps21Δ and ypt72Δ mutants had defects in filamentous growth, while the double mutant was completely deficient in polarized growth. The defects in hyphal growth were not suppressed by an “active” RIM101 allele or loss of the hyphal repressor encoded by TUP1. In addition, both single mutants had significant attenuation in a mouse model of hematogenously disseminated candidiasis, while the double mutant was rapidly cleared. Histological examination confirmed that the vps21Δ and ypt72Δ mutants are deficient in hyphal growth in vivo. We suggest that the PVC and vacuole are required on two levels during C. albicans infection: (i) stress resistance functions required for survival within tissue and (ii) a role in filamentous growth which may aid host tissue invasion.
Candida albicans is a commensal organism that resides upon mucosal surfaces of many healthy individuals, including the oral cavity and gastrointestinal and vaginal tracts. Under conditions where host immunity is compromised, C. albicans can invade the underlying tissues to initiate infection. There are two main categories of candidiasis, infections of mucosal membranes and serious systemic disease involving dissemination and invasion of deeper organs. Mucosal infections include oropharyngeal candidiasis and esophageal candidiasis, which are signature markers of human immunodeficiency virus infection (19), as well as vaginitis (61). Systemic candidiasis is often fatal, with mortality rates of greater than 30% (23, 65, 68).
The ability of C. albicans to cause disease is intimately associated with its capacity to switch among yeast, pseudohyphal, and true hyphal forms (42, 57). It has been proposed that yeast cells are suited for dissemination between host tissues or through the bloodstream, while the polarized hyphal form facilitates tissue penetration and damage (57). Transition to the hyphal mode of growth involves the induction of a hypha-specific gene transcription program, the establishment of a polarized cytoskeleton, and localized secretion to the extending hyphal apex. This necessitates the transport of secretory vesicles to the hyphal apex, followed by exocytosis to facilitate localized cell expansion. However, recent findings with several filamentous fungi have established that there is a countermovement of membrane material through trafficking from the hyphal apex to the distal regions of the cell and that this is also required to maintain polarized hyphal growth (59, 62). Furthermore, in C. albicans, germ tube emergence is accompanied by a dramatic increase in vacuolar volume in the subapical regions (3, 29, 30). As the germ tube extends, “empty”-looking subapical compartments can be observed that are composed almost entirely of vacuole, as the protoplasm migrates in the hyphal tip compartment. At present, the mechanism underlying hyphal cell vacuolation, its regulation, and its significance with respect to virulence remain obscure. Furthermore, the importance of other vacuolar functions during infection is unknown. However, a similar phenomenon has been observed in the plant pathogen Ustilago maydis (41, 63). Formation of invasive dikaryotic hyphae occurs on the host leaf surface and, as in C. albicans, involves migration of the cytoplasm with the apical cell compartment and the formation of “empty” subapical cells. The similarity of these events in C. albicans and U. maydis suggests that hyphal cell vacuolation may be an important part of a “host invasion strategy.”
The fungal vacuole is a highly dynamic organelle, with established functions in stress resistance and cellular homeostasis (33). At least four pathways deliver material to the vacuole through vesicle-mediated transport (16). This includes transport from the cell surface via endocytosis, the Golgi apparatus via CPY (carboxypeptidase Y) and alkaline phosphatase trafficking pathways, and the cytoplasmic degradation pathway known as autophagy (13). Material delivered to the vacuole through endocytosis and the CPY pathway first transits through an intermediate late endosomal compartment known as the prevacuolar compartment (PVC). The dramatic increase in vacuolar volume that occurs during C. albicans germ tube emergence necessitates the incorporation of new membrane into the vacuole. However, the trafficking events which facilitate hyphal cell vacuolation remain to be established.
Rab GTPases are a conserved family of small GTPases that are key regulators of membrane trafficking and organelle biogenesis in eukaryotes (18). Eukaryotes encode multiple members of the Rab GTPase family, each localizing to a distinct cell compartment and functioning at a specific transport step (54). We have identified two Rab GTPases which function in C. albicans PVC and vacuolar biogenesis. Functional analysis has demonstrated that these Rab GTPases are required for C. albicans filamentous growth and virulence.
MATERIALS AND METHODS
Growth conditions.
Strains were routinely grown at 30°C on YPD (1% yeast extract, 2% Bacto peptone, 2% dextrose) supplemented with uridine (50 μg ml−1) when necessary (31). For growth curves, overnight cultures were subcultured to 20 ml fresh YPD medium to an optical density at 600 nm (OD600) of 0.2 and incubated at 30°C with shaking. The OD600 was determined from samples taken hourly. Transformants were selected on minimal medium (YNB; 6.75 g liter−1 yeast nitrogen base plus ammonium sulfate and without amino acids, 2% dextrose, 2% Bacto agar) supplemented with the appropriate auxotrophic requirements, as described for Saccharomyces cerevisiae (14), except for uridine, which was added at 50 μg ml−1.
Stress phenotypes.
For agar plate growth assays, C. albicans was grown overnight in YPD at 30°C, cells were washed in sterile distilled water, the cell density was adjusted to 107/ml, and serial 1:5 dilutions were performed in a 96-well plate. Cells were then applied to agar with a sterile multipronged applicator. Resistance to temperature stress was determined on YPD agar at 37 and 42°C, and resistance to osmotic stress was determined on YPD agar plus 2.5 M glycerol or 1.5 M NaCl. Other stresses included YPD plus 5 mM caffeine, 10 nM rapamycin, or 5 mM sodium orthovanadate. Carbon source utilization was tested with 3% ethanol, glycerol, or potassium acetate instead of glucose. Secreted protease activity was examined on bovine serum albumin (BSA) plus YE agar at 37°C (17). Resistance to nitrogen starvation was determined as follows. Each strain was grown in YNB broth for 48 h at 30°C and washed twice in SD-N (0.17% yeast nitrogen base without ammonium sulfate or amino acids, 2% glucose), and 107 cells were resuspended in 2 ml SD-N (46). Viability was determined at time intervals as the number of CFU on YPD agar. Sensitivity to H2O2 was determined by measuring growth in YPD medium supplemented with 0 to 12 mM H2O2.
Morphogenesis assays.
Filamentous growth on M199 (pH 7.5) and 10% fetal bovine serum (FBS) agar was performed as previously described (51). Cells from overnight cultures were washed twice in distilled water and induced to filament in 10% or 1% FBS (in distilled H2O) at 37°C after inoculation at 106 cells ml−1. Samples were examined microscopically at intervals and scored for the presence of germ tubes. Statistical significance was determined with the unpaired Student t test. Filamentous growth was also stimulated by embedding C. albicans cells within a YPS agar matrix or sandwiching cells between two layers of YPS agar (11). Chlamydospores were induced on cornstarch-Tween agar (49).
Fluorescence microscopy and localization.
Vacuole and endosome morphology was visualized with the lipophilic fluorescent dye FM4-64 (64). Cells grown overnight were subcultured 1:100 in fresh YPD and grown for 4 h at 30°C with shaking. Approximately 2 × 107 cells were pelleted, resuspended in 50 μl of YPD plus 1 μl of a 100 μM stock of FM4-64 (in dimethyl sulfoxide), and labeled for 20 min. Cells were then “chased” in 1 ml fresh YPD for a further 20 to 120 min. Cells were then harvested, washed twice in distilled water, applied to a poly-l-lysine-coated slide, and observed with an epifluorescence microscope with a tetramethyl rhodamine isocyanate (TRITC) filter set. For colocalization of green fluorescent protein (GFP) fusion proteins, a fluorescein isothiocyanate filter set was also used.
Sequence analysis.
Protein sequences were aligned by using the EMBOSS pairwise align algorithm (http://www.ebi.ac.uk/emboss/).
Strain construction.
The strains used in this study are described in Table 1. Gene deletion strains were constructed by the PCR-based approach described by Wilson et al. with ura3− his1− arg4− mutant strain BWP17 (70), kindly provided by A. Mitchell (Carnegie Mellon University). C. albicans was transformed by the lithium acetate procedure (26). VPS21 and YPT72 deletion cassettes were amplified by PCR with pARGΔSpe, pGEMHIS1, or pDDB57 (which has a recyclable URA3-dpl200 marker) (69, 70) as the template with primers VPS21DISF and VPS21DISR or YPT72DISF and YPT72DISR (Table 2), respectively. Sequential deletion of each allele was achieved for either gene by using ARG4 and HIS1 to generate ura3− vps21Δ/Δ and ypt72Δ/Δ gene deletion strains. A vps21Δ/Δ ypt72Δ/Δ double mutant was made by the use of the recyclable URA3 deletion cassette. The regeneration of ura3− recombinants was selected on YNB medium supplemented with uridine and 1 μg/ml 5-fluoroorotic acid (8). Correct integration of either cassette was confirmed at each step by diagnostic PCR with primer ARG4DET2 and either primer VPS21AMPR or YPT72AMPR2 (ARG4 integration), primer HIS1F1268 and either primer VPS21AMPF or YPT72AMPF3 (HIS1 integration), or primer URA3-5 and either primer VPS21AMPF or YPT72AMPF3 (URA3 integration) (Table 2). Southern blot analysis was also performed with specific probes to the 5′ untranslated region (UTR) of either gene, amplified with primers VPS21AMPF and VPS21PBR or YPT72AMPF3 and YPT72PBR. Correct gene deletion resulted in replacement of the entire 651-bp VPS21 open reading frame (ORF) or the entire 651-bp YPT72 ORF. Finally, a wild-type copy of VPS21 or YPT72, including the 5′ and 3′ flanking sequences, was introduced into the deletion strains on pLUXVPS21 or pLUXYPT72 to produce prototrophic “reconstituted” strains. Prototrophic gene deletion strains were produced by transforming the mutant strains with plasmid vector pLUX alone. Either plasmid was digested with NheI prior to transformation to target integration into (and reconstitute) the URA3 loci. Strains expressing GFP-VPS21 and GFP-YPT72 were produced by linearizing the pGFPVPS21 and pGFPYPT72 constructs with NheI and transforming them into the vps21Δ/Δ and ypt72Δ/Δ mutant strains, respectively. The presence of the GFP fusion was confirmed by PCR analysis with primer GFPSEQF and either primer VPS21SEQR2 or YPT72SEQR. The hyphal repressor TUP1 was deleted by a strategy identical to that outlined above, with the use of a recyclable tup1Δ:URA3dpl200 deletion cassette amplified with primers TUP1DISF and TUP1DISR and 5-fluoroorotic acid counterselection. Correct integration and URA3 excision were confirmed by diagnostic PCR with primers TUP1DETF and TUP1DETR and primers TUP1DETF and URA3-5. To produce strains TR1/3 and DTR1-3 with a single copy of TUP1 under the control of the MET3 promoter, plasmid pREGTUP1 was linearized within the 3′-truncated TUP1 ORF with AvaI to target integration into the remaining wild-type copy of a TUP1/tup1Δ heterozygous strain. Correct integration was confirmed by diagnostic PCR with primers MET3DETF and TUP1DETR2 to confirm the presence of a MET3-TUP1 fusion, and the absence of endogenous TUP1 was confirmed with TUP1DETF and TUP1DETR or TUP1DETR2. Plasmids harboring the wild-type (pEM-31) or “active” (pEM16-1 and pEM16-3) RIM101 allele (20) were linearized with HpaI to target integration into the RIM101 locus and transformed into ura3− vps21Δ/Δ and ypt72Δ/Δ mutant strains or CAI4 (21), and Ura+ transformants were selected.
TABLE 1.
C. albicans strains used in this study
| Strain | Relevant genotype | Source |
|---|---|---|
| SC5314 | VPS21/VPS21 YPT72/YPT72 | 27 |
| YJB6284 | VPS21/VPS21 YPT72/YPT72 | 6 |
| BWP17 | VPS21/VPS21 YPT72/YPT72 ura3Δ/Δ his1Δ/Δ arg4Δ/Δ | 70 |
| CAI4 | VPS21/VPS21 YPT72/YPT72 ura3Δ/Δ | 21 |
| VD1-6 | ura3Δ/Δ his1Δ/Δ arg4Δ/Δ vps21Δ:ARG4/vps21Δ:HIS1 | This study |
| VR1-6 | ura3Δ/Δ:URA3:VPS21 his1Δ/Δ arg4Δ/Δ vps21Δ:ARG4/vps21Δ:HIS1 | This study |
| YD1-6 | ura3Δ/Δ his1Δ/Δ arg4Δ/Δ ypt72Δ:ARG4/ypt72Δ:HIS1 | This study |
| YR1-6 | ura3Δ/Δ:URA3:VPS21 his1Δ/Δ arg4Δ/Δ ypt72Δ:ARG4/ypt72Δ:HIS1 | This study |
| DDL1/10 | ura3Δ/Δ his1Δ/Δ arg4Δ/Δ vps21Δ:dpl200/vps21Δ:HIS1 ypt72Δ:URA3/ypt72Δ:ARG4 | This study |
| 21R1-3 | ura3Δ/Δ:URA3:VPS21 his1Δ/Δ arg4Δ/Δ vps21Δ:dpl200/vps21Δ:HIS1 ypt72Δ:dpl200/ypt72Δ:ARG4 | This study |
| 7R1-3 | ura3Δ/Δ:URA3:YPT72 his1Δ/Δ arg4Δ/Δ vps21Δ:dpl200/vps21Δ:HIS1 ypt72Δ:dpl200/ypt72Δ:ARG4 | This study |
| VG1/32 | ura3Δ/Δ:URA3:GFP-VPS21 his1Δ/Δ arg4Δ/Δ vps21Δ:ARG4/vps21Δ:HIS1 | This study |
| YG1-3 | ura3Δ/Δ:URA3:GFP-YPT72 his1Δ/Δ arg4Δ/Δ ypt72Δ:ARG4/ypt72Δ:HIS1 | This study |
| VTD1-3 | ura3Δ/Δ his1Δ/Δ arg4Δ/Δ vps21Δ:dpl200/vps21Δ:HIS1 tup1Δ:URA3/tup1Δ:ARG4 | This study |
| YTD1-3 | ura3Δ/Δ his1Δ/Δ arg4Δ/Δ ypt72Δ:dpl200/ypt72Δ:HIS1 tup1Δ:URA3/tup1Δ:ARG4 | This study |
| TH1A2 | ura3Δ/Δ his1Δ/Δ arg4Δ/Δ tup1Δ:HIS1/tup1Δ:ARG4 | This study |
| TR1/3 | ura3Δ/Δ his1Δ/Δ arg4Δ/Δ tup1Δ:HIS1/tup1Δ:URA3:MET3-TUP1 | This study |
| DTR1-3 | ura3Δ/Δ his1Δ/Δ arg4Δ/Δ vps21Δ:dpl200/vps21Δ:HIS1 ypt72Δ:dpl200/ypt72Δ:ARG4 tup1Δ:dpl200/tup1Δ:URA3:MET3-TUP1 | This study |
TABLE 2.
Oligonucleotides used in this study
| Primer | Sequence 5′→3′a |
|---|---|
| VPS21DISF | CCATCATTATTTCTACTATTTGTATTGCATTGTCGCATTCAGGATCAAAGAGCCAAAATACATACTAGCATGTGGAATTGTGAGCGGATA |
| VPS21DISR | ACCATGGAGATCTGTACACACTTGCCTATTTGTATATTACCTTAACTTCAGTCTCCTTGTTTATTTATATTTTCCCAGTCACGACGTT |
| VPS21AMPF | TCATCAGGATCCCCGAAGAAAAGGGGAAGGAGA |
| VPS21AMPR | TCATCAGGATCCGTAAAATTGCACGACTGTGCC |
| VPS21AMPRP | AAAACTGCAGGTAAAATTGCACGACTGTGCC |
| VPS21PBR | TTGAGTTAAAAACGCAGCTCC |
| VPS21SEQF | AATACTAGTAGTATCACCATC |
| VPS21SEQR2 | GGAGATCTGTACACACTTGCC |
| VPS21EAGMUTF | GAGCCAAAATACATACTAGCACGGCCGAGTCAACATCCAGCCCCCGC |
| VPS21EAGMUTR | GCGGGGGCTGGATGTTGACTCGGCCGTGCTAGTATGTATTTTGGCTC |
| YPT72DISF | AACGATACAGAGTTTAATTTAATTTAATTCAATTCAATTTAATTAATTAATCATATACACTTAATTTTCATGTGGAATTGTGAGCGGATA |
| YPT72DISR | AAACGAATTCGTGATATCTATTTGTTCTCTTTCTTCACCTGTGTATTTGAAATTGATTGTTATATTATTATTTCCCAGTCACGACGTT |
| YPT72AMPF3 | TCATCAGGATCCCCAAAAATATAGAAATGTTGCG |
| YPT72AMPR2 | TCATCAGGATCCATTAAAACTGGTAATGATTTGG |
| YPT72AMPRP | AAAACTGCAGATTAAAACTGGTAATGATTTGG |
| YPT72PBR | TTTACCAACACCAGAATCTCC |
| YPT72SEQF | GAAGATAATTAAAAGGCTGAC |
| YPT72SEQR | TTACTCCATCTTTGGCACTGG |
| YPT72SEQR2 | CAACAATTATAAACGAATTCG |
| YPT72XMAMUTF | TAATTAATCATATACACTTAATTTTCACCCGGGTCATCATCATCTAGAAAGAAAACA |
| YPT72XMAMUTR | TGTTTTCTTTCTAGATGATGATGACCCGGGTGAAAATTAAGTGTATATGATTAATTA |
| GFPXMAF | TCCCCCCGGGATGTCTAAAGGTGAAGAATTATTC |
| GFPXMAR | TCCCCCCGGGTTTGTACAATTCATCCATACC |
| GFPEAGF | TCACGGCCGATGTCTAAAGGTGAAGAATTATTC |
| GFPEAGR | TCACGGCCGTTTGTACAATTCATCCATACC |
| GFPSEQF | TACTTATCCACTCAATCTGCC |
| TUP1DISF | ATGTCCATGTATCCCCAACGCACCCAGCACCAACAACGTTTGACAGAGTTGTTGGATGCAATCAAAACTGTGTGGAATTGTGAGCGGATA |
| TUP1DISR | TTATTTTTTGGTCCATTTCCAAATTCTGGCTTTACAATCGCCACTACCTGTAGCGAAGATACCTTCGGTTGTTTTCCCAGTCACGACGTT |
| TUP1DETF | ATTTGAAAAAAGCGCACCCCC |
| TUP1DETF2 | GAACAAATCGCTTCCGGGTCC |
| TUP1DETR | GGTGCTGTGACAGAAGCAGAG |
| TUP1DETR2 | CCAAGAGTTTTCCGTCAGGGG |
| TUP1REGF | TCATCAGGATCCCTTCTTCCATCCCCACCAGCA |
| TUP1REGR | TCATCACTGCAGTTATACAACACATAGTATTCGAGG |
| ARG4DET2 | ATCAATTAACACAGAGATACC |
| HIS1F1268 | CCGCTACTGTCTCTACTTTG |
| URA3-5 | CCTATGAATCCACTATTGAACC |
| MET3DETF | CATCAAGTATACGTAATCTCC |
Engineered restriction enzyme sites are underlined.
Plasmid construction.
Plasmids pGEMURA3, pGEMHIS1, and pRSARGΔSpe (70) were provided by A. Mitchell (Carnegie Mellon University). Plasmid pLUX (56) was provided by W. Fonzi (Georgetown University). Plasmids pEM31, pEM16-1, and pEM16-3 (20) were provided by F. Mühlschlegel (University of Kent, United Kingdom). Plasmids pLUXVPS21 and pLUXYPT72 were made as follows. Either ORF was amplified with 5′ and 3′ UTR sequences from SC5314 genomic DNA with HiFi Platinum Taq (Invitrogen) and primer set VPS21AMPF and VPS21AMPR or YPT72AMPF3 and YPT72AMPR2 (Table 2), each of which has a BamHI site engineered at its 5′ end. Purified products were then cloned into the BamHI site of vector pLUX. Plasmid pGFPVPS21 was derived from pLUXVPS21 through the introduction of an EagI site in place of the ATG start codon of VPS21 with primers VPS21EAGMUTF and VPS21EAGMUTR and the QuikChange site-directed mutagenesis kit (Stratagene). C. albicans-optimized GFP was then amplified from pGFPURA3 (24) with primers GFPEAGF and GFPEAGR and cloned into the EagI site of the modified pLUXVPS21 plasmid. pGFPYPT72 was produced by a similar strategy, except that an XmaI site was introduced with primers YPT72XMAMUTF and YPT72XMAMUTR and GFP was amplified with GFPXMAF and GFPXMAR with subsequent XmaI cloning. All constructs were sequenced to confirm the correct introduction of the restriction sites, the correct orientation and in-frame cloning of the insert, and the absence of unintended mutations. pMET3-TUP1 was produced by amplifying the first 548 bp of the TUP1 ORF with the 21-nucleotide 5′ UTR with primers TUP1REGF and TUP1REGR and cloning the product between the BamHI and PstI sites in pCaDIS (15).
Mouse model of hematogenously disseminated candidiasis.
All procedures were approved by the Institutional Animal Care and Use Committee of the Louisiana State University Health Sciences Center, New Orleans, and complied with all relevant federal guidelines. C. albicans was grown overnight in YPD cultures at 30°C (200 rpm). Cells were washed twice in sterile phosphate-buffered saline (PBS), and cell density was determined with a hemocytometer. Each strain was then diluted to 5 × 106 cells/ml in sterile PBS. Viable cell counts were confirmed by plating appropriate dilutions of each inoculum onto YPD agar plates and counting the colonies that formed after 48 h. A 0.1-ml volume of each cell suspension was then inoculated into the lateral tail veins of BALB/c mice (Charles River Laboratories, Frederick, MD). Mice were monitored for 28 days, and those showing distress were sacrificed. Kaplan-Meier survival curves were plotted, and statistical significance was determined with the log rank test. Forty-eight-hour colonization experiments were performed as described above, but all of the mice were sacrificed at 48 h postinfection. Kidneys were collected at the time of sacrifice and cut longitudinally. The fungal burden was determined for one half of each organ as the number of CFU per gram of tissue. The two half kidneys from each mouse were weighed and homogenized in PBS, and serial dilutions of homogenate were plated onto YPD agar. The number of colonies on each plate was determined after 48 h at 30°C, and the number of CFU per gram of tissue was calculated. Aliquots of homogenate were also mixed with an equal volume of 60% KOH, incubated for 2 h at 30°C, and washed in sterile water, and fungal morphology was scored by using the previously described morphology index (MI) (43, 47) as follows: MI of 1, spherical and nearly spherical cells; MI of 2, ovoid cells with a length of up to twice the cell width; MI of 3, pseudohyphal cells with obvious constrictions; MI of 4, parallel-sided true hyphal cells. The other half of each kidney was fixed in 10% formalin for histological analysis. This consisted of paraffin embedment, sectioning, and staining with either hematoxylin and eosin or Grocott methenamine silver (GMS) (55). Stained sections were examined microscopically for (i) Candida infiltration and morphology, (ii) alteration of normal tissue structure, and (iii) immune cell infiltration.
RESULTS
Identification of C. albicans Vps21p and Ypt72p Rab GTPases.
In S. cerevisiae, VPS21 encodes a Rab GTPase which localizes to the PVC and is required for endosomal trafficking (25), while YPT7 encodes a Rab GTPase required for vacuole biogenesis (58, 67). In order to conduct a functional analysis of the PVC and vacuolar compartment, we identified C. albicans VPS21 and YPT7 homologs through BLASTP searches of the Candida genome database (www.candidagenome.org/). In each case, one clear homolog was identified. The predicted gene product of C. albicans VPS21 is 216 amino acids and is 66.8% identical to ScVps21p. The C. albicans YPT7-like ORF encodes a 217-amino-acid protein which is 61.2% identical to S. cerevisiae Ypt7p. Interestingly, the Candida genome database has named a different ORF YPT7. This ORF encodes a larger Rab GTPase-like protein of 288 amino acids that is 30.4% identical to ScYpt7p. We therefore named the YPT7-like ORF that we identified YPT72. As the VPS21- and YPT72-encoded products were the most like S. cerevisiae PVC and vacuolar Rab proteins, we focused on these genes for further investigation.
Sequence analysis of PCR-amplified YPT72 revealed the presence of three polymorphisms within the ORF, none of which altered the protein coding sequence. No polymorphism was detected within the VPS21 ORF. Vps21p (XXCC) and Ypt72p (XXCXC) each have two C-terminal cysteine residues which conform to the dual geranylgeranylation consensus sequences characteristic of the Rab family of GTPases (52). Guanine base binding (G1-3) and phosphate binding (PM1-3) motifs were highly conserved in both protein sequences, strongly supporting the notion that they are GTPases. Furthermore, the presence of five short diagnostic sequences (RabF1 to -5) (52) supports the idea that both of these proteins belong to the Rab family of GTPases.
The two alleles of either gene were sequentially deleted by a PCR-based approach in the BWP17 strain background with combinations of the ARG4, HIS1, and recyclable URA3 selectable markers. In addition, we constructed a double-mutant strain with both VPS21 and YPT72 deleted. Reconstituted strains were made by reintroducing wild-type alleles of VPS21 and YPT72 into the respective mutant strains on the vector pLUX, which integrates at and reconstitutes the URA3 locus, thus circumventing the well-described positional effects of URA3 integration (39). All of the mutant phenotypes described in the following sections were complemented by reintroduction of the corresponding wild-type allele. Furthermore, phenotypic analysis was conducted on multiple independently constructed isolates of each genotype.
The vps21Δypt72Δ mutant exhibits a synthetic growth defect.
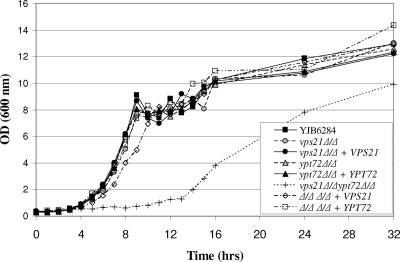
In order to determine if the mutant strains were affected in growth, we assessed propagation in YPD medium at 30°C. Both the vps21Δ and ypt72Δ mutants grew at the same rate as VPS21/VPS21 YPT72/YPT72 control strain YJB6284 (Fig. 1). Strikingly, the vps21Δypt72Δ double-deletion strain had a prolonged lag phase and a drastically reduced growth rate in liquid culture, indicating a synthetic growth defect. However, each mutant was able to utilize a range of carbon (including glucose, glycerol, ethanol, and acetate) and nitrogen (including ammonium sulfate, glutamine, and BSA) sources for growth.
FIG. 1.
The C. albicans vps21Δ and ypt72Δ mutants have a normal growth rate. Each strain was subcultured to fresh YPD medium and grown at 30°C. Samples were removed at intervals, and the OD600 was determined. Data representative of three experiments are shown.
Vps21p and Ypt72p influence stress resistance.
The fungal vacuole is known to have important functions during adaptive responses and for stress resistance (33). In order to establish the role of these GTPases in C. albicans stress resistance, we subjected the vps21Δ, ypt72Δ, and vps21Δypt72Δ mutants to a variety of stresses (Fig. 2A). Neither single mutant was sensitive to temperature stress at either 37°C or 42°C; however, while the vps21Δypt72Δ double mutant grew well at 37°C, it was unable to grow at 42°C. Similarly, both single mutants grew well under conditions of osmotic stress (1.5 M NaCl and 2.5 M glycerol), while the growth of the double mutant was severely affected. These data support the notion of a synthetic defect in the double mutant. Both single mutants exhibited sensitivity to the TOR kinase inhibitor rapamycin and, to a lesser extent, sodium orthovanadate, which interferes with phosphate metabolism; the double mutant was completely unable to grow under either of these conditions. The ypt72Δ mutant also exhibited sensitivity to caffeine, which is known to have a range of effects, including inhibition of the TOR kinase (35). Neither single mutant was sensitive to hydrogen peroxide; however, the double mutant was approximately fourfold more sensitive than YJB6284 (data not shown).
FIG. 2.
Vps21p and Ypt72p function in resistance to cellular stresses. (A) Cell suspensions of each strain were prepared by serial dilution, applied to YPD agar, and incubated at 30 or 42°C or applied to YPD agar supplemented with 2.5 M glycerol (osmotic stress), 1.5 M sodium chloride (osmotic/ionic stress), 5 mM caffeine, 10 nM rapamycin, or 5 mM sodium orthovanadate and incubated at 30°C. (B) To determine resistance to nitrogen starvation, each strain was incubated in SD-N medium. Cell viability was determined as the number of CFU from samples taken at intervals and expressed as a percentage of the number of CFU at time zero. Data shown are the means and standard deviations from three independent experiments.
The vacuole also has a major role in resisting periods of nitrogen starvation through storing metabolites including amino acids (33) and recycling cytoplasmic macromolecules through autophagic degradation (73). We therefore examined survival in medium lacking a nitrogen source. Control strains underwent two or three cell divisions in SD-N medium prior to growth arrest and maintained viability for >30 days (Fig. 2B). The vps21Δ mutant exhibited minimal sensitivity to nitrogen starvation, which only become apparent after 30 days, while the ypt72Δ mutant was much more sensitive, steadily loosing viability. The majority of double-mutant cells were nonviable by 24 h. Thus, both the vps21Δ and ypt72Δ mutants are sensitive to a subset of cellular stresses, with the double mutant exhibiting extreme sensitivity to a broad range of stresses.
C. albicans Vps21p and Ypt72p function in endosomal trafficking and biogenesis.
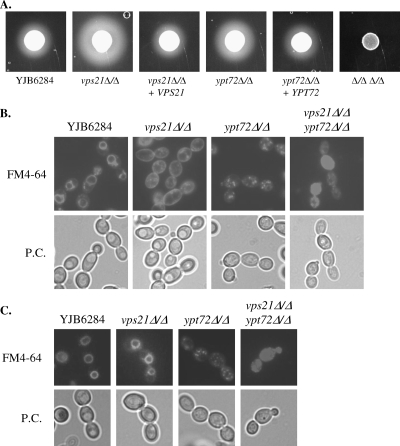
Endosomal trafficking within the mutant strains was examined through pulse-labeling with the lipophilic dye FM4-64 (64), which binds to the plasma membrane. Following internalization by endocytosis, the dye passes through early and late endosomal compartments (PVC) before accumulating within the vacuole. Following a 30-min chase period, most of the FM4-64 localized to one or two large, ring-like structures in cells of the control strain (Fig. 3B) typical of the vacuole compartment. However, a significant amount of the dye remained at the cell surface of the vps21Δ mutant, indicating a reduction in endosomal trafficking from the cell surface, presumably due to a block in trafficking through the PVC. Nonetheless, following a longer chase period of 2 h, the dye localized to an internal ring-like structure (Fig. 3C), indicating a largely normal vacuole morphology. The ypt72Δ mutant internalized FM4-64 with kinetics similar to those of the control strain; however, it accumulated within a heterogeneous mixture of cellular compartments, indicating a highly fragmented vacuole morphology. Some intermediate-size compartments were discernible, which are likely to be early endosomal compartments and the PVC. The double-mutant strain exhibited characteristics of either mutant, with significant cell surface labeling, as well as both punctate and diffuse staining within the cytoplasm. The highly fragmented vacuole morphology of the ypt72Δ and vps21Δypt72Δ mutants was confirmed with a second fluorescent dye, CMAC (60), which does not depend upon endocytic trafficking for vacuolar localization (data not shown). This confirmed that Vps21p functions in endosomal trafficking from the cell surface to the vacuole and that Ypt72p is required for vacuolar biogenesis.
FIG. 3.
The C. albicans vps21Δ and ypt72Δ mutants are defective in endosomal trafficking and vacuole biogenesis. (A) Secreted protease activity was detected on BSA plus YE agar after 48 h of incubation at 37°C. The opaque halo around each colony is the result of protease-mediated BSA hydrolysis. (B) Each strain was pulse-labeled with the endocytic marker FM4-64 and chased for 30 min in fresh medium at 30°C. Cells were then observed with an epifluorescence microscope with a TRITC filter (top) or by phase-contrast (P.C.) microscopy (bottom). (C) Cells were labeled with FM4-64 as described above but chased for 2 h. All data are representative of three or more repeat experiments.
Blocking vacuolar trafficking leads to missorting of vacuolar proteases to the cell surface (1). Lee and colleagues found that a C. albicans vps4Δ mutant (40) deficient in multivesicular body trafficking within endosomes has greater secreted protease activity than do control strains. This is attributed to the missorting of vacuolar CPY to the cell surface. An agar plate-based BSA hydrolysis assay revealed that the vps21Δ mutant, and to a lesser extent the ypt72Δ mutant, also had greater secreted protease activity (Fig. 3A). No detectable BSA hydrolysis occurred within the double-mutant strain.
Vps21p and Ypt72p localize to distinct endosomal compartments.
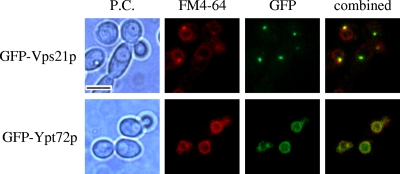
In order to determine the site of action of these two Rab GTPases, we produced GFP-VPS21 and GFP-YPT72 gene fusions expressed from the VPS21 and YPT72 5′ UTRs, respectively. Each fusion construct fully complemented all of the defects associated with the respective mutant strains (data not shown), confirming that the constructs encoded functional fusion proteins. Initial observation revealed that GFP-Vps21p accumulated in one or two intensely labeled small compartments within the cell, while GFP-Ypt72p seemed to label a much larger subcellular compartment less vividly. Each strain was pulse-labeled with FM4-64, followed by different chase periods. Following a short chase period of 20 min, FM4-64 colocalized with the GFP-Vps21p compartment (Fig. 4). However, at later time points, FM4-64 was less associated with GFP-Vps21p and localized to the GFP-Ypt72p-labeled compartment. This is consistent with Vps21p localizing to the PVC and Ypt72p localizing to the vacuole.
FIG. 4.
Vps21p and Ypt72p localize to endosomal and vacuolar compartments, respectively. Cells expressing the GFP-VPS21 or GFP-YPT72 fusion were pulse-labeled with FM4-64 and chased in fresh medium for 20 min (top) to label endosomes or for 60 min (bottom) to label vacuoles. Cells were then observed by phase-contrast (P.C.) microscopy (left column) and epifluorescence microscopy with a TRITC filter for FM4-64 (second column from the left) or a fluorescein isothiocyanate filter for GFP (third column). The combined FM4-64-GFP images are also shown (fourth column). FM4-64 colocalized with GFP-Vps21p and GFP-Ypt72p in all cells with detectable levels of these fusion proteins (>95% of the cells had detectable GFP-Vps21p expression; >90% of the cells had detectable levels of GFP-Ypt72p). Bar = 5 μm.
Vps21p and Ypt72p influence morphogenesis.
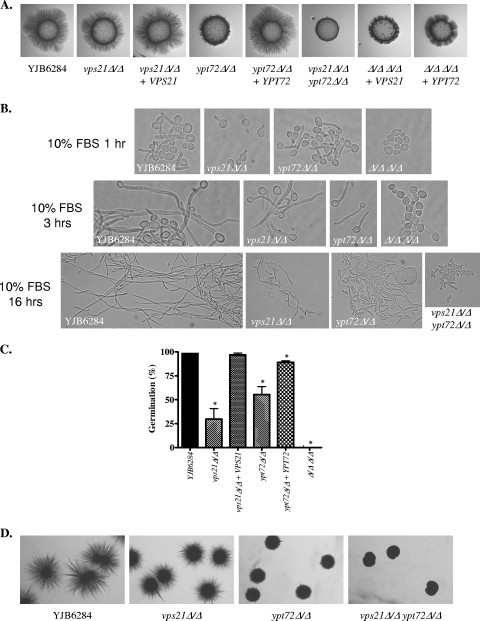
Several previous studies have reported that mutants defective in endosomal and vacuolar biogenesis are defective in yeast-hypha differentiation (4, 7, 12, 22, 48). Initially, we examined filamentous growth on 10% FBS (Fig. 5A) and M199 (data not shown) agar at 37°C. Under these conditions, the vps21Δ mutant had a mild reduction in filamentous growth relative to that of the control and the ypt72Δ mutant was more severely affected, while the vps21Δypt72Δ double mutant was completely afilamentous, even after prolonged incubation. In liquid 10% FBS medium, the germ tubes of the vps21Δ mutant emerged approximately 30 min later than that of the control strain (Fig. 5B) and at later time points (3 to 5 h) its hyphae were approximately two hyphal compartments shorter than control hyphae. Germ tube emergence in the ypt72Δ mutant occurred with normal kinetics in the majority of the cells; however, at later time points, the hyphae were consistently two or three cell compartments shorter than those of control cells. The hyphae produced by both single mutants after 24 h remained significantly shorter than those produced by the control strain. The vps21Δ hyphae were noticeably more branched than control hyphae, and the ypt72Δ mutant demonstrated a tendency to revert to a budding mode of growth (Fig. 5B), which was not observed in the control strain. In 1% FBS liquid culture, the difference between the mutant and control strains was much more striking. Both the vps21Δ and ypt72Δ mutants had reduced efficiency of germ tube emergence relative to that of the YJB6284 control (Fig. 5C). Also, those germ tubes that did emerge from these mutants were delayed by 30 to 60 min and were shorter than those of the control strain. The vps21Δypt72Δ mutant was completely defective in polarized growth in either 1% or 10% FBS but continued to divide slowly as yeast or rounded pseudohyphae (Fig. 5B).
FIG. 5.
Vps21p and Ypt72p are required for filamentous growth. (A) Cell suspensions of each strain were applied as spots to 10% FBS agar and incubated at 37°C for 4 days. (B) Each strain was induced to filament in 10% FBS at 37°C for 1 h (top), 3 h (middle), or 16 h (bottom). (C) Each strain was subcultured to 1% FBS at 37°C, and the number of cells with germ tubes was determined microscopically after 3 h. Results are the mean and standard deviation of four separate experiments. *, P < 0.005 compared to the YJB6284 control. (D) Cells were sandwiched between two layers of YPS agar and incubated at room temperature for 3 days.
A number of mutants with reduced filamentous growth under the above-described conditions exhibit increased hyphal growth relative to that of the control strain when embedded in an agar matrix (22, 28). However, when sandwiched between or embedded within YPS agar, both the vps21Δ and ypt72Δ mutants exhibited a marked decrease in filamentous growth compared with that of the YJB6284 control (Fig. 5D). The double mutant was again afilamentous under these conditions. All of the mutant strains grew well under hypoxic conditions (data not shown); thus, increased susceptibility to hypoxia does not account for the reduced filamentous growth observed in the YPS matrix.
Previous studies have demonstrated that vps23Δ, vps28Δ, and vps36Δ mutants, defective in the ESCRT-mediated multivesicular body sorting pathway, have decreased filamentous growth due to a defect in the proteolytic activation of the Rim101p transcription factor (34, 71). The defects in filamentous growth could be suppressed by the expression of a C-terminally truncated active RIM101 allele (71). However, introduction of active RIM101 alleles (20) did not suppress the defects in hyphal growth of either the vps21Δ or the ypt72Δ mutant on solid or in liquid medium (data not shown). This suggests that the defects in hyphal growth are not due to defective Rim101p processing. We further considered that the defects in yeast-hypha morphogenesis could result from either a mechanical deficiency in polarized growth or perhaps an indirect effect on hyphal signaling pathways, leading to reduced efficiency of hyphal induction. To address this, we examined the consequences of derepressing hyphal growth in the vps21Δ, ypt72Δ, and double-mutant strains through deletion of the hyphal repressor TUP1 (10). As previously described, deletion of TUP1 from parental strain BWP17 led to constitutive filamentous growth, even on rich growth medium such as YPD (Fig. 6A). However, there was significantly less filamentous growth in either the vps21Δ or the ypt72Δ strain background. Thus, even when hyphal growth is derepressed, the defects in filamentous growth persist. While deletion of one allele of TUP1 was readily achieved in the vps21Δypt72Δ mutant, we were unable to replace the second allele. None of 72 transformants had the second TUP1 allele deleted. In order to test if deletion of TUP1 in the vps21Δypt72Δ mutant was a lethal event, we placed the second allele under the control of the MET3 regulatable promoter. Repression of TUP1 transcription through the addition of cysteine and methionine was confirmed by reverse transcription-PCR (data not shown) but was not lethal in the vps21Δypt72Δ strain background. Furthermore, repression of TUP1 did not lead to the formation of filamentous colonies (Fig. 6B) or polarized hyphal forms in liquid culture (data not shown).
FIG. 6.
Derepression of filamentous growth does not restore normal hypha formation in the vps21Δ, ypt72Δ, and vps21Δypt72Δ mutants. (A) The hyphal repressor TUP1 was deleted from the vps21Δ and ypt72Δ mutants. tup1Δ/Δ mutant strains were streaked onto YPD medium and grown at 30°C for 2 days. (B) In the vps21Δypt72Δ double mutant, one copy of TUP1 was deleted and the second copy was placed under the control of the MET3 promoter. Strains were then streaked onto YNB agar, and TUP1 transcription was repressed by the addition of cysteine (Cys) and methionine (Met) to the medium. Images were taken after 2 days at 30°C.
We previously found that a vps11Δ mutant deficient in PVC and vacuole biogenesis was unable to produce chlamydospores (50). However, both the vps21Δ and ypt72Δ mutants were able to form chlamydospores, although the filamentous supporting structures were much shorter for the ypt72Δ mutant (data not shown). Similar to the vps11Δ strain, the vps21Δypt72Δ mutant was completely blocked in chlamydospore induction, lacking either filamentous supporting structures, suspensor cells, or chlamydospores (data not shown).
Vps21p and Ypt72p are required for C. albicans virulence in a mouse model of disseminated disease.
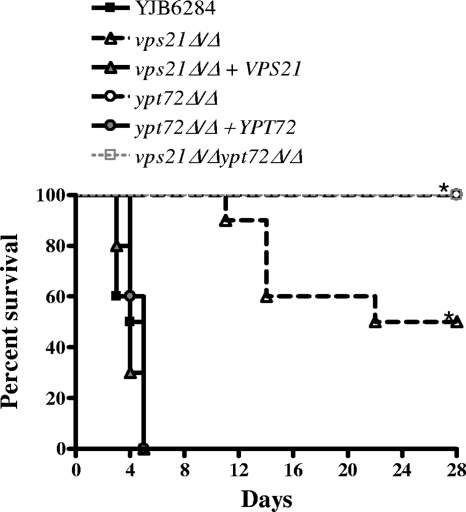
In order to establish the role of endosomal and vacuolar biogenesis during host interaction, we examined the virulence of the vps21Δ, ypt72Δ, and vps21Δypt72Δ mutants in a mouse model of hematogenously disseminated infection. BALB/c mice were inoculated with approximately 5 × 105 CFU of each C. albicans strain, and their survival was followed for 28 days. All of the mice inoculated with “wild-type” control strain YJB6284 succumbed by day 5 (Fig. 7). Mice infected with either the vps21Δ or the ypt72Δ mutant strain survived significantly longer. The ypt72Δ mutant failed to kill any of the mice by day 28, while the vps21Δ mutant only achieved 50% mortality. Virulence was fully restored in the reconstituted strains. Not surprisingly, the vps21Δypt72Δ double mutant was completely avirulent, with the infected animals failing to exhibit any symptoms. In order to determine if the mutant strains were cleared from the surviving mice, we quantified the C. albicans present within kidney tissue. Three of the five surviving vps21Δ mutant-infected mice had no detectable C. albicans, suggesting that the mutant was cleared, and the remaining two had low numbers of CFU. One of 10 mice surviving infection with the ypt72Δ mutant had no detectable CFU, while the fungal burden in the other 9 was highly variable (data not shown). Interestingly, some of these animals had very high fungal burdens, including one animal with a kidney CFU count more than 10-fold higher than that of mice that succumbed to the control strain. This suggests that even when the ypt72Δ mutant is not cleared from mouse tissues, it is significantly less lethal. No C. albicans was isolated from the kidneys of mice infected with the vps21Δypt72Δ double mutant.
FIG. 7.
The C. albicans vps21Δ and ypt72Δ mutants have severely attenuated virulence. Ten BALB/c mice were inoculated with approximately 5 × 105 cells of each C. albicans strain via the lateral tail vein. Kaplan-Meier survival curves were plotted for a 28-day period. Note that the survival curves of the ypt72Δ and vps21Δypt72Δ mutant-challenged mice overlap. *, P < 0.0001 versus the YJB6284 control. A repeat experiment was performed with five mice per C. albicans strain, and similar results (not shown) were obtained.
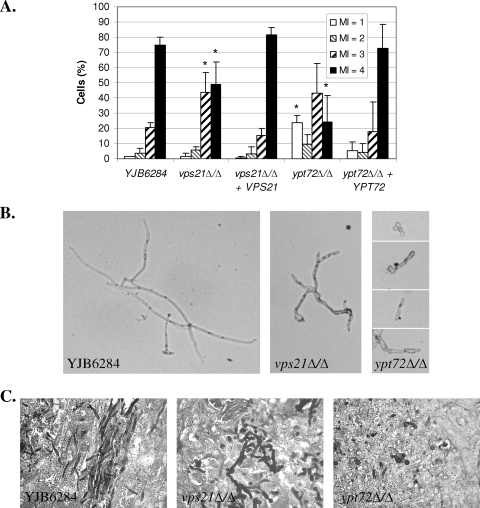
We next analyzed the fungal burdens and morphology in kidneys from mice sacrificed at 48 h postinoculation. There was a significantly lower number of CFU of the ypt72Δ mutant at 48 h than of the wild-type control (1.4 ± 0.7 × 105/g [mean ± standard error of the mean] versus 2.8 ± 0.8 × 106/g; n = 5, P = 0.0089), while the double mutant was undetectable in all animals, indicating rapid clearance in vivo. Neither the vps21Δ mutant nor the reconstituted strain had a significant difference in the number of CFU from that of the YJB6284 control strain at 48 h postinoculation. However, significant differences were observed in the fungal morphology of both the vps21Δ and ypt72Δ mutants within infected kidney tissue (Fig. 8A). The vps21Δ mutant had significantly fewer cells in the true hyphal form (MI = 4) than did the control strain, and the hyphae that were observed were shorter and more branched than those of the control strain (Fig. 8B). The ypt72Δ mutant also had fewer true hyphal cells than did the control, and those present tended to be very short. GMS staining of kidney sections at 48 h postinfection (Fig. 8C) also revealed that YJB6284 was primarily found as true hyphae, the vps21Δ mutant was largely pseudohyphal, and the ypt72Δ mutant was mostly yeast and pseudohyphal. These data confirm that both the vps21Δ and ypt72Δ mutants have defects in hyphal growth in vivo. Hematoxylin and eosin staining of kidneys extracted at 48 h postinoculation revealed a strong inflammatory response to all of the strains except the double mutant. The cellular infiltrate consisted predominantly of neutrophils in each case (data not shown). The apparent absence of inflammation in mice infected with the vps21Δypt72Δ mutant suggests that it may be cleared before such a response is mounted.
FIG. 8.
C. albicans VPS21 and YPT72 affect in vivo morphogenesis. Mice inoculated with approximately 5 × 105 cells of each C. albicans strain were sacrificed at 48 h postinfection. (A) Kidney tissue was homogenized and treated with KOH to dissolve host cells. Fungal cell morphology was observed microscopically and scored according to the MI described by Merson-Davies and Odds (43, 47) as follows: MI of 1, spherical and nearly spherical cells; MI of 2, ovoid cells with a length of up to twice the cell width; MI of 3, pseudohyphal cells with obvious constrictions; MI of 4, parallel-sided true hyphal cells. Data are the means of three or more animals for each strain; error bars represent standard deviations. *, P < 0.05 versus the YJB6284 control. (B) Representative images from KOH-treated kidney homogenate. (C) Sections of kidney tissue stained with GMS to reveal in vivo fungal morphology.
DISCUSSION
The vacuole has been observed to play a cytologically prominent role during C. albicans yeast-hypha differentiation, a transition which is crucial for virulence. Germ tube emergence is accompanied by the formation of extensively vacuolated subapical compartments, with cytoplasmic material migrating within the apical cell compartment. However, the significance of hyphal cell vacuolation with respect to pathogenesis remains unknown. Here we report the identification of two C. albicans Rab GTPases, Vps21p and Ypt72p, required for endosomal trafficking and vacuole biogenesis, respectively. Vps21p appears to localize to a late endosomal compartment, and Ypt72p appears to localize to the vacuole itself. Loss of either of these GTPases impacted the production of the invasive hyphal form, as well as the virulence, of C. albicans.
The first significant observation was that while the vps21Δ and ypt72Δ mutants grow with wild-type kinetics, the vps21Δypt72Δ mutant has a striking growth defect. In addition, the two single mutants, for the most part, had mild defects in stress resistance, with either resistant to osmotic, temperature, and oxidative stresses. However, the double mutant was hypersensitive to a diverse array of cellular stresses, indicating a synthetic defect in the double mutant. The exquisite specificity of Rab GTPases for their target organelle (54) makes functional redundancy of the Vps21p and Ypt72p GTPases unlikely. However, these results could suggest that there is some redundancy between the function of the PVC and vacuolar compartment in C. albicans. A similar synthetic defect was described in an S. cerevisiae mutant deficient in the PVC and vacuolar SNARE proteins Pep12p and Vam3p (53). Thus, perhaps loss of PVC (vps21Δ) or vacuole (ypt72Δ) function alone can be partially compensated by the other compartment.
Loss of Vps21p or Ypt72p also results in defects in hyphal growth, while loss of both completely abrogated polarized growth, suggesting that PVC and vacuolar biogenesis is crucial during hyphal growth. This again may suggest that there is some functional overlap between the PVC and vacuole in the establishment and maintenance of polarized hyphal growth. Several distinct signaling pathways are known to control hyphal development in C. albicans, with each responsive to a different subset of inducing conditions (66). However, the degree of hyphal retardation in the vps21Δ, ypt72Δ, and vps21Δypt72Δ mutants is similar irrespective of the inducing conditions used. Furthermore, derepression of hyphal growth through deletion of TUP1, which bypasses much of the upstream signal transduction pathways normally regulating yeast-hypha morphogenesis, did not correct this defect. This supports a mechanistic defect in polarized growth in either mutant, rather than a defect in hyphal signaling. Deletion of TUP1 also enabled us to compare the hyphal growth of the vps21Δ and ypt72Δ mutants on rich YPD medium. Reduced hyphal growth of the mutants under these conditions rules out the possibility that reduced hyphal growth is simply the result of a diminished capacity to utilize complex or suboptimal nutrient sources found in many hyphal inducing media. An inviting explanation is that the vacuolar expansion observed in subapical hyphal compartments fulfills a physical requirement for apical extension. Hyphal cell vacuolation necessitates the transport and incorporation of new membrane material into the vacuole and perhaps the PVC. Thus, loss of PVC and vacuolar Rab GTPases, which facilitate membrane trafficking and organelle biogenesis, may block vacuolation and therefore impede apical extension. We envisage several potential mechanisms by which vacuolar expansion may aid polarized hyphal growth. One major advantage of hyphal cell vacuolation may be that it requires minimal amounts of de novo cytoplasmic biosynthesis, while permitting rapid germination and apical extension. The vacuole is significantly less dense than cytoplasm with respect to protein and other macromolecule content (72). Hyphal cell vacuolation may therefore overcome the rate-limiting step of cytoplasmic biosynthesis and enable hyphae to emerge under nutrient-poor conditions. Second, the fungal vacuole has a central role in regulating the osmotic potential of the cell (2, 37, 38), and modulation of vacuolar volume occurs in response to osmotic stress through homotypic fission/fusion events (9, 36, 45). Thus, the sudden increase in vacuolar volume may increase turgor pressure within the emerging C. albicans germ tube. The opposing force of the rigid cell wall would lead to exertion of the turgor pressure at the wall's weakest point, usually the site of active growth, i.e., the hyphal apex. Increased turgor pressure may thus aid polarized growth and apical extension by providing a mechanical force to push out the hyphal tip (44). This, in turn, may enable the fungus to exert force upon host tissues at a focal point (i.e., the hyphal tip). A number of plant fungal pathogens form specialized invasion structures such as appresoria to penetrate the plant's tough surface layers (5, 32). These cells generate huge osmotic pressure, which is channeled into a narrow projection, enabling the fungal cell to exert significant force at a focal point on the plant surface. We do not envisage that penetration of mucosal surfaces by C. albicans involves the extreme pressures generated in these plant pathogens. However, it is possible that elevated turgor pressure is produced in the emerging germ tube and this may aid polarized growth or enable C. albicans which is tightly bound to the epithelium to exert some force on host tissue at the hyphal apex. Either way, the observation of a similar developmental program of subapical vacuolation in the plant-invasive form of U. maydis (41) may indicate that this is a shared strategy for host invasion.
Several previous studies have linked the vacuole to C. albicans virulence in various models of infection, including a macrophage challenge model and a mouse model of disseminated candidiasis. However, the mutants used were typically affected in both PVC and vacuole biogenesis (50) or were deficient in proteins known to function in diverse pathways (12). We found that loss of either Vps21p or Ypt72p severely attenuated virulence in a mouse model of disseminated candidiasis. This established that loss of either endosome or vacuole biogenesis is sufficient to cause major defects in virulence. The ypt72Δ mutant had a decreased number of CFU in mouse kidneys at 48 h postinfection, suggesting a defect in tissue colonization. However, even when present at very high fungal burdens, the ypt72Δ mutant did not cause death. This suggests that this mutant is deficient on two levels: (i) colonization of host tissues in some animals and (ii) inflicting tissue damage which contributes to death. Both the vps21Δ and ypt72Δ mutants had an impaired capacity to form true hyphae in vivo, similar to that observed in vitro, with the vps21Δ mutant forming shorter, more branched hyphae and the ypt72Δ mutant forming fewer and shorter hyphae than the control strain. Thus, it is not clear if the decreased colonization of the ypt72Δ mutant at 48 h postinfection is due to a decreased capacity to survive within the host or if the decreased capacity to form hyphae limits fungal propagation within host tissue. Segregation of Vps21p and Ypt72p functions in stress resistance and hyphal growth should elucidate the contribution of these GTPases to virulence and remains a key goal for future studies.
We previously reported that a C. albicans vps11Δ mutant with defects in both PVC and vacuole biogenesis is deficient in both survival within and the ability to cause lysis of the J774A.1 mouse macrophage-like cell line (50). In order to establish the role of Vps21p and Ypt72p during this interaction, we repeated these macrophage challenge experiments with the vps21Δ, ypt72Δ, and double-mutant strains. This revealed that both the vps21Δ and ypt72Δ mutants were able to cause macrophage lysis, similar to the control strain. However, the vps21Δypt72Δ mutant caused no detectable loss of macrophage viability (data not shown), similar to the previously reported vps11Δ mutant. Thus, according to this simple in vitro model, despite severe defects in the mouse model, neither the vps21Δ nor the ypt72Δ mutant has profound defects in phagocytic cell interaction.
The results presented here and elsewhere have clearly established that the fungal PVC and vacuole are crucial during host interaction. We suggest that the PVC and vacuole may be required on two levels during C. albicans infection, (i) stress resistance functions required for adaptation and survival within host tissues and (ii) a role in filamentous growth which may aid host tissue invasion and injury. Segregation of the vacuole's role in fungal morphogenesis from its functions in stress resistance and cellular homeostasis will be crucial in defining the role it plays in pathogenesis and thus the therapeutic potential of disrupting vacuolar integrity.
Acknowledgments
This publication was made possible by grant P20RR020160 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH). This work was also supported by NIH grant R21AI60371 to J.S.
The contents of this publication are solely our responsibility and do not necessarily represent the official view of NCRR or NIH.
We thank Paul Fidel for assistance with the animal studies and Aaron Mitchell (Carnegie Mellon University) and Fritz Mühlschlegel (University of Kent) for strains and plasmid constructs. We give special thanks to Seth Pincus, Research Institute at Children's Hospital, New Orleans, and Jim Cutler for housing and supporting our research activities in the dark days following Hurricane Katrina.
Editor: A. Casadevall
Footnotes
Published ahead of print on 13 April 2009.
REFERENCES
- 1.Bankaitis, V. A., L. M. Johnson, and S. D. Emr. 1986. Isolation of yeast mutants defective in protein targeting to the vacuole. Proc. Natl. Acad. Sci. USA 839075-9079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banta, L. M., J. S. Robinson, D. J. Klionsky, and S. D. Emr. 1988. Organelle assembly in yeast: characterization of yeast mutants defective in vacuolar biogenesis and protein sorting. J. Cell Biol. 1071369-1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barelle, C. J., E. A. Bohula, S. J. Kron, D. Wessels, D. R. Soll, A. Schafer, A. J. Brown, and N. A. Gow. 2003. Asynchronous cell cycle and asymmetric vacuolar inheritance in true hyphae of Candida albicans. Eukaryot. Cell 2398-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barelle, C. J., M. L. Richard, C. Gaillardin, N. A. Gow, and A. J. Brown. 2006. Candida albicans VAC8 is required for vacuolar inheritance and normal hyphal branching. Eukaryot. Cell 5359-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bastmeyer, M., H. B. Deising, and C. Bechinger. 2002. Force exertion in fungal infection. Annu. Rev. Biophys. Biomol. Struct. 31321-341. [DOI] [PubMed] [Google Scholar]
- 6.Bensen, E. S., S. G. Filler, and J. Berman. 2002. A Forkhead transcription factor is important for true hyphal as well as yeast morphogenesis in Candida albicans. Eukaryot. Cell 1787-798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bernardo, S. M., Z. Khalique, J. Kot, J. K. Jones, and S. A. Lee. 2008. Candida albicans VPS1 contributes to protease secretion, filamentation, and biofilm formation. Fungal Genet. Biol. 45861-877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boeke, J. D., F. LaCroute, and G. R. Fink. 1984. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet. 197345-346. [DOI] [PubMed] [Google Scholar]
- 9.Bone, N., J. B. Millar, T. Toda, and J. Armstrong. 1998. Regulated vacuole fusion and fission in Schizosaccharomyces pombe: an osmotic response dependent on MAP kinases. Curr. Biol. 8135-144. [DOI] [PubMed] [Google Scholar]
- 10.Braun, B. R., and A. D. Johnson. 1997. Control of filament formation in Candida albicans by the transcriptional repressor TUP1. Science 277105-109. [DOI] [PubMed] [Google Scholar]
- 11.Brown, D. H., Jr., A. D. Giusani, X. Chen, and C. A. Kumamoto. 1999. Filamentous growth of Candida albicans in response to physical environmental cues and its regulation by the unique CZF1 gene. Mol. Microbiol. 34651-662. [DOI] [PubMed] [Google Scholar]
- 12.Bruckmann, A., W. Kunkel, A. Hartl, R. Wetzker, and R. Eck. 2000. A phosphatidylinositol 3-kinase of Candida albicans influences adhesion, filamentous growth and virulence. Microbiology 146(Pt. 11)2755-2764. [DOI] [PubMed] [Google Scholar]
- 13.Bryant, N. J., and T. H. Stevens. 1998. Vacuole biogenesis in Saccharomyces cerevisiae: protein transport pathways to the yeast vacuole. Microbiol. Mol. Biol. Rev. 62230-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burke, D., D. Dawson, and T. Stearns. 2000. Methods in yeast genetics: a Cold Spring Harbor Laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 15.Care, R. S., J. Trevethick, K. M. Binley, and P. E. Sudbery. 1999. The MET3 promoter: a new tool for Candida albicans molecular genetics. Mol. Microbiol. 34792-798. [DOI] [PubMed] [Google Scholar]
- 16.Conibear, E., and T. H. Stevens. 1995. Vacuolar biogenesis in yeast: sorting out the sorting proteins. Cell 83513-516. [DOI] [PubMed] [Google Scholar]
- 17.Crandall, M., and J. E. Edwards, Jr. 1987. Segregation of proteinase-negative mutants from heterozygous Candida albicans. J. Gen. Microbiol. 133(Pt. 10)2817-2824. [DOI] [PubMed] [Google Scholar]
- 18.Deneka, M., M. Neeft, and P. van der Sluijs. 2003. Regulation of membrane transport by rab GTPases. Crit. Rev. Biochem. Mol. Biol. 38121-142. [DOI] [PubMed] [Google Scholar]
- 19.de Repentigny, L., D. Lewandowski, and P. Jolicoeur. 2004. Immunopathogenesis of oropharyngeal candidiasis in human immunodeficiency virus infection. Clin. Microbiol. Rev. 17729-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.El Barkani, A., O. Kurzai, W. A. Fonzi, A. Ramon, A. Porta, M. Frosch, and F. A. Mühlschlegel. 2000. Dominant active alleles of RIM101 (PRR2) bypass the pH restriction on filamentation of Candida albicans. Mol. Cell. Biol. 204635-4647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fonzi, W. A., and M. Y. Irwin. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134717-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Franke, K., M. Nguyen, A. Hartl, H. M. Dahse, G. Vogl, R. Wurzner, P. F. Zipfel, W. Kunkel, and R. Eck. 2006. The vesicle transport protein Vac1p is required for virulence of Candida albicans. Microbiology 1523111-3121. [DOI] [PubMed] [Google Scholar]
- 23.Fraser, V. J., M. Jones, J. Dunkel, S. Storfer, G. Medoff, and W. C. Dunagan. 1992. Candidemia in a tertiary care hospital: epidemiology, risk factors, and predictors of mortality. Clin. Infect. Dis. 15414-421. [DOI] [PubMed] [Google Scholar]
- 24.Gerami-Nejad, M., J. Berman, and C. A. Gale. 2001. Cassettes for PCR-mediated construction of green, yellow, and cyan fluorescent protein fusions in Candida albicans. Yeast 18859-864. [DOI] [PubMed] [Google Scholar]
- 25.Gerrard, S. R., N. J. Bryant, and T. H. Stevens. 2000. VPS21 controls entry of endocytosed and biosynthetic proteins into the yeast prevacuolar compartment. Mol. Biol. Cell 11613-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gietz, D., A. St Jean, R. A. Woods, and R. H. Schiestl. 1992. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 201425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gillum, A. M., E. Y. Tsay, and D. R. Kirsch. 1984. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol. Gen. Genet. 198179-182. [DOI] [PubMed] [Google Scholar]
- 28.Giusani, A. D., M. Vinces, and C. A. Kumamoto. 2002. Invasive filamentous growth of Candida albicans is promoted by Czf1p-dependent relief of Efg1p-mediated repression. Genetics 1601749-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gow, N. A., and G. W. Gooday. 1984. A model for the germ tube formation and mycelial growth form of Candida albicans. Sabouraudia 22137-144. [DOI] [PubMed] [Google Scholar]
- 30.Gow, N. A., and G. W. Gooday. 1982. Vacuolation, branch production and linear growth of germ tubes in Candida albicans. J. Gen. Microbiol. 128(Pt. 9)2195-2198. [DOI] [PubMed] [Google Scholar]
- 31.Guthrie, C., and G. Fink. 1991. Guide to yeast genetics and molecular biology. Academic Press, New York, NY.
- 32.Howard, R. J., and B. Valent. 1996. Breaking and entering: host penetration by the fungal rice blast pathogen Magnaporthe grisea. Annu. Rev. Microbiol. 50491-512. [DOI] [PubMed] [Google Scholar]
- 33.Klionsky, D. J., P. K. Herman, and S. D. Emr. 1990. The fungal vacuole: composition, function, and biogenesis. Microbiol. Rev. 54266-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kullas, A. L., M. Li, and D. A. Davis. 2004. Snf7p, a component of the ESCRT-III protein complex, is an upstream member of the RIM101 pathway in Candida albicans. Eukaryot. Cell 31609-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuranda, K., V. Leberre, S. Sokol, G. Palamarczyk, and J. Francois. 2006. Investigating the caffeine effects in the yeast Saccharomyces cerevisiae brings new insights into the connection between TOR, PKC and Ras/cAMP signalling pathways. Mol. Microbiol. 611147-1166. [DOI] [PubMed] [Google Scholar]
- 36.LaGrassa, T. J., and C. Ungermann. 2005. The vacuolar kinase Yck3 maintains organelle fragmentation by regulating the HOPS tethering complex. J. Cell Biol. 168401-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Latterich, M., and M. D. Watson. 1993. Evidence for a dual osmoregulatory mechanism in the yeast Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 1911111-1117. [DOI] [PubMed] [Google Scholar]
- 38.Latterich, M., and M. D. Watson. 1991. Isolation and characterization of osmosensitive vacuolar mutants of Saccharomyces cerevisiae. Mol. Microbiol. 52417-2426. [DOI] [PubMed] [Google Scholar]
- 39.Lay, J., L. K. Henry, J. Clifford, Y. Koltin, C. E. Bulawa, and J. M. Becker. 1998. Altered expression of selectable marker URA3 in gene-disrupted Candida albicans strains complicates interpretation of virulence studies. Infect. Immun. 665301-5306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee, S. A., J. Jones, Z. Khalique, J. Kot, M. Alba, S. Bernardo, A. Seghal, and B. Wong. 2007. A functional analysis of the Candida albicans homolog of Saccharomyces cerevisiae VPS4. FEMS Yeast Res. 7973-985. [DOI] [PubMed] [Google Scholar]
- 41.Lehmler, C., G. Steinberg, K. M. Snetselaar, M. Schliwa, R. Kahmann, and M. Bolker. 1997. Identification of a motor protein required for filamentous growth in Ustilago maydis. EMBO J. 163464-3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lo, H. J., J. R. Kohler, B. DiDomenico, D. Loebenberg, A. Cacciapuoti, and G. R. Fink. 1997. Nonfilamentous C. albicans mutants are avirulent. Cell 90939-949. [DOI] [PubMed] [Google Scholar]
- 43.Merson-Davies, L. A., and F. C. Odds. 1989. A morphology index for characterization of cell shape in Candida albicans. J. Gen. Microbiol. 1353143-3152. [DOI] [PubMed] [Google Scholar]
- 44.Money, N. P. 1997. Wishful thinking of turgor revisited: the mechanics of hyphal growth. Fungal Genet. Biol. 21173-187. [Google Scholar]
- 45.Nass, R., and R. Rao. 1999. The yeast endosomal Na+/H+ exchanger, Nhx1, confers osmotolerance following acute hypertonic shock. Microbiology 145(Pt. 11)3221-3228. [DOI] [PubMed] [Google Scholar]
- 46.Noda, T., J. Kim, W. P. Huang, M. Baba, C. Tokunaga, Y. Ohsumi, and D. J. Klionsky. 2000. Apg9p/Cvt7p is an integral membrane protein required for transport vesicle formation in the Cvt and autophagy pathways. J. Cell Biol. 148465-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Odds, F. C., L. Van Nuffel, and N. A. Gow. 2000. Survival in experimental Candida albicans infections depends on inoculum growth conditions as well as animal host. Microbiology 146(Pt. 8)1881-1889. [DOI] [PubMed] [Google Scholar]
- 48.Palmer, G. E., A. Cashmore, and J. Sturtevant. 2003. Candida albicans VPS11 is required for vacuole biogenesis and germ tube formation. Eukaryot. Cell 2411-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Palmer, G. E., K. J. Johnson, S. Ghosh, and J. Sturtevant. 2004. Mutant alleles of the essential 14-3-3 gene in Candida albicans distinguish between growth and filamentation. Microbiology 1501911-1924. [DOI] [PubMed] [Google Scholar]
- 50.Palmer, G. E., M. N. Kelly, and J. E. Sturtevant. 2005. The Candida albicans vacuole is required for differentiation and efficient macrophage killing. Eukaryot. Cell 41677-1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Palmer, G. E., and J. E. Sturtevant. 2004. Random mutagenesis of an essential Candida albicans gene. Curr. Genet. 46343-356. [DOI] [PubMed] [Google Scholar]
- 52.Pereira-Leal, J. B., and M. C. Seabra. 2000. The mammalian Rab family of small GTPases: definition of family and subfamily sequence motifs suggests a mechanism for functional specificity in the Ras superfamily. J. Mol. Biol. 3011077-1087. [DOI] [PubMed] [Google Scholar]
- 53.Peterson, M. R., and S. D. Emr. 2001. The class C Vps complex functions at multiple stages of the vacuolar transport pathway. Traffic 2476-486. [DOI] [PubMed] [Google Scholar]
- 54.Pfeffer, S., and D. Aivazian. 2004. Targeting Rab GTPases to distinct membrane compartments. Nat. Rev. Mol. Cell Biol. 5886-896. [DOI] [PubMed] [Google Scholar]
- 55.Qian, Q., and J. E. Cutler. 1997. Gamma interferon is not essential in host defense against disseminated candidiasis in mice. Infect. Immun. 651748-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ramón, A. M., and W. A. Fonzi. 2003. Diverged binding specificity of Rim101p, the Candida albicans ortholog of PacC. Eukaryot. Cell 2718-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saville, S. P., A. L. Lazzell, C. Monteagudo, and J. L. Lopez-Ribot. 2003. Engineered control of cell morphology in vivo reveals distinct roles for yeast and filamentous forms of Candida albicans during infection. Eukaryot. Cell 21053-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schimmöller, F., and H. Riezman. 1993. Involvement of Ypt7p, a small GTPase, in traffic from late endosome to the vacuole in yeast. J. Cell Sci. 106(Pt. 3)823-830. [DOI] [PubMed] [Google Scholar]
- 59.Seiler, S., M. Plamann, and M. Schliwa. 1999. Kinesin and dynein mutants provide novel insights into the roles of vesicle traffic during cell morphogenesis in Neurospora. Curr. Biol. 9779-785. [DOI] [PubMed] [Google Scholar]
- 60.Shoji, J. Y., M. Arioka, and K. Kitamoto. 2006. Vacuolar membrane dynamics in the filamentous fungus Aspergillus oryzae. Eukaryot. Cell 5411-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sobel, J. D. 2007. Vulvovaginal candidosis. Lancet 3691961-1971. [DOI] [PubMed] [Google Scholar]
- 62.Steinberg, G. 2007. On the move: endosomes in fungal growth and pathogenicity. Nat. Rev. Microbiol. 5309-316. [DOI] [PubMed] [Google Scholar]
- 63.Steinberg, G., M. Schliwa, C. Lehmler, M. Bolker, R. Kahmann, and J. R. McIntosh. 1998. Kinesin from the plant pathogenic fungus Ustilago maydis is involved in vacuole formation and cytoplasmic migration. J. Cell Sci. 111(Pt. 15)2235-2246. [DOI] [PubMed] [Google Scholar]
- 64.Vida, T. A., and S. D. Emr. 1995. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J. Cell Biol. 128779-792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wey, S. B., M. Mori, M. A. Pfaller, R. F. Woolson, and R. P. Wenzel. 1988. Hospital-acquired candidemia. The attributable mortality and excess length of stay. Arch. Intern. Med. 1482642-2645. [DOI] [PubMed] [Google Scholar]
- 66.Whiteway, M., and C. Bachewich. 2007. Morphogenesis in Candida albicans. Annu. Rev. Microbiol. 61529-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wichmann, H., L. Hengst, and D. Gallwitz. 1992. Endocytosis in yeast: evidence for the involvement of a small GTP-binding protein (Ypt7p). Cell 711131-1142. [DOI] [PubMed] [Google Scholar]
- 68.Wilson, L. S., C. M. Reyes, M. Stolpman, J. Speckman, K. Allen, and J. Beney. 2002. The direct cost and incidence of systemic fungal infections. Value Health 526-34. [DOI] [PubMed] [Google Scholar]
- 69.Wilson, R. B., D. Davis, B. M. Enloe, and A. P. Mitchell. 2000. A recyclable Candida albicans URA3 cassette for PCR product-directed gene disruptions. Yeast 1665-70. [DOI] [PubMed] [Google Scholar]
- 70.Wilson, R. B., D. Davis, and A. P. Mitchell. 1999. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J. Bacteriol. 1811868-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xu, W., F. J. Smith, Jr., R. Subaran, and A. P. Mitchell. 2004. Multivesicular body-ESCRT components function in pH response regulation in Saccharomyces cerevisiae and Candida albicans. Mol. Biol. Cell 155528-5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yokoyama, K., and K. Takeo. 1983. Differences of asymmetrical division between the pseudomycelial and yeast forms of Candida albicans and their effect on multiplication. Arch. Microbiol. 134251-253. [DOI] [PubMed] [Google Scholar]
- 73.Yorimitsu, T., and D. J. Klionsky. 2005. Autophagy: molecular machinery for self-eating. Cell Death Differ. 12(Suppl. 2)1542-1552. [DOI] [PMC free article] [PubMed] [Google Scholar]