Abstract
Nontypeable Haemophilus influenzae is a commensal that frequently causes otitis media and respiratory tract infections. The lex2 locus encodes a glycosyltransferase that is phase variably expressed and contributes to the significant intrastrain heterogeneity of lipopolysaccharide (LPS) composition in H. influenzae. In serotype b strains, Lex2B adds the second β-glucose in the oligosaccharide extension from the proximal heptose of the triheptose inner core backbone; this extension includes a digalactoside that plays a role in resistance of the bacteria to the killing effect of serum. As part of our studies of the structure and genetics of LPS in nontypeable H. influenzae, we show here that there are allelic polymorphisms in the lex2B sequence that correlate with addition of either a glucose or a galactose to the same position in the LPS molecule across strains. Through exchange of lex2 alleles between strains we show that alteration of a single amino acid at position 157 in Lex2B appears to be sufficient to direct the alternative glucosyl- or galactosyltransferase activities. Allelic exchange strains express LPS with altered structure and biological properties compared to the wild-type LPS. Thus, Lex2B contributes to both inter- and intrastrain LPS heterogeneity through its polymorphic sequences and phase-variable expression.
Haemophilus influenzae is a gram-negative bacterium that is an obligate commensal in the human respiratory tract. This organism, however, has a propensity to spread contiguously, and the predominant unencapsulated (nontypeable H. influenzae [NTHi]) strains can cause diseases such as respiratory tract infections and otitis media (OM). Lipopolysaccharide (LPS), the major cell surface glycolipid, significantly influences host-microbe interactions and is a virulence determinant. From the well-conserved triheptose-containing inner core backbone, oligosaccharides (OS) comprising mainly hexose sugars and various nonsugar substituents extend, and these OS vary in composition between strains (31, 32, 36). The number of sugars and substituent groups comprising these OS can vary at a high frequency within individual strains, and the reversible loss and gain of epitopes is called phase variation (30). Phase variation of LPS in H. influenzae is mediated by switching in the expression of LPS-specific loci that contain tetranucleotide repeats (microsatellites) in the 5′ end of the reading frame (17, 19, 21, 38). These repeat tracts undergo mutation via DNA slippage during replication (25), leading to loss or gain of repeat units that place the reading frame in or out of frame with the translational initiation codon. Thus, depending on the number of tetranucleotide units in the tract, each phase-variable gene can be correctly translated (“on”) or not correctly translated (“off”). For any H. influenzae strain having multiple phase-variable loci, a population of cells can simultaneously generate a range of LPS glycoforms, allowing the organism to adapt to different microenvironments of the host and helping it escape from host immune responses.
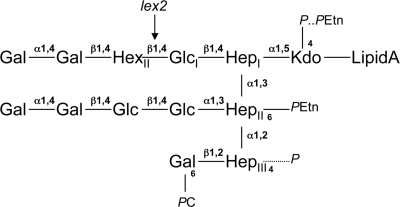
The lex2 locus, comprising lex2A and lex2B, was first identified as an H. influenzae phase-variable LPS biosynthetic locus in a serotype b strain, strain DL42 (21). A tract of multiple, tandem 5′-GCAA repeats is located in the 5′ end of the lex2A reading frame (21). Disruption of lex2B in strain DL42 led to a loss of reactivity of this strain with an LPS-specific monoclonal antibody (MAb), MAb 5G8 (21). Structural analysis of LPS isolated from mutant H. influenzae serotype b strains revealed that Lex2B functions as a glucosyltransferase that adds a second glucose in a β-(1-4) linkage to the first β-glucose linked to the first heptose of the LPS of H. influenzae (14) (Fig. 1). Both reading frames, lex2A and lex2B, are required for Lex2B activity (14). The lex2 locus permits further OS extensions that include a digalactoside that binds MAb 5G8 (13) (Fig. 1) and that play a role in resistance of the bacteria to the killing effect of serum (12).
FIG. 1.
Schematic representation of the structure of H. influenzae LPS, showing the relative position in biosynthesis that is influenced by the lex2 locus. HexII indicates the position of the sugar added by Lex2B (linkage indicated by an arrow) to GlcI. The example shown is the structure for a serotype b strain, RM7004 (29), where HexII is a glucose. The following molecules are present in the structure: Kdo, 2-keto-3-deoxyoctulosonic acid; Hep, l-glycero-d-manno-heptose; Hex, hexose (Glc or Gal); Glc, d-glucose; Gal, d-galactose; PEtn, phosphoethanolamine; P, phosphate; PC, phosphocholine.
H. influenzae exhibits significant LPS structural variation both within and between strains. In addition to phase variation, factors that contribute to LPS heterogeneity are differences in the repertoire of LPS biosynthesis genes between strains and gene sequence polymorphisms that direct alternative transferase functions and additions to the LPS molecule. Depending on the allele present, the lic1D gene can direct addition of phosphocholine to one of several locations in the LPS (26), and the lpsA gene product can add either glucose or galactose in one of two alternative linkages to the same position in the LPS molecule (6).
As part of our ongoing studies of the LPS structure and genetics of a set of 26 NTHi strains from OM patients (3), detailed analysis of the LPS from NTHi strains 432 and 1124 (40; unpublished data) has shown that the second hexose in the OS extension from HepI is a galactose rather than the glucose described previously at this position (14). We wanted to determine whether this sugar specificity was determined by lex2B alone, in a way analogous to that seen for the lpsA gene, or whether an alternative gene is involved. In the present study, we further characterize the lex2 locus of H. influenzae and show that the lex2B gene exhibits sequence polymorphism between strains that direct the attachment of either a β-glucose or a β-galactose as the second hexose in the OS extension from the proximal heptose of H. influenzae LPS. Lex2B is a glycosyltransferase that contributes to LPS structural heterogeneity through both its phase-variable expression and allelic polymorphisms that direct addition of alternative sugars.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The 26 NTHi strains used in this study are a subset of the strains which were isolated from children with OM by the Finnish Otitis Media Study group (3). H. influenzae strain RM118 (Rd; strain KW20) (19), serotype b strain RM153 (Eagan) (2), and NTHi strain 2019 (11, 31) have been described previously. H. influenzae strains were grown in brain heart infusion (BHI) broth supplemented with 10 μg ml−1 hemin, 2 μg ml−1 NAD, or agar (1%, wt/vol) at 37°C. When required, kanamycin was added at 10 μg ml−1.
Escherichia coli strain DH5α (15) was used for DNA cloning and propagation of plasmid DNA and was cultured in Luria broth or on agar plates at 37°C. When necessary, isopropyl-β-d-thiogalactopyranoside (IPTG) (40 μg ml−1), 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) (40 μg ml−1), ampicillin (100 μg ml−1), or kanamycin (50 μg ml−1) was added to the medium.
DNA procedures.
Restriction enzymes and DNA-modifying enzymes were purchased from Roche or New England Biolabs and were used according to the manufacturers' instructions. Plasmid DNA was prepared from E. coli strains by alkaline lysis (34). Chromosomal DNA was prepared from H. influenzae by a method described elsewhere (33).
For Southern blot analysis (34), DNA probes for hybridization were labeled using the GE Healthcare ECL system according to the manufacturer's instructions. The membranes were prehybridized at 42°C for 1 to 4 h in hybridization buffer and then hybridized in the same buffer with labeled DNA probe overnight at 42°C. Filters were subsequently washed twice in primary wash buffer (6 M urea, 0.4% sodium dodecyl sulfate [SDS], 0.5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate]) for 20 min at 42°C and then once in 2× SSC for 5 min at 42°C before they were developed with detection reagents and hybridized bands were visualized by autoradiography using ECL film.
PCR amplification and DNA sequencing.
The oligonucleotide primers used for PCR amplification and DNA sequencing are listed in Table 1 and were designed using the lex2 sequences available in the databases. PCR amplification was routinely carried out as described previously (18). Prior to DNA sequencing, the lex2 genes were amplified by PCR using primers blex2for and lex2CHP1. The DNA sequences of the templates were determined using primers blex2for, lex2CHP1, oligo1, and oligo2, a BigDye DNA sequencing kit (PE Applied Biosystems), and an automated ABI 3730 sequencer. DNA sequences were collated and analyzed using the GCG sequence analysis program (7). Sequence homologies were determined using the GenBank DNA and protein sequence databases through the National Center for Biotechnology Information BLAST network server (1).
TABLE 1.
Oligonucleotide primers used for lex2 genes and the adjacent sequence in this study
| Primer | Sequence |
|---|---|
| blex2for | 5′-AAATGAATAGGTAGGGG |
| lex2CHP1 | 5′-GGAATGGGCAACTTATGGCG |
| lex2RegForA | 5′-GAAGCGCGTGTGAGTTTAG |
| lex2RegRevB2 | 5′-CCGATGCAACCAGTAAGTGC |
| oligo1 | 5′-GCGGTTGAATGCAAAGGG |
| oligo2 | 5′-ATGGCTAGCATAACAGCC |
Cloning of lex2 for allelic exchange and mutation.
A PCR product containing the lex2A and lex2B genes and 500 bp of upstream and downstream flanking DNA sequence was amplified from chromosomal DNA of NTHi strains 432, 1008, 1124, 1247, 1292, and 2019 using primers lex2RegForA and lex2RegRevB2 (Table 1) and then cloned into plasmid pCR2.1TOPO (Invitrogen). pCR2.1-TOPO carries an ampicillin resistance cassette as well as a kanamycin resistance (Kanr) cassette, so the insert was subcloned into plasmid pT7Blue (Novagen) to enable a Kanr cassette to be inserted into the cloned DNA to permit selection following subsequent transformation into H. influenzae. The lex2 inserts were excised from pCR2.1TOPO by digestion with BamHI and NsiI, the pT7Blue plasmid was digested with BamHI and PstI, and then the two DNAs were ligated together before transformation into E. coli.
A Kanr cassette, released from pUC4Kan by digestion with HincII, was inserted 110 bp upstream of the lex2 locus in the plasmid clone from each strain at an SnaBI restriction site. Following ligation and transformation into E. coli, correct plasmid constructs were confirmed by restriction digestion and PCR amplification. Plasmid clones plex2Gal432, plex2Glc1008, plex2Gal1124, plex2Glc1247, plex2Glc1292, and plex2Gal2019, representing the sequences from the target strains, were selected for subsequent transformation into H. influenzae.
To inactivate the lex2B gene, the Kanr cassette from pUC4Kan was inserted into a unique StyI site located 209 bp downstream of the initiation codon of lex2B in plasmid pCR2.1lex2 (14). This plasmid construct was introduced into E. coli, and a correct plasmid clone (pDL1) was verified by restriction digestion and PCR amplification.
Transformation of H. influenzae.
For allelic exchange, plasmid DNA containing the cloned lex2 genes and flanking sequences was linearized by restriction digestion and then transformed into selected H. influenzae strains by the MIV method (16). Putative transformed colonies were regrown on BHI agar plates containing kanamycin. PCR amplification products obtained using primers lex2RegForA and lex2CHP1 were sequenced to confirm that correct and complete allelic exchange of the lex2 locus had occurred in the target strain.
The lex2 locus was inactivated in strains 432, 1008, 1124, 1247, 1292, and 2019 by transformation with linearized plasmid pDL1. Following transformation and regrowth on kanamycin-containing medium, correct mutant strains were confirmed by PCR amplification of extracted DNA with primers blex2for and lex2CHP1.
LPS phenotypic analysis.
For gel fractionation of LPS, whole-cell lysates were prepared from H. influenzae strains grown overnight as described by Serino and Virgi (37). The lysates were fractionated by Tricine-SDS- polyacrylamide gel electrophoresis (PAGE) (24), and LPS profiles were detected by staining with silver (Quicksilver; GE Healthcare).
For immunoblotting, H. influenzae strains grown overnight in sectors on BHI agar plates were incubated with the appropriate LPS-specific MAb as described previously (9). MAb 4C4 was provided by E. J. Hansen (University of Texas), and MAb LLA5 was provided by J. Richards (National Research Council, Canada).
LPS structural analysis.
The LPS structures from NTHi transformant strains 1008lex2Gal1124 and 1124lex2Glc1008 were elucidated using a combination of mass spectrometry and nuclear magnetic resonance (NMR) techniques and a protocol outlined previously (40). Briefly, cells from 5-liter batch cultures (five 1-liter lots) were harvested after overnight growth, and LPS was extracted from lyophilized bacteria using phenol-chloroform-light petroleum. Reduced core OS material was obtained after mild acid hydrolysis of LPS. The results of multiple-step tandem electrospray ionization-mass spectrometry ESI-MS (ESI-MSn) experiments performed with permethylated and dephosphorylated OS samples were recorded in the positive ion mode with a Finnigan LCQ iontrap mass spectrometer (Finnigan-MAT, San Jose, CA) coupled to a 2690 Waters high-performance liquid chromatography system (Waters, Milford, MA) using a microbore C18 column [Phenomenex LUNA 5 μm C18(2)] at 21°C. The mobile phase gradient used had constant concentrations of 1% acetic acid and 0.1 mM sodium acetate and was changed gradually over time so that it contained methanol (solvent A) and water (solvent B) at a flow rate of 0.08 ml min−1. The gradient was as follows: 50% solvent A at 0 min, 100% solvent A at 50 min, 100% solvent A at 60 min, and 50% solvent A at 70 min.
NMR spectra were recorded for OS samples in deuterium oxide (D2O) at 22°C and 25°C. Spectra were acquired with a JEOL 500-MHz spectrometer using standard pulse sequences as previously described (40). For interresidue correlation, two-dimensional nuclear Overhauser effect (NOE) spectroscopy experiments with a mixing time of 250 ms were used. Total correlation spectroscopy experiments were done using a mixing time of 180 ms.
Nucleotide sequence accession numbers.
The GenBank accession numbers for sequences determined in this study are FJ376687 to FJ376710.
RESULTS
In H. influenzae type b strains, lex2B has been shown to encode the glycosyltransferase required to add the second glucose in the OS extension from HepI of the LPS (Fig. 1). In studies of the LPS structure of NTHi strains, a galactose has been identified at the same position in a minority of strains, including NTHi strains 1124 (40) and 2019 (11, 31). To test the hypothesis that the alternative additions of glucose and galactose were dependent on the lex2 locus, we sequenced the lex2A and lex2B genes of 25 of the 29 H. influenzae strains examined. The locus was not amplified by PCR in four strains (NTHi strains 723, 1158, 1159, and Rd), and Southern blot analysis confirmed that the locus was not present in strains 1158, 1159, and Rd, although it was present in strain 723 (data not shown). The lack of PCR amplification of the lex2 locus in strain 723 could have resulted from polymorphisms in the primer binding regions or from the locus being present in a different chromosomal location. The sequence of each gene was highly similar between strains (there was at least 97.9% DNA sequence identity between strains), and translated amino sequences are shown aligned in Fig. 2. The size of the combined lex2A and lex2B reading frames is approximately 1,060 bp. lex2A is the smaller gene and has a reading frame around 320 bp long that contains the tetranucleotide repeat tract, in which the number of 5′-GCAA repeats varies from 3 to 41 between strains (Table 2). The lex2A and lex2B reading frames are separated by a small 13-bp intergenic region. It has been reported that the glycosyltransferase encoded by lex2B is functional only if lex2A is expressed (“on”) (14, 21); this occurs when there are 3+3n repeats in lex2A. Thus, in 9 of the 25 strains lex2 expression would be predicted to be “on,” and in the rest of the strains it would be predicted to be “off” (Table 2). The Lex2A sequence shares no significant homology to the sequences of other proteins in the databases. It is interesting that the lex2A reading frames of NTHi strains 162, 1003, and 1247 contained a 5-bp deletion that introduced a frameshift, likely resulting in an inactive Lex2A protein. The lex2B reading frame is approximately 740 bp long and encodes a glycosyltransferase (14). The lex2B gene of strain 162 contains a 2-bp deletion 200 bp into the reading frame, resulting in a frameshift. Based on the assumption that Lex2B adds a sugar in an equivalent position in the LPS in strains, we hypothesized from structural data available for the LPS of our 29 strains that Lex2B was able to add either a glucose or a galactose depending on its sequence.
FIG. 2.
Alignment of a segment of the translated amino acid sequences encoded by the lex2B gene for 25 strains. Shading indicates polymorphisms between the sequences, in particular, at the position of amino acid 157, which can be either a threonine (T) or an alanine (A). Strain 162 has a predicted truncated open reading frame due to a 2-bp deletion.
TABLE 2.
Number of tetranucleotide repeats, expression status of lex2A, and predicted specificity of hexose addition in our test strains
| Strain | No. of repeats in lex2A | Lex2A expressiona | DNA sequenceb | Amino acid in sequencec | Predicted hexose addedd |
|---|---|---|---|---|---|
| 162 | 25 | Offe | GCT | A | (Glc) |
| 176 | 23 | Off | GCT | A | (Glc) |
| 285 | 41 | Off | ACT | T | (Gal) |
| 375 | 19 | Off | ACT | T | (Gal) |
| 432 | 18 | On | ACT | T | Gal |
| 477 | 15 | On | GCT | A | Glc |
| 486 | 22 | Off | GCT | A | (Glc) |
| 667 | 25 | Off | GCT | A | Glc |
| 723 | NDf | ND | Present | ||
| 981 | 12 | On | GCT | A | Glc |
| 1003 | 14 | Offe | GCT | A | (Glc) |
| 1008 | 9 | On | GCT | A | Glc |
| 1124 | 3 | On | ACT | T | Gal |
| 1158 | Absent | ||||
| 1159 | Absent | ||||
| 1180 | 18 | On | GCT | A | Glc |
| 1181 | 18 | On | GCT | A | Glc |
| 1200 | 17 | Off | GCT | A | (Glc) |
| 1207 | 25 | Off | ACT | T | (Gal) |
| 1209 | 26 | Off | ACT | T | (Gal) |
| 1231 | 20 | Off | GCT | A | Glc |
| 1232 | 20 | Off | GCT | A | Glc |
| 1233 | 26 | Off | ACT | T | (Gal) |
| 1247 | 15 | (On)e | GCT | A | Glc |
| 1268 | 18 | On | GCT | A | (Glc) |
| 1292 | 18 | On | GCT | A | Glc |
| 2019 | 19 | Off | ACT | T | Gal |
| Rd | NP | ||||
| Eagan | 20 | Off | GCT | A | (Glc) |
The locus is predicted to be expressed (“on”) for 3+3n repeats.
DNA sequence encoding amino acid 157 in the lex2B gene. Present, present as determined by Southern blotting; absent, absent as determined by Southern blotting; NP, not present in the genome sequence.
Amino acid at position 157 for each lex2B gene.
Bold type indicates that the predicted hexose agrees with the known LPS structure; parentheses indicate that the predicted hexose is not present due to a lack of lex2A expression or an alternative extension in the LPS; and underlining indicates that the LPS structure is unknown.
The lex2A sequence contains a 5-bp deletion.
ND, not determined.
When the translated sequences of lex2B were aligned, there were a total of 21 polymorphic sites. A correlation between the specific Lex2B sequence and the hexose (HexII in Fig. 1) that is known to occur at the second position in the OS extension from HepI in the LPS from the strain was observed (Table 2). The specificity of addition of either a glucose or a galactose correlated with a single amino acid change at position 157 in Lex2B (Fig. 2), a position in the glycosyltransferase sequence similar to that observed for a related characteristic in previous studies of LpsA (6). Eight of 25 strains contain a threonine (T) at position 157, corresponding with the predicted potential to express a galactose in the LPS at the HexII position (Table 2). Conversely, the other strains contain an alanine (A) at position 157, consistent with Lex2B being able to add a glucose.
Based on the Lex2B sequence data and our knowledge of the LPS structures of the strains, we chose six NTHi strains, strains 432, 1008, 1124, 1247, 1292, and 2019, for further study. Lex2B from strains 432, 1124, and 2019 is predicted to add a galactose, and LPS structural studies have confirmed that a galactose residue is indeed found as HexII in each of these strains (11, 40; unpublished data). For the other three strains, Lex2B is predicted to add a glucose, a finding that is also supported by LPS structural data available for each strain (E. K. H. Schweda, unpublished data).
To confirm our prediction of different functions for allelic variants of lex2, plasmid constructs containing the lex2 locus cloned from strains in which the locus normally directs addition of a glucose (strains 1008, 1247, and 1292) were used for transformation to exchange the lex2 genes by allelic replacement in strains that naturally add a galactose (strains 432, 1124, and 2019). Reciprocal transformations were also carried out using the glucose-specific lex2 plasmid clones and the target strains that naturally express a galactose in the LPS. As a control for the possibility that insertion of the Kanr cassette into the intergenic region adjacent to lex2 influenced LPS structure, lex2 plasmid constructs were also transformed into the homologous strains. Transformed H. influenzae strains were designated as follows: recipient strain followed by lex2 followed by the sugar added by the donor strain, with the donor strain indicated by a subscript. For example, if strain 1008 was transformed with the lex2 locus from 1124 (a strain that normally adds a galactose as HexII [Fig. 1]), the resulting strain was designated 1008lex2Gal1124. The DNA sequence of the lex2 locus in each transformed strain was determined to ascertain that the complete locus had been replaced by reciprocal recombination and that the number of tetranucleotide repeat units was compatible with Lex2 expression. Table 3 lists the transformed strains. In later analyses we focused on transformed strains made using plasmid constructs derived from strains 432, 1008, 1124, and 1292, and each transformed strain was shown by DNA sequencing to contain a lex2 locus with an “on” number of repeats. The lex2 locus was also inactivated in each of the six strains chosen for study (Table 3).
TABLE 3.
Relative levels of MAb binding
| Strain | Level of binding toa:
|
|
|---|---|---|
| MAb 4C4 | MAb LLA5 | |
| 1008 | ++++ | + |
| 1008lex2Glc1008b | ++++ | + |
| 1008lex2Gal1124 | + | + |
| 1008lex2B | + | + |
| 1124 | ++ | + |
| 1124lex2Gal1124 | ++ | + |
| 1124lex2Glc1008 | ++ | + |
| 1124lex2B | ++ | + |
| 1247 | ++++ | ++ |
| 1247lex2Glc1247 | ++++ | ++ |
| 1247lex2Gal432 | ++ | ++ |
| 1247lex2Gal1124 | ++ | ++ |
| 1247lex2Glc1008 | ++++ | ++ |
| 1247lex2B | ++ | ++ |
| 2019 | +++ | ++++ |
| 2019lex2Gal2019 | ++ | ++++ |
| 2019lex2Glc1247 | ++ | ++++ |
| 2019lex2Glc1292 | ++ | ++ |
| 2019lex2B | ++ | +++ |
Relative levels of MAb binding to strains compared to the corresponding parent and lex2 mutant strains. ++++, strong binding; +++, intermediate binding; ++, weak binding; +, background.
Bold type indicates allelic exchange strains investigated in this study.
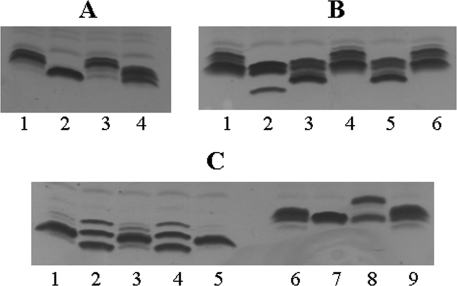
The LPS phenotypes of allelic exchange and mutant strains were first investigated by comparing the electrophoretic profiles to those of the corresponding wild-type strains following Tricine-SDS-PAGE (Fig. 3). For strain 1008, a strain in which Lex2B would normally be predicted to add a glucose as HexII (Fig. 1), the LPS profile of strain 1008lex2Glc1008 was the same as that of the parent strain (Fig. 3A). In contrast, the 1008 lex2B mutant strain had a truncated LPS profile, consistent with the loss of one or two sugars from the major glycoforms (Fig. 3A). The LPS profile for the allelic exchange strain 1008lex2Gal1124, in which the lex2 locus was replaced by the locus from strain 1124 and was expected to add a galactose, is intermediate between the lex2B mutant and parent patterns. This is consistent with inclusion of a galactose in LPS glycoforms at a position that might lead to shorter oligosaccharide chains from HepI. Strain 1247, another strain in which lex2 naturally adds a glucose, displayed conservation of the LPS profile similar to that of the parent strain following allelic exchange in strains 1247lex2Glc1008 and 1247lex2Glc1247 (Fig. 3B), indicating that homologous and heterologous Glc-specific lex2B alleles encode the same function. When transformed with a galactose-specific allele, strains 1247lex2Gal432 and 1247lex2Gal1124 have truncated LPS glycoforms (Fig. 3B). This indicates that independent Gal-specific lex2B genes have equivalent functions once they are inserted into the genome of strain 1247. The LPS from mutant strain 1247lex2B displayed the most truncated LPS profile, which is consistent with it having no hexose extension off GlcI in the major glycoforms (Fig. 3B).
FIG. 3.
Electrophoretic profiles of LPS from the NTHi parent and corresponding allelic exchange and mutant strains. (A) Strain 1008 and corresponding mutant and allelic exchange strains. Lane 1, 1008; lane 2, 1008lex2B; lane 3, 1008lex2Glc1008; lane 4, 1008lex2Gal1124. (B) Strain 1247 and corresponding mutant and allelic exchange strains. Lane 1, 1247; lane 2, 1247lex2B; lane 3, 1247lex2Gal432; lane 4, 1247lex2Glc1008; lane 5, 1247lex2Gal1124; lane 6, 1247lex2Glc1247. (C) Strains 2019 and 1124 and corresponding mutant and allelic exchange strains. Lane 1, 2019; lane 2, 2019lex2B; lane 3, 2019lex2Glc1247; lane 4, 2019lex2Glc1292; lane 5, 2019lex2Gal2019; lane 6, 1124; lane 7, 1124lex2B; lane 8, 1124lex2Glc1008; lane 9, 1124lex2Gal1124.
Strain 1124, a strain in which Lex2B would normally be predicted to add a galactose, showed an LPS profile for the lex2 mutant strain different from that of the parent strain, with the upper band of the two major bands absent (Fig. 3C). As expected, the allelic exchange involving the homologous gene (1124lex2Gal1124) resulted in an LPS profile that is highly similar to that of the parent strain. For strain 1124lex2Glc1008 expressing the different functional category of the lex2 locus (i.e., glucose specific), there is an increase in the size of some LPS glycoforms compared to the parent strain, likely due to addition of a glucose as HexII and perhaps further extensions from this glucose. For 2019, another strain where lex2 naturally adds a galactose, the situation is somewhat more complex. There is a clear difference between the LPS profile of the lex2 mutant strain and that of the parent, and both lower- and higher-molecular-weight glycoforms are present. Strain 2019lex2Gal2019 displayed an LPS profile equivalent to that of the parent strain, and the allelic exchange strain 2019lex2Glc1292 expressed an LPS pattern similar to that seen for the lex2 mutant. This observation was explained after sequencing of the lex2A repeat tract in the transformed strain. A number of 5′-GCAA repeats inappropriate for expression of lex2A were present in the genome following transformation and culture; a lex2 “off” phenotype was expected to be equivalent to that of a lex2 mutant strain. For 2019lex2Glc1247, the LPS pattern was similar to that of the wild type, indicating that some lex2-dependent glycoform extension had occurred in this strain.
Immunoblotting analysis of this set of strains using MAbs 4C4 and LLA5 revealed some differences in binding between isogenic strains. MAb 4C4 interacts with a digalactoside epitope when it is present at some positions in the LPS of H. influenzae (14), particularly when it is located at the terminus of a globotetraose extension from either HepI or HepII (Fig. 1). A globotetraose extension is found in GlcI of the LPS of NTHi strain 1008 (E. K. H. Schweda, unpublished). Consistent with this, the wild-type strain reacted with MAb 4C4, while the lex2 mutant strain and the allelic exchange strain 1008lex2Gal1124 did not react with MAb 4C4, confirming that the corresponding epitopes had been altered in the LPS of these strains. A similar observation was made for NTHi strain 1247; the wild type (in which lex2 incorporates a Glc) reacted with MAb 4C4, yet two independent allelic exchange strains, 1247lex2Gal432 and 1247lex2Gal1124, did not react with this MAb. With MAb LLA5, an antibody that reacts with the high-molecular-weight extensions that can be attached to GlcI (Fig. 1) in some strains (20), only wild-type strain 2019 had a significant level of reactivity (Table 3). This reactivity was not greatly diminished in the lex2 mutant strain but was significantly reduced in the allelic exchange strain 2019lex2Glc1292. This is assumed to reflect an alteration of the balance of alternative OS extensions that can be synthesized from HepI in this strain background, which exhibits a complex LPS expression pattern.
The structure of OS material from NTHi strains 1124lex2Glc1008 and 1008lex2Gal1124 was investigated in detail by using ESI-MSn and NMR, and the resulting data were compared with the data for the corresponding parent strains. Detailed information on the structure of LPS glycoforms for wild-type strains 1124 (40) and 1008 (E. K. H. Schweda, unpublished) is available from previous studies. ESI-MSn performed with dephosphorylated and permethylated OS is a powerful tool for providing sequence and branching information for the various isomeric glycoforms present in heterogeneous mixtures. Our focus was to profile OS extensions from HepI.
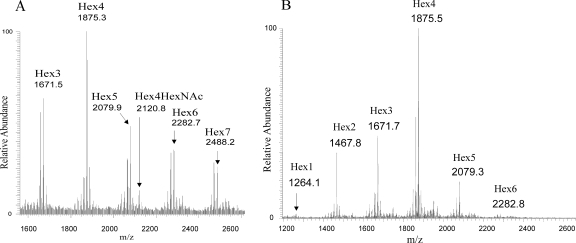
In the ESI-MS spectrum of 1124lex2Glc1008, six sodiated adduct ions ([M+Na]+) were observed at m/z 1671.5, 1875.3, 2079.9, 2120.8, 2282.7, and 2488.2, corresponding to Hex3, Hex4, Hex5, Hex4·HexNAc, Hex6, and Hex7 glycoforms (Fig. 4A). In order to obtain sequence and branching information, these molecular ions were further fragmented in MS2 and MS3 experiments. One Hex7 glycoform was observed by fragmenting the molecular ion at m/z 2488.2. The isomer was defined by the resulting fragment ions at m/z 2269.9, 2065.7, 1861.7, 1613.6, and 1365.9 corresponding to consecutive loss of residues from tHex-Hex-Hex-HepIII-HepII. The structure was confirmed in MS3 experiments with m/z 1613.6, which resulted in ions at m/z 1396.0, 1192.8, 987.3, and 785.4 due to loss of terminal tHex-Hex-Hex-Hex. By analogy, structures were determined for the remaining glycoforms (data not shown). It could be concluded that strain 1124lex2Glc1008 expresses LPS glycoforms similar to those of wild-type NTHi strain 1124 (40) except that they do not have HexNAc-Hex-Hex-Hex but rather have Hex-Hex-Hex-Hex extending from HepI.
FIG. 4.
High-performance liquid chromatography-ESI-MS full-scan spectra (positive mode) for dephosphorylated and permethylated OS of 1124lex2Glc1008 (A) and 1008lex2Gal1124 (B). The singly charged sodiated ions observed correspond to the following molecules: m/z 1264.1, Hex1·Hep3·reduced anhydro-2-keto-3-deoxyoctulosonic acid (AnKdo-ol); m/z 1467.8, Hex2·Hep3·AnKdo-ol; m/z 1671.7, Hex3·Hep3·AnKdo-ol; m/z 1875.5, Hex4·Hep3·AnKdo-ol; m/z 2079.3, Hex5·Hep3·AnKdo-ol; m/z 2120.8, HexNAc·Hex4·Hep3·AnKdo-ol; m/z 2282.8, Hex6·Hep3·AnKdo-ol; and m/z 2486.6, Hex7·Hep3·AnKdo-ol.
In the ESI-MS spectrum of 1008lex2Gal1124, six sodiated adduct ions ([M+Na]+) were observed at m/z 1264.1, 1467.8, 1671.7, 1875.5, 2079.3, and 2282.8 corresponding to Hex1 to Hex6 glycoforms (Fig. 4B). One Hex6 glycoform was observed by fragmenting the molecular ion at m/z 2282.8. The isomer was defined by the resulting fragment ions at m/z 2065.6, 1861.2, 1657.6, 1409.9, and 1161.7 corresponding to consecutive loss of residues from tHex-Hex-Hex-HepIII-HepII. The structure was confirmed in MS3 experiments for m/z 1409.9, which resulted in ions at m/z 1192, 987.8, and 783.6 due to loss of terminal tHex-Hex-Hex. Similarly, structures were obtained for the remaining glycoforms (data not shown). It was concluded that 1008lex2Gal1124 expresses LPS glycoforms that are structurally similar to those of NTHi wild-type strain 1124 (40) but not to LPS glycoforms containing HexNAc. 1008lex2Gal1124 did not express any of the Hex7 glycoforms that NTHi wild-type strain 1008 displays (E. K. H. Schweda, unpublished data), nor does it have any tetrasaccharide units off HepI in its LPS.
The identity of the hexose linked to GlcI in OS was established by one-dimensional and two-dimensional NMR experiments as described previously (40). In the 1H NMR spectra, the anomeric proton resonances from HepI, HepII, and HepIII residues were typically recognized between 5.0 and 6.0 ppm. As described previously, the anomeric protons of HepI and HepIII have close chemical shifts occurring between 5.0 and 5.2 ppm, whereas the anomeric proton of HepII occurs further downfield between 5.5 and 5.9 ppm (35).
The anomeric proton of GlcI is easily identified by interresidue NOE connectivities between proton pairs GlcI H1 and HepI H4/H6 (35). The chemical shift of the anomeric proton of Glc linked to O-2 of HepIII (GlcII) is easily identified due to interresidue NOE connectivities between H1 GlcII H1 and HepIII H1/H2. The chemical shift of the anomeric proton of Gal/Glc (GlcIII) linked to O-4 of Glc I has previously been shown to resonate exceptionally downfield at δ 4.8 in glycoforms in which HepII does not have oligosaccharide extensions and HepIII is replaced by globotetraose (40; unpublished data). In glycoforms with truncated versions of globotetraose linked to HepIII, the anomeric proton of Gal linked to GlcI shifts more upfield and occurs in a region between δ 4.77 and 4.64 (40).
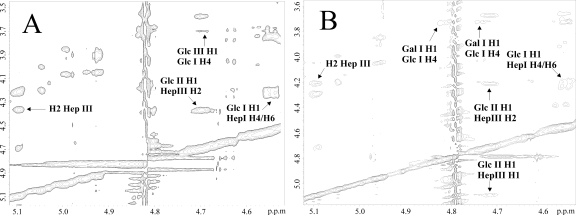
In the 1H NMR spectrum of 1124lex2Glc1008, anomeric resonances corresponding to the triheptosyl moiety (HepI to HepIII) were identified at δ 5.04, 5.68, and 5.09, respectively. The typical resonances of a Gal residue linked to GlcI as observed by Yildirim and colleagues were absent (40). The anomeric proton of GlcI was assigned to δ 4.54 based on its interresidue NOE to H4/H6 of HepI (Fig. 5A). H-4 of GlcI was found at δ 3.70 (data not shown). The anomeric proton of GlcII was assigned to δ 4.69 based on its interresidue NOE to H1/H2 of HepIII (Fig. 5A). We concluded that the anomeric proton of GlcIII overlapped that of GlcII at δ 4.69 based on the occurrence of an interresidue NOE to H-4 of GlcI at δ 3.70. This was corroborated by an heteronuclear multiple-bond correlation experiment in which H-4 of GlcI showed connectivity to C-1 of GlcIII at δ 103.00 (data not shown). Thus, in strain 1124lex2Glc1008 a Glc residue replaced O-4 of GlcI instead of Gal, as has been described previously for the LPS of the parent strain (40).
FIG. 5.
Selected regions of two-dimensional gradient NOE spectroscopy spectra (mixing time, 250 ms) of OS material derived from strains 1124lex2Glc1008 (A) and 1008lex2Gal1124 (B). Cross peaks of significant interresidue NOE connectivities are labeled for GlcI, GlcII, GalI, and GlcIII.
In the 1H NMR spectrum of 1008lex2Gal1124, anomeric resonances corresponding to the triheptosyl moiety (HepI to HepIII) were identified at δ 5.05, 5.62, and 5.09, respectively. Interestingly, two anomeric protons of Gal residues observed at δ 4.82 and 4.74 were absent in the corresponding spectra of the wild-type strain (unpublished data). The anomeric proton of GlcI was assigned to δ 4.55 based on its interresidue NOE to H4/H6 of HepI (Fig. 5B). H-4 of GlcI was observed at δ 3.72 (data not shown). The anomeric proton of GlcII was assigned to δ 4.71 based on its interresidue NOE to H1/H2 of HepIII (Fig. 5B). The anomeric protons at δ 4.82 and 4.74 both showed interresidue NOE to H-4 of GlcI (Fig. 5B). Thus, Gal replaced O-4 of GlcI in the LPS of strain 1008lex2Glc1124, unlike the Glc found at the same position in the parent strain. The occurrence of two anomeric signals of this Gal residue is in agreement with the observation of longer and shorter extensions from HepIII.
Some biological significance for the differential function of the lex2 genes was sought for these allelic exchange strains through the bactericidal assay; changes in LPS glycoforms often correlate to altered susceptibility of the expressing strain to serum killing. For strain 1247, the serum resistance of the allelic exchange strains1247lex2Glc1247 and 1247lex2Glc1008, each with a glucose included as HexII, was very similar to that of the parent strain (data not shown). For the lex2 mutant and the galactose-specific exchange strains 1247lex2Gal1124 and 1247lex2Gal432, serum resistance was somewhat reduced compared to that of the parent strain (data not shown), a finding consistent with truncation of the LPS glycoforms observed upon SDS-PAGE. For strain 1124, the homologous exchange strain again showed resistance to killing equivalent to that of the parent strain, and the lex2 mutant derivative showed some reduction in serum resistance (data not shown). The allelic exchange strain 1124lex2Glc1008, with a Gal swapped for a Glc, was less resistant to killing; this correlates with an apparent increased glycoform size for LPS compared to the wild type (Fig. 3C). For NTHi strains 1008 and 2019 there was little apparent change in serum resistance within each set of isogenic strains tested (data not shown).
DISCUSSION
A characteristic of the LPS of H. influenzae is the intrastrain heterogeneity of structure generated through phase variation by frequent and reversible “on”-“off” switching of translation of LPS biosynthetic loci containing tetranucleotide repeat tracts. The combinatorial effect of having multiple phase-variable genes in one strain provides a potential for extensive heterogeneity of glycoform structure. We have now shown that one of these loci, lex2, is the source of further heterogeneity of LPS structure through allelic sequence variation. Lex2B is the first glycosyltransferase in H. influenzae whose expression is phase variable and that is able to add alternative sugars at the same position in the H. influenzae LPS molecule.
Lex2B is a member of family 25 of the glycosyltransferase A superfamily of glycosyltransferases that transfer an activated monosaccharide residue (from UDP-Glc or UDP-Gal) to an acceptor molecule, which in this case is LPS. This is the same glycosyltransferase family that contains another H. influenzae LPS glycosyltransferase, LpsA, which has been extensively studied in our laboratory and shown to add either a glucose or a galactose in a β1-2 or β1-3 linkage in LPS biosynthesis (6). The glycosyltransferase A superfamily 25 glycosyltransferases possess a DXD motif, found at positions 99 to 101 in the Lex2B sequence (data not shown), a conserved amino acid motif that binds Mn2+ and is part of the catalytic domain for the donor sugar. Lex2B is predicted to use an inverting catalytic mechanism in which the α-linked donor sugar is transferred to the LPS to form a β-linked product. GlcI, the sugar residue which acts as the acceptor for Lex2B, is universally present in all H. influenzae LPS structures elucidated to date. It is therefore not unsurprising that the lex2 locus is found in a majority of NTHi strains tested, although this is not the only means by which sugars can be added to GlcI. GlcI can also be replaced by either an ld-Hep residue (28) or a dd-Hep residue (27) or by addition of two alternative four-sugar units (5), making the OS extensions from HepI the most heterogeneous OS extensions in structure from the H. influenzae LPS inner core.
The specificity of Lex2B for adding a glucose or a galactose is predicted to be at amino acid 157, where there is an absolute correlation between the presence of a T directing a galactose addition and the presence of an A directing a glucose addition. This is a location similar to that of the amino acid at position 151 that was shown by site-directed mutagenesis to direct specificity for LpsA, where again a T was required for galactose incorporation (6). Indeed, this specificity is likely conserved in family 25 glycosyltransferases across a range of bacterial and other species. Position 157 of Lex2B is the only variant residue in a region of the protein sequence that is absolutely conserved in all NTHi sequences investigated (Fig. 2).
Adding the second glucose in the extension from HepI of the LPS inner core backbone, as seen for Lex2B in serotype b strains in previous studies (14, 21) and NTHi strain 1008 in our study, facilitates further oligosaccharide extension that includes digalactoside-containing moieties (Gal-α-1 to 4-Gal-β-1 to 4-Glc-β-1 to 4-Glc-α) that bind MAbs 4C4 and 5G8 (13, 14, 22). The presence of these digalactoside moieties can have biological consequences, and several studies have shown that the digalactoside on H. influenzae LPS can enhance the ability of the organism to escape the bactericidal activity of serum in in vitro assays and facilitates intravascular survival in the infant rat model of H. influenzae infection (4, 10, 12, 19, 22, 23, 39). The importance to the bacterium of varying the expression of the Gal-Gal-Glc moiety added to GlcI can be inferred from the fact that addition of each of these three consecutive sugars is dependent upon the action of an independent phase variably expressed glycosyltransferase (12).
The mechanism by which the lex2A reading frame and the repeats in this reading frame affect the phase-variable expression of the Lex2B phenotype is not clear. A relationship between the numbers of repeat units in the lex2A tract and incorporation of the digalactoside (measured by MAb 5G8 binding) has previously been shown (21). The genes are cotranscribed (14), and it has been shown that in Helicobacter pylori expression of cotranscribed genes can co-phase vary through transcriptional-translational coupling (8). However, in the study of Griffin et al., translational fusions with a reporter gene indicated that each gene is translated independently, and a nonpolar mutation of lex2A resulted in an inactive Lex2B phenotype (14). In our study, strain 1247 contained a lex2A sequence with a 5-bp deletion, which implies that Lex2A was inactive, but this does not correspond with LPS structural data that indicate that Lex2B is active (i.e., the LPS contains a hexose attached to GlcI [unpublished]). Data base searches with the Lex2A sequence reveal no significant matches (data not shown), and thus the contribution of this sequence, which is unique to H. influenzae glycosyltransferases, to the function of the lex2 locus remains unclear and requires further study.
Acknowledgments
M.E.D., D.W.H., and E.R.M. were supported by the U.K. Medical Research Council.
Editor: J. N. Weiser
Footnotes
Published ahead of print on 16 March 2009.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 253389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, P., R. B. Johnston, Jr., and D. H. Smith. 1972. Human serum activities against Hemophilus influenzae, type b. J. Clin. Investig. 5131-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cody, A. J., D. Field, E. J. Feil, S. Stringer, M. E. Deadman, A. G. Tsolaki, B. Gratz, V. Bouchet, R. Goldstein, D. W. Hood, and E. R. Moxon. 2003. High rates of recombination in otitis media isolates of non-typeable Haemophilus influenzae. Infect. Genet. Evol. 357-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cope, L. D., R. Yogev, J. Mertsola, J. C. Argyle, G. H. McCracken, Jr., and E. J. Hansen. 1990. Effect of mutations in lipooligosaccharide biosynthesis genes on virulence of Haemophilus influenzae type b. Infect. Immun. 582343-2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cox, A. D., D. W. Hood, A. Martin, K. M. Makepeace, M. E. Deadman, J. Li, J. R. Brisson, E. R. Moxon, and J. C. Richards. 2002. Identification and structural characterization of a sialylated lacto-N-neotetraose structure in the lipopolysaccharide of Haemophilus influenzae. Eur. J. Biochem. 2694009-4019. [DOI] [PubMed] [Google Scholar]
- 6.Deadman, M. E., S. L. Lundström, E. K. Schweda, E. R. Moxon, and D. W. Hood. 2006. Specific amino acids of the glycosyltransferase LpsA direct the addition of glucose or galactose to the terminal inner core heptose of Haemophilus influenzae lipopolysaccharide via alternative linkages. J. Biol. Chem. 28129455-29467. [DOI] [PubMed] [Google Scholar]
- 7.Devereux, J., P. Haeberli, and O. Smithies. 1984. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Vries, N., D. Duinsbergen, E. J. Kuipers, R. G. Pot, P. Wiesenekker, C. W. Penn, A. H. van Vliet, C. M. Vandenbroucke-Grauls, and J. G. Kusters. 2002. Transcriptional phase variation of a type III restriction-modification system in Helicobacter pylori. J. Bacteriol. 1846615-6623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dixon, K., C. D. Bayliss, K. Makepeace, E. R. Moxon, and D. W. Hood. 2007. Identification of the functional initiation codons of a phase-variable gene of Haemophilus influenzae, lic2A, with the potential for differential expression. J. Bacteriol. 189511-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Erwin, A. L., S. Allen, D. K. Ho, P. J. Bonthuis, J. Jarisch, K. L. Nelson, D. L. Tsao, W. C. Unrath, M. E. Watson, Jr., B. W. Gibson, M. A. Apicella, and A. L. Smith. 2006. Role of lgtC in resistance of nontypeable Haemophilus influenzae strain R2866 to human serum. Infect. Immun. 746226-6235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gaucher, S. P., M. T. Cancilla, N. J. Phillips, B. W. Gibson, and J. A. Leary. 2000. Mass spectral characterization of lipooligosaccharides from Haemophilus influenzae 2019. Biochemistry 3912406-12414. [DOI] [PubMed] [Google Scholar]
- 12.Griffin, R., C. D. Bayliss, M. A. Herbert, A. D. Cox, K. Makepeace, J. C. Richards, D. W. Hood, and E. R. Moxon. 2005. Digalactoside expression in the lipopolysaccharide of Haemophilus influenzae and its role in intravascular survival. Infect. Immun. 737022-7026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Griffin, R., A. D. Cox, K. Makepeace, J. C. Richards, E. R. Moxon, and D. W. Hood. 2005. Elucidation of the monoclonal antibody 5G8-reactive, virulence-associated lipopolysaccharide epitope of Haemophilus influenzae and its role in bacterial resistance to complement-mediated killing. Infect. Immun. 732213-2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Griffin, R., A. D. Cox, K. Makepeace, J. C. Richards, E. R. Moxon, and D. W. Hood. 2003. The role of lex2 in lipopolysaccharide biosynthesis in Haemophilus influenzae strains RM7004 and RM153. Microbiology 1493165-3175. [DOI] [PubMed] [Google Scholar]
- 15.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166557-580. [DOI] [PubMed] [Google Scholar]
- 16.Herriott, R. M., E. M. Meyer, and M. Vogt. 1970. Defined nongrowth media for stage II development of competence in Haemophilus influenzae. J. Bacteriol. 101517-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.High, N. J., M. E. Deadman, and E. R. Moxon. 1993. The role of a repetitive DNA motif (5′-CAAT-3′) in the variable expression of the Haemophilus influenzae lipopolysaccharide epitope alpha Gal(1-4)beta Gal. Mol. Microbiol. 91275-1282. [DOI] [PubMed] [Google Scholar]
- 18.Hood, D. W., M. E. Deadman, A. D. Cox, K. Makepeace, A. Martin, J. C. Richards, and E. R. Moxon. 2004. Three genes, lgtF, lic2C and lpsA, have a primary role in determining the pattern of oligosaccharide extension from the inner core of Haemophilus influenzae LPS. Microbiology 1502089-2097. [DOI] [PubMed] [Google Scholar]
- 19.Hood, D. W., M. E. Deadman, M. P. Jennings, M. Bisercic, R. D. Fleischmann, J. C. Venter, and E. R. Moxon. 1996. DNA repeats identify novel virulence genes in Haemophilus influenzae. Proc. Natl. Acad. Sci. USA 9311121-11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hood, D. W., G. Randle, A. D. Cox, K. Makepeace, J. Li, E. K. Schweda, J. C. Richards, and E. R. Moxon. 2004. Biosynthesis of cryptic lipopolysaccharide glycoforms in Haemophilus influenzae involves a mechanism similar to that required for O-antigen synthesis. J. Bacteriol. 1867429-7439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jarosik, G. P., and E. J. Hansen. 1994. Identification of a new locus involved in expression of Haemophilus influenzae type b lipooligosaccharide. Infect. Immun. 624861-4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kimura, A., and E. J. Hansen. 1986. Antigenic and phenotypic variations of Haemophilus influenzae type b lipopolysaccharide and their relationship to virulence. Infect. Immun. 5169-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kimura, A., C. C. Patrick, E. E. Miller, L. D. Cope, G. H. McCracken, Jr., and E. J. Hansen. 1987. Haemophilus influenzae type b lipooligosaccharide: stability of expression and association with virulence. Infect. Immun. 551979-1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lesse, A. J., A. A. Campagnari, W. E. Bittner, and M. A. Apicella. 1990. Increased resolution of lipopolysaccharides and lipooligosaccharides utilizing tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis. J. Immunol. Methods 126109-117. [DOI] [PubMed] [Google Scholar]
- 25.Levinson, G., and G. A. Gutman. 1987. Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol. Biol. Evol. 4203-221. [DOI] [PubMed] [Google Scholar]
- 26.Lysenko, E., J. C. Richards, A. D. Cox, A. Stewart, A. Martin, M. Kapoor, and J. N. Weiser. 2000. The position of phosphorylcholine on the lipopolysaccharide of Haemophilus influenzae affects binding and sensitivity to C-reactive protein-mediated killing. Mol. Microbiol. 35234-245. [DOI] [PubMed] [Google Scholar]
- 27.Månsson, M., D. W. Hood, E. R. Moxon, and E. K. Schweda. 2003. Structural characterization of a novel branching pattern in the lipopolysaccharide from nontypeable Haemophilus influenzae. Eur. J. Biochem. 2702979-2991. [DOI] [PubMed] [Google Scholar]
- 28.Månsson, M., D. W. Hood, E. R. Moxon, and E. K. Schweda. 2003. Structural diversity in lipopolysaccharide expression in nontypeable Haemophilus influenzae. Identification of l-glycerol-d-manno-heptose in the outer-core region in three clinical isolates. Eur. J. Biochem. 270610-624. [DOI] [PubMed] [Google Scholar]
- 29.Masoud, H., A. Martin, P. Thibault, E. R. Moxon, and J. C. Richards. 2003. Structure of extended lipopolysaccharide glycoforms containing two globotriose units in Haemophilus influenzae serotype b strain RM7004. Biochemistry 424463-4475. [DOI] [PubMed] [Google Scholar]
- 30.Moxon, E. R., P. B. Rainey, M. A. Nowak, and R. E. Lenski. 1994. Adaptive evolution of highly mutable loci in pathogenic bacteria. Curr. Biol. 424-33. [DOI] [PubMed] [Google Scholar]
- 31.Phillips, N. J., M. A. Apicella, J. M. Griffiss, and B. W. Gibson. 1992. Structural characterization of the cell surface lipooligosaccharides from a nontypable strain of Haemophilus influenzae. Biochemistry 314515-4526. [DOI] [PubMed] [Google Scholar]
- 32.Phillips, N. J., M. A. Apicella, J. M. Griffiss, and B. W. Gibson. 1993. Structural studies of the lipooligosaccharides from Haemophilus influenzae type b strain A2. Biochemistry 322003-2012. [DOI] [PubMed] [Google Scholar]
- 33.Preston, A., D. Maskell, A. Johnson, and E. R. Moxon. 1996. Altered lipopolysaccharide characteristic of the I69 phenotype in Haemophilus influenzae results from mutations in a novel gene, isn. J. Bacteriol. 178396-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 35.Schweda, E. K., and J. C. Richards. 2003. Structural profiling of short-chain lipopolysaccharides from Haemophilus influenzae. Methods Mol. Med. 71161-183. [DOI] [PubMed] [Google Scholar]
- 36.Schweda, E. K., J. C. Richards, D. W. Hood, and E. R. Moxon. 2007. Expression and structural diversity of the lipopolysaccharide of Haemophilus influenzae: implication in virulence. Int. J. Med. Microbiol. 297297-306. [DOI] [PubMed] [Google Scholar]
- 37.Serino, L., and M. Virji. 2000. Phosphorylcholine decoration of lipopolysaccharide differentiates commensal Neisseriae from pathogenic strains: identification of licA-type genes in commensal Neisseriae. Mol. Microbiol. 351550-1559. [DOI] [PubMed] [Google Scholar]
- 38.Weiser, J. N., J. M. Love, and E. R. Moxon. 1989. The molecular mechanism of phase variation of H. influenzae lipopolysaccharide. Cell 59657-665. [DOI] [PubMed] [Google Scholar]
- 39.Weiser, J. N., A. Williams, and E. R. Moxon. 1990. Phase-variable lipopolysaccharide structures enhance the invasive capacity of Haemophilus influenzae. Infect. Immun. 583455-3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yildirim, H. H., J. Li, J. C. Richards, D. W. Hood, E. R. Moxon, and E. K. Schweda. 2005. An alternate pattern for globoside oligosaccharide expression in Haemophilus influenzae lipopolysaccharide: structural diversity in nontypeable strain 1124. Biochemistry 445207-5224. [DOI] [PubMed] [Google Scholar]