Abstract
Two-partner secretion systems of gram-negative organisms are utilized in adherence, invasion, and biofilm formation. The HrpAB proteins of Neisseria meningitidis are members of a two-partner secretion system, and HrpA is established as being important to adherence and intracellular escape. This study set out to determine the expression pattern of members of the hrpBAS putative operon and to find a functional role for the HrpA protein. The upregulation of these genes was found in situations of anaerobiosis and cell contact. These observations prompted the study of the function of HrpA in biofilms on human bronchial epithelial cells. HrpA mutants in encapsulated and unencapsulated NMB strains demonstrated biofilm growth equivalent to that of the wild-type strain at 6 h but a decreased ability to form biofilms at 48 h. Biofilms formed by hrpA mutants for 48 h on collagen-coated coverslips demonstrated significant reductions compared to those of wild-type strains. Taken together, these observations imply a role for HrpA in the biofilm structure. Further analysis demonstrated the presence of HrpA on the surface of the bacterium.
Neisseria meningitidis is the causative agent of meningococcal meningitis. It is a common isolate of the respiratory tract, living as a commensal in 5 to 10% of the population during periods in which infection is endemic (4). The primary adhesin for N. meningitidis is the type IV pilus, which is utilized as a long-range adherence factor. The pilus retracts after attachment to the host tissue, resulting in the intimate contact of the bacterium with the host respiratory tissue (53). Nonfimbrial adhesins such as Opa, Opc, NhhA, and NadA are important for intimate adherence to the host membrane (3, 41, 52).
Recently, a type V secretion system was described in N. meningitidis (42, 46, 48). There are two categories of type V secretion in gram-negative organisms. The first is the autotransporter, and the second is the two-partner secretion system. Both utilize the Sec transport system to cross the inner membrane. The autotransporter has its own secretion domain that embeds in the outer membrane and catalyzes its own secretion across the outer membrane (20). The two-partner system consists of two proteins generically referred to as TpsA and TpsB, in which the transporter TpsB embeds in the outer membrane and serves as a dedicated transporter for TpsA (27). This secreted protein often is a large adhesin. The best-characterized two-partner secretion system is the FhaBC system in Bordetella pertussis, in which the adhesin, FhaB, is secreted by the outer membrane transporter, FhaC (10, 17). FhaB is processed at the N-terminal end when it passes through the Sec pathway in the inner membrane and is processed again at the C-terminal end as it exits the outer membrane by SphB1, an autotransporter subtilisin-like protease (8, 22). There is some evidence that proteases in addition to SphB1 are involved in the final processing event (30). Completely processed FhaB is referred to as FHA. Common characteristics of the TpsA adhesins are a high molecular weight, an extended signal peptide, a predicted beta-solenoid shape for at least one domain of the protein, and a hemagglutination domain (2, 6, 22, 27).
The N. meningitidis two-partner secretion system consists of HrpAB, where HrpA is the putative adhesin and HrpB is the outer membrane transporter. HrpB secretes HrpA, which functions as an adhesin in an unencapsulated and lipooligosaccharide mutant background (42). HrpA also is essential for N. meningitidis escape from the intracellular space after invasion (46). FHA of B. pertussis is known to have multiple roles, which include heparin binding, carbohydrate binding, CR3 and adenylate cyclase interaction, and involvement in adherence, invasion, proinflammatory response, and antiapoptotic characteristics (1, 5, 19, 24, 25, 31, 32, 35, 38, 55). Therefore, it is plausible that HrpA has multiple roles in N. meningitidis pathogenesis.
We recently showed that N. meningitidis could form biofilms on transformed human bronchial epithelial (HBE) cells (34). Our current work set out to determine if hrpA was regulated and if HrpA was important in biofilm formation on HBE cells in a flow cell system. Nonfimbrial adhesins are important for mature biofilm formation in other organisms, including the FhaBC system in Bordetella bronchiseptica (23). Our work demonstrated the importance of HrpA in biofilm formation on HBE cells in both wild-type and unencapsulated strains of N. meningitidis. We further demonstrated the regulation of the hrpA gene and the localization of the HrpA protein to the outer membrane.
MATERIALS AND METHODS
Strains and growth conditions.
N. meningitidis strains NMB, NMB siaA-D (ST4609; ST-8 complex, cluster A4), MC58 siaD (ST-32 ET-5), C311 siaD (unassigned multilocus sequence type), and their derivatives were used in this study (see Table S1 in the supplemental material) (37, 45, 51). All strains contained the Neisseria pGFP plasmid with the chloramphenicol cassette (11). Chloramphenicol resistance has been reported to occur naturally (26, 44), and the use of this as the selection is in accordance with NIH recombinant DNA guidelines (Section IV-C-2-a). Strains were grown from frozen stock on GC agar (Becton Dickinson, Sparks, MD) supplemented with 1% (vol/vol) IsoVitaleX. Kanamycin (75 μg/ml) and chloramphenicol (5 μg/ml) were utilized as needed (Sigma, St. Louis, MO). Escherichia coli strains were grown in Luria-Bertani (LB) broth (tryptone, 10 g/liter; yeast extract, 5 g/liter; NaCl, 10 g/liter) or on LB agar (LB broth with agar, 15 g/liter) (Becton Dickinson, Sparks, MD). Kanamycin (50 μg/ml) and ampicillin (125 μg/ml) were utilized in E. coli cultures as needed (Sigma, St. Louis, MO). Bacteria processed for protein or RNA were grown in RPMI 1640 with 1% (vol/vol) IsoVitaleX (Gibco, Grand Island, NY).
Generation of mutants.
Genomic DNA was isolated from strains NMB siaA-D and MC58 siaD using ArchivePure DNA cell/tissue kits (5 Prime, Gaithersburg, MD). Digoxigenin-labeled probes for Southern blottings were generated from strain MC58 siaD using primers unique to regions of the five different hrpA-like genes in that strain (see Table S1 in the supplemental material) (Roche, Mannheim, Germany). Five different Southern blottings were performed to determine which of the five hrpA-like genes (NMB0493, NMB0497, NMB1214, NMB1768, and NMB1779) were present in the NMB genome. A deletion mutant was made of the putative hrpA gene in strain NMB by the replacement of the gene with a kanamycin-sacB cassette from pJJ260 in both the encapsulated and unencapsulated strain NMB backgrounds. The pJJ260 cassette has the P2 promoter from Haemophilus influenzae driving the expression of the sacB cassette; however, the P2 promoter is repressed by the tetR repressor. The mutation construct was assembled in pBluescript SK−. The assembled construct was PCR amplified and transformed into N. meningitidis strain NMB. Complementation was performed by knock-in to replace the kan-sacB cassette with the original gene through the transformation of genomic DNA from the wild-type organism and selection on 10% sucrose on LB agar with 1% IsoVitaleX and heat-inactivated chlortetracycline (1 μg/ml). Knock-in complementation was described previously for Neisseria gonorrhoeae (43). Southern and Western blottings were performed to verify the mutation, chromosomal complementation, and expected HrpA production from these strains. All primers utilized in this study are in Table S1 in the supplemental material.
Real-time RT-PCR.
RNA was isolated from N. meningitidis grown under the conditions listed in Table 1 using the RNeasy kit (Qiagen, Valencia, CA). RNA integrity was determined on an Agilent bioanalyzer (Agilent, Santa Clara, CA) and used only if it had a relative integrity number above 8.0 on a scale of 1 to 10. Bacterial samples were grown on three separate occasions in the appropriate conditions, and RNA was isolated for use in real-time reverse transcriptase PCR (RT-PCR). Primer/probe sets for genes hrpB, hrpA, and hrpS1 were ordered from Applied Biosystems (Austin, TX), and the primer probe set for the rmpM transcript was ordered from Integrated DNA Technologies (Coralville, IA). Standard curves were analyzed to determine the efficiency of amplification and the ΔΔCT method, with the efficiency correction as defined by Pfaffl, was used for the analysis of the level (n-fold) of change (36). Normalized natural log threshold cycle values of the control and test populations were analyzed by two-sample t test for statistical significance. The efficiency of both primer sets was the same, so no correction was needed for the two-sample t tests.
TABLE 1.
Results of real-time RT-PCR analyses of hrpB, hrpA, and hrpS1a
| RT-PCR condition | hrpB | hrpA | hrpS1 | P value of HrpA |
|---|---|---|---|---|
| Iron, 10 μm | −1.177 ± 0.19 | −1.326 ± 0.69 | −2.301 ± 0.12 | 0.1731 |
| Desferol, 30 μm | 1.238 ± 0.71 | 1.319 ± 0.79 | −1.287 ± 0.22 | 0.7499 |
| Serum, 5% | −1.458 ± 0.27 | 1.169 ± 0.43 | −1.129 ± 0.19 | 0.6490 |
| Serum, 5%, plus Desferol, 30 μm | 1.319 ± 0.34 | −1.268 ± 1.07 | −1.252 ± 0.06 | 0.9561 |
| Anaerobic, 1 h | 4.479 ± 0.48 | 3.968 ± 2.82 | 2.437 ± 0.36 | 0.0100* |
| Anaerobic, 2 h | 8.753 ± 0.69 | 5.097 ± 2.18 | 7.099 ± 0.73 | 0.0005* |
| Anaerobic, 24 h | 4.539 ± 3.44 | 2.840 ± 1.73 | 3.001 ± 0.27 | 0.0618 |
| Biofilm, 48 h | 1.054 ± 0.71 | 1.179 ± 0.84 | −1.084 ± 0.06 | 0.9982 |
| Biofilm, planktonic | 14.445 ± 0.51 | 11.344 ± 16.45 | 1.357 ± 0.65 | 0.0747 |
| Cell association, 5 h | 11.064 ± 3.43 | 5.105 ± 4.89 | 2.885 ± 0.97 | 0.0259* |
Data are the level of change (n-fold) from that of bacteria grown in aerobic broth conditions. The expression of the constitutive rmp gene was used to normalize the hrpB, hrpA, and hrpS real-time RT-PCR results. P values from nonparametric two-sample t tests of the hrpA data are given (an asterisk indicates P ≤ 0.05).
Polyclonal antibody development.
The first 900 bp (positions 220 to 1119) after the predicted encoding signal sequence of the hrpA gene was amplified and cloned into pET15B (Novagen, EMD Biosciences, Madison, WI) and transformed into BL21 pLysS chemically competent cells (Invitrogen, Carlsbad, CA). Cultures were grown in 500 ml LB broth until an optical density (OD) of 0.3 at 600 nm was reached. The cultures were induced with 1 mM isopropyl-â-d-thiogalactopyranoside (IPTG) and grown for three additional hours. The bacteria were pelleted at 3,000 × g for 15 min, and the pellet was resuspended in 50 ml 8 M urea, 20 mM sodium phosphate [pH 7.8], 500 mM NaCl buffer and sonicated for three 20-s bursts. The lysed solution was spun at 3,000 × g for 30 min, and the supernatant was filtered through a 0.2-μm filter. The filtered material then was placed in a 50-ml GE superloop (GE Healthcare, Upsalla, Sweden) on a GE fast-performance liquid chromatograph (GE Healthcare, Upsala Sweden) and chromatographed through a HisTrap Fast Flow nickel affinity column (GE Healthcare, Upsalla, Sweden) at a rate of 2 ml per min, washed with 10 column volumes of 8 M urea buffer [pH 6.0], washed with another 10 column volumes with 8 M urea buffer (pH 5.2), and finally eluted in 8 M urea buffer (pH 4.0). The eluted material then was concentrated to 0.5 ml in a Centricon YM10 filter at 3,000 × g and dialyzed into 4 M urea. This material was passed through a HiLoad 16/60 Superdex 75-pg gel filtration column in 4 M urea buffer to obtain a single species of protein as visualized by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and continuous UV analysis of the fractionated material. Stepwise dialysis in 2, 1, and 0.5 M urea was performed, and the resulting purified protein was immunized into New Zealand White rabbits for polyclonal antibody production, which was performed by the Iowa State University Hybridoma Facility (Ames, IA).
Western blot analyses.
Bacterial pellets were lysed in 4% (mol/vol) SDS in H2O, and lysates were sheared through 23-gauge needles. The protein concentration was determined by colorimetric assay (Pierce, Rockford, IL), and 5 μg of lysate was electrophoresed in each lane of NuPage 4 to 12% Bis-Tris polyacrylamide gels (Invitrogen, Carlsbad, CA). Proteins were transferred to an Immobilon-P polyvinylidene difluoride membrane (Millipore, Bedford, MA) at 150 mA per gel for 16 h in Western Breeze transfer buffer (Invitrogen, Carlsbad, CA) with 10% (vol/vol) methanol at 4°C. Membranes were blocked with Western Breeze blocking agent (Invitrogen, Carlsbad, CA), anti-HrpA polyclonal antibody was applied at a 1:500 dilution, and goat anti-rabbit immunoglobulin G horseradish peroxidase (HRP) secondary (Bio-Rad, Hercules, CA) was used at 1:10,000. The chemiluminescence of the HRP secondary conjugate was activated with Pico-West super signal (Pierce, Rockford, IL) and developed on Kodak MR film (Eastman Kodak, Rochester, NY). Mouse monoclonal antibody 2C3 against the H.8 protein was used as a loading control with goat anti-mouse HRP secondary (Bio-Rad, Hercules, CA) at 1:10,000.
Tissue culture conditions.
Simian virus 40-transformed HBE cells (ATCC number CRL-9609) were cultured in minimal essential medium (MEM) tissue culture medium with 10% (vol/vol) heat-inactivated fetal calf serum (FCS) and 2 mM l-glutamine at 37°C in the presence of 5% CO2 (Gibco, Grand Island, NY). HBE cells were trypsinized and placed onto 22- by 50-mm coverslips coated with 0.5 mg/ml bovine collagen for use in biofilm chambers after the cells had matured to at least 90% confluence as determined by visual examination.
Biofilms on HBE cells.
N. meningitidis was cultured for 16 h as described above and then suspended in 0.1× HBE tissue culture medium (MEM plus 10% FCS and 2 mM l-glutamine) diluted in 1× phosphate-buffered saline (PBS), pH 7.4, supplemented with IsoVitaleX (1:100, vol/vol) and 100 μM sodium nitrite. Cultures were vortexed for 15 s at high speed and then shaken at 150 rpm for 30 min to disperse the bacteria (as verified by microscopic inspection) and adjusted to 1 × 108 organisms per ml (28). Bacterial suspensions were added to biofilm chambers containing the HBE cell monolayers and allowed to incubate at 37°C for 1 h before the initiation of medium flow. The flow rate was 150 μl per min (14). Biofilms were allowed to mature for either 6 or 48 h before microscopy and Comstat analysis.
Biofilm apparatus.
To assess biofilm growth on tissue culture cells, we constructed a flowthrough chamber that permitted the insertion of a coverslip coated with live HBE cells. The chamber was composed of three pieces. The bottom was a 35- by 70- by 4-mm piece of acrylic with a 17-mm-wide, 45-mm long, and 2-mm deep chamber machined out of the block. Medium inflow and outflow ports were centered on the ends of the chamber at a 2-mm depth. A 1-mm-thick rubber gasket the size of the acrylic block with a 17- by 45-mm section removed from the center was placed on top of the chamber bottom. The top piece was acrylic, with the same 17- by 45-mm middle section removed. The underside of the top piece has a 22- by 50-mm section machined out to the same depth as the coverslip to accommodate the tissue culture cells. Ten screws held the apparatus together.
Confocal microscopy.
Tissue culture cells were labeled with cell tracker orange (Molecular Probes, Eugene, OR) prior to chamber inoculation. The use of the Nikon Eclipse 80I confocal microscope, lasers, Comstat analysis, and Volocity image rendering were described previously (29). We performed five separate experiments for each growth condition and three separate Z-series per flow cell.
Statistical analysis.
For each biofilm condition, Z-series were taken on the laser-scanning confocal microscope and analyzed by Comstat software for average height and total biomass (MathWorks, Natick, MA) (21). Comstat analysis is a comprehensive digital analysis of a Z-series of images from a confocal microscope. It allows the statistical analysis of the biomass and average height of the biofilm across the entire Z-series of images. To accomplish this, each Z-series photomicrograph is saved as a series of TIFF images that are converted into 8-bit grayscale images in Comstat for pixel analysis (21). GraphPad Prism (San Diego, CA) was used to calculate a nonparametric one-way analysis of variance (ANOVA) with a Dunn's multiple-comparison post test to compare biofilms and generate graphs from a 12 to 18 Z-series. A P value of ≤0.05 was considered significant.
ELISA analysis.
To determine the degree of piliation at the time of chamber inoculation, we coated the enzyme-linked immunosorbent assay (ELISA) plates with 100 μl of an inoculum (OD at 600 nm of 0.1) and probed them with a rabbit polyclonal antibody against PilE (a gift from Michael Jennings, University of Queensland, Brisbane, Australia). The wild-type and complemented strains were compared to the hrpA mutants as well as a pilE mutant of strain C311. Control ELISA analysis against H8 with monoclonal antibody 2C3 was utilized to demonstrate equal loading between strains.
HrpA localization.
Biofilm inoculum bacteria were spun down and washed with PBS and suspended to an OD of 0.3. A drop was placed on a microscope slide and dispersed around a circle 1 cm in diameter and dried to completion. The bacteria were blocked with 5% normal goat serum (Sigma, St. Louis, MO) and labeled either with anti-HrpA antibody (1:100) and Cy5 conjugate secondary antibody (1:250) (Jackson ImmunoResearch, West Grove, PA) or with the secondary antibody alone. Image analysis was performed on an Olympus BX-51 light microscope with fluorescent capabilities.
RESULTS
Strain NMB has paralogs to the MC58 genes NMB0497 and NMB1779 as determined by Southern blot analysis (data not shown). The cloning and sequencing of these regions demonstrated that the NMB0497 paralog was identical to NMC0444 of the sequenced strain Fam18 (ST-11 complex/ET-37 complex) and is referred to as hrpA for this study. Further analysis demonstrated that the NMB1779 paralog was truncated from the 5′ end with the first 3,850 bp of the total 5,988 bp present in the MC58 strain deleted. The remaining 3′ fragment initiated 557 bp downstream of the stop codon of the hrpA gene in strain NMB with a 510-bp open reading frame located between the two hrpA-related sequences. This truncated hrpA is referred to as hrpS1 (NMC0446 homologue) for the duration. Sequence analysis demonstrated greater than 99% sequence identity from upstream of hrpB through downstream of the hrpS3 gene (NMC0450 homologue), corresponding to the NMC0440-NMC0451 region of the Fam18 genome (GenBank accession number FJ644951). A detailed view of this genetic region in Fam18, Z2491, and MC58 was published previously (49). A review of secretion systems in N. meningitidis comments on the phenomenon of predicted pseudogenes or predicted noncoding regions that are 1,500 to 3,000 bp in length with sequence similarities to the full-length hrpA genes (49). These pseudogenes are scattered downstream of the predicted hrpA gene in all currently sequenced genomes of N. meningitidis. This phenomenon is postulated to be similar to the pilE and pilS genetic loci in the Neisseria sp. genomes, in which the pilS sequences are randomly incorporated into pilE by homologous recombination for the antigenic variation of the pilus (49). We refer to these hrpA pseudogenes as hrpS regions to maintain continuity to the pilE and pilS system.
Real-time RT-PCR was performed on RNA samples from bacteria grown under conditions known to activate regulatory pathways in N. meningitidis to determine if the hrpB, hrpA, or hrpS1 gene was regulated (Table 1) (15, 16, 18, 39). These conditions were iron excess and iron depletion, the presence of serum, anaerobic growth, biofilm growth, and cell association. There was an upregulation of all three genes in anaerobic conditions, between 2.4- to 8.7-fold depending on the time spent in the anaerobic chamber. Interestingly, there was an upregulation of all three transcripts after 5 h of cell association in 24-well tissue culture plates. We also observed the upregulation of bacteria detaching from biofilms on HBE cells in which hrpB was upregulated 14.4-fold and hrpA was upregulated 11.34-fold, while hrpS1 was not upregulated. This is interesting, considering that hrpBA are operonic and hrpS1 appears to be operonic based on genetic structure (42). The lack of the upregulation of the hrpS1 transcript under this condition may be explained by the size and stability of the large transcript or by alternative terminator sites. Real-time RT-PCR results of the hrpA transcript from anaerobic conditions (1 h, P = 0.01; 2 h, P = 0.0005) and cell association (P = 0.025) were statistically upregulated compared to levels for the hrpA transcript of broth-grown organisms (Table 1). Significance was not reached in the 24-h anaerobic conditions and the biofilm planktonic conditions due to the large variation between samples. We are unsure of the reasons for this variation at this time, but it could be due to instability of the mRNA.
Western blots demonstrated HrpA at both 220 and 180 kDa in lysates of aerobically grown cultures (Fig. 1). This doublet band was described previously in Western blots of HrpA (48). Growth in anaerobic conditions consistently demonstrated a significant decrease in the intensity of the 220-kDa band (Fig. 1A). This may indicate a more efficient processing of the 220-kDa protein in low-oxygen conditions. When densitometry measurements of the 180-kDa band for wild-type strains grown aerobically or grown for 1, 2, or 24 h anaerobically in RPMI medium were normalized to densitometry measurements of the constitutively expressed H.8 band, statistically significant increases in the 180-kDa band at 24 h (P = 0.04) were observed compared to results for normalized aerobic conditions (Fig. 1A). Densitometry was performed on Western blots from three separate experiments. Strains MC58 siaA-D and C311 and the complemented NMB hrpA strain demonstrated the production of HrpA antibody-reactive bands (Fig. 1B). The MC58 strain also produced a third high-molecular-weight band that was HrpA antibody reactive. This is not surprising, considering that MC58 has five hrpA paralogs.
FIG. 1.
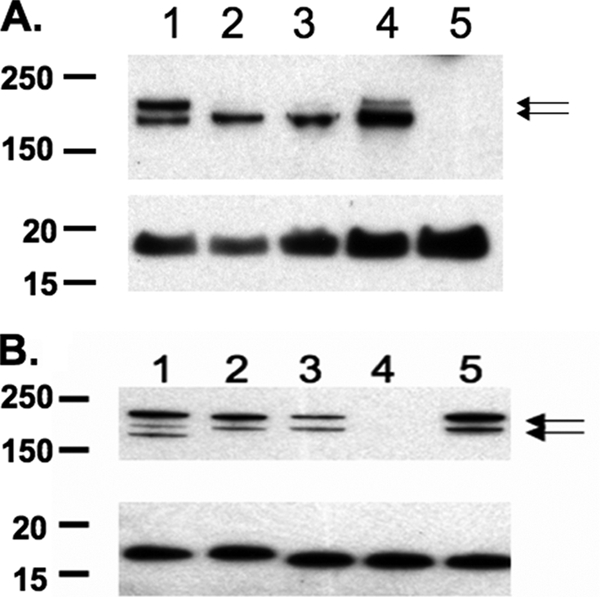
Western blots using a rabbit anti-HrpA antibody and monoclonal antibody 2C3 that binds to the outer membrane protein H.8. H.8 was used as the loading control for the individual samples (A and B, lower images). (A) Lysates of strain NMB siaA-D grown under aerobic conditions (lane 1) or anaerobic conditions for 1 (lane 2), 2 (lane 3), and 24 h (lane 4); lane 5 shows the growth of NMB siaA-D hrpA. The normalized 180-kDa band in lane 4 was significantly upregulated compared to the normalized 180-kDa band in lane 1. (B) Lysates of strains MC58 siaD (lane 1), C311 (lane 2), NMB siaA-D (lane 3), NMB siaA-D hrpA, (lane 4), and a lysate of NMB siaA-D hrpA complemented with hrpA (lane 5) grown aerobically. Arrows indicate the HrpA-specific (220- and 180-kDa) bands.
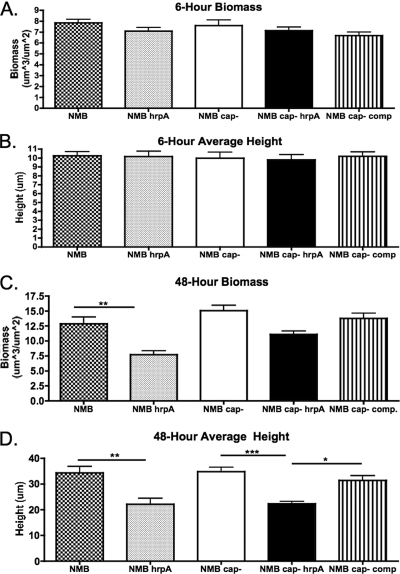
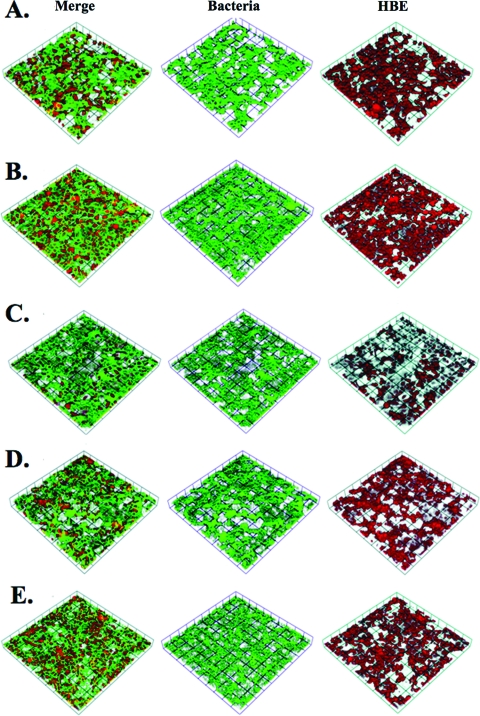
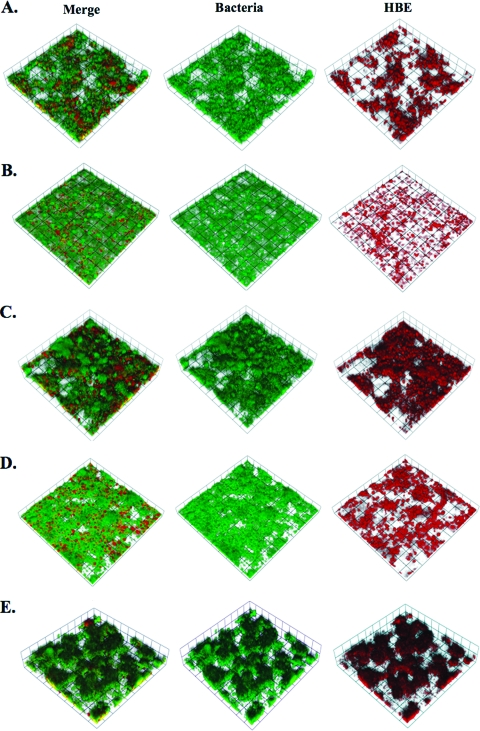
We did not observe changes in the transcript of any gene in the hrpBAS operon in biofilms collected at 48 h (Table 1). However, an increase in the transcript of hrpBA after 5 h of cell association as well as anaerobic conditions led us to determine if HrpA had a role in biofilm formation on HBE cells. The biofilm system allows for biofilm formation under conditions of shear stress on HBE cells to mimic conditions in association with human airway epithelial cells. First, growth curves with NMB siaA-D, NMB siaA-D hrpA, and NMB siaA-D hrpA with a chromosomal complementation of hrpA were performed, and no growth defect was demonstrated in the mutant or complemented strains (data not shown). We next screened the strains used in biofilm analyses by ELISA against whole bacteria for the presence of pilus. All strains were equally piliated. Images of 4′,6′-diamidino-2-phenylindole-stained HBE cells and adherent bacteria were taken after the 1-h static incubation period and indicated that initial attachment was not statistically different for the wild type or hrpA mutant. There was an average of 19 ± 7 bacteria per cell across all strains utilized in this study at the time of flow initiation. Mutants of hrpA in both the capsule-positive and capsule-negative strain of NMB demonstrated no difference in average height and biomass at a 6-h time point (Fig. 2A and B). However, 48-h biofilms on HBE cells demonstrated significantly reduced biofilm formation in the hrpA mutants when analyzed for biomass (encapsulated strains only) and average height (encapsulated and unencapsulated strains) by confocal microscopy and Comstat analysis (Fig. 2C and D). Images of the 6-h biofilms can be seen in Fig. 3 and demonstrate that the bacteria preferentially adhere to the HBE cells, as seen by gaps in the HBE monolayer and a few bacteria filling these gaps. The 48-h images (Fig. 4) demonstrate the growth of the biofilm compared to that of the 6-h images and clearly demonstrate the decrease in the biofilm in the hrpA mutant compared to that of the wild type. In addition, Fig. 4 demonstrates that the 48-h biofilm is affecting HBE cell confluence and cell morphology compared to the level of confluence of HBE cells at 6 h postinfection in Fig. 3. Uninfected HBE monolayers incubated in flow cells for 48 h demonstrated 80% survival and normal cell morphology. HBE cells infected with strain NMB or NMB siaA-D had 60 and 70% survival, respectively, at 48 h. The fact that there was 20% cell death in the uninfected chambers indicates that the chamber environment is not optimal for the growth of the HBE cells, and the bacterial infection could exacerbate this effect.
FIG. 2.
Graphs of Comstat analyses of 6-h biomass (A) and average height (B), the same results for 48-h biomass (C), and the average height of biofilms formed on HBE cells (D). A total of 12 to 15 Z-series were analyzed by ANOVA for the 6-h biofilm data, and 14 to 18 Z-series were analyzed for the 48-h biofilm data for each condition tested from five to six different experiments. Data from the 6-h conditions were not significant by ANOVA; however, data for both 48-h biomass and average height were significant by ANOVA (P < 0.0001). Significance bars are results from ANOVA post tests. *, P < 0.05; **, P < 0.01; ***, P < 0.0001. cap−, siaA-D capsular mutant; cap− comp, NMB siaA-D hrpA with a chromosomal complementation of hrpA.
FIG. 3.
Volocity-rendered confocal Z-series of 6-h biofilms on HBE cells infected with strains NMB (A), NMB hrpA (B), NMB siaA-D (C), NMB siaA-D hrpA (D), and NMB siaA-D hrpA complemented with hrpA (E). Bacteria are green fluorescent protein labeled, and HBE cells are labeled with cell tracker orange (red channel).
FIG. 4.
Volocity-rendered confocal Z-series of 48-h biofilms on HBE cells infected with strains NMB (A), NMB hrpA (B), NMB siaA-D (C), NMB siaA-D hrpA (D), and NMB siaA-D hrpA complemented with hrpA (E). Bacteria are green fluorescent protein labeled, and HBE cells are labeled with cell tracker orange (red channel).
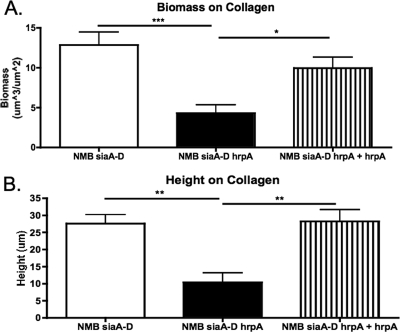
HrpA is a factor that is involved in escape from invaded cells (46). We grew biofilms for 48 h on collagen-coated coverslips to determine if the reason for reduced biofilm formation on HBE cells at 48 h by the hrpA mutant was due to an inability of the organism to escape from the cells and then contribute to the biofilm structure. Biofilms of NMB siaA-D and the NMB siaA-D hrpA complemented strain formed significantly larger biofilms than the NMB siaA-D hrpA mutant (Fig. 5). This suggests that egressing organisms from the HBE cells do not make significant contributions to the biofilm structure. Also, since the 6-h biofilms on HBE cells were equivalent, this implies a role for HrpA in the evolving structure of the biofilm and not necessarily as an adhesin in our model system (42).
FIG. 5.
Comstat analyses of 48-h biofilms grown on collagen-coated coverslips in the same flow cells and media as biofilms produced on HBE cells. A total of 10 Z-series were analyzed from a minimum of four experiments for each condition. *, P < 0.05; **, P < 0.01; ***, P < 0.0001.
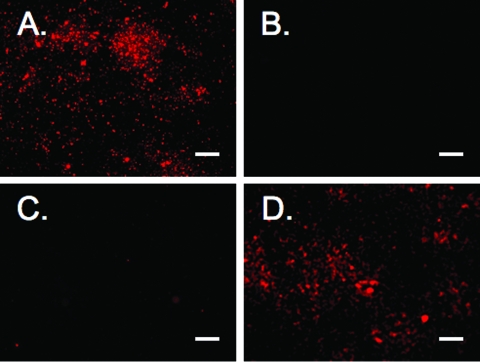
In contrast to previous publications, our Western blot studies of strains NMB, MC58, and C311 failed to demonstrate the secretion of HrpA into the supernatant of aerobically shaken broth cultures (42, 48). We performed fluorescence microscopy to determine if the HrpA protein was present on the surface of strain NMB. Figure 6 demonstrates the presence of HrpA on the surface of the wild-type NMB and the hrpA-complemented NMB siaA-D hrpA strain but not the hrpA mutant or wild-type strain reacted with the Cy5 conjugate secondary antibody alone.
FIG. 6.
Fluorescence microscopy of whole bacteria immunolabeled with anti-HrpA with a Cy5 secondary antibody. (A) NMB siaA-D; (B) NMB siaA-D with secondary antibody alone; (C) NMB siaA-D hrpA; and (D) NMB siaA-D hrpA complemented with hrpA. Scale bar, 10 μm.
DISCUSSION
Two-partner secretion systems support adherence to tissue in B. pertussis, Moraxella catarrhalis, and enterotoxigenic E. coli (2, 7, 13). The HrpAB two-partner secretion system of N. meningitidis has a role in adherence to tissue culture cells but only in the absence of capsule and with a lipooligosaccharide truncation (42). HrpA also is important in escape from HeLa cells (46). We used an expression-based approach to determine if hrpA was regulated. If regulation occurred, we hoped that this would point to a role for HrpA in N. meningitidis pathogenesis. Culture after 5 h in the presence of HBE cells as well as culture in anaerobic conditions resulted in the upregulation of hrpA transcript. Interestingly, the significant upregulation of transcript in anaerobic conditions at 1 and 2 h (Table 1) resulted in a significant increase in HrpA protein only at 24 h of anaerobic growth (for the 180-kDa band, P = 0.04) compared to that of organisms grown in aerobic conditions (Fig. 1). The reason for this is not known at this time, but it could be due to the inefficient transfer of HrpA onto the membrane for the Western blots or other unknown reasons. The production of the 180-kDa form of HrpA grown anaerobically is significantly enhanced, based on the densitometry analysis of Western blots, compared to that of the 180-kDa form during growth in aerobic conditions. This may point to its function in the biofilm process during conditions of low oxygen. Alternatively, it is possible that a sequence-specific protease is more active in the anaerobic conditions and cleaves HrpA to the 180-kDa form.
The results of the real-time RT-PCR experiments directed us to evaluate the role of HrpA in biofilm formation. A significant reduction in biofilm biomass and average height for hrpA mutants was demonstrated in the 48-h biofilms but not in the 6-h biofilms. There are three possible explanations for the hrpA mutant phenotype. The initial finding that HrpA is an adhesin in an unencapsulated lipooligosaccharide truncation strain of N. meningitidis supports the first possibility, that biofilms are reduced because of decreased adherence to HBE cells. If fewer numbers of initial bacteria bound to the HBE cells, this would result in fewer bacteria being available to initiate microcolony formation and biofilm development. However, as stated above, the levels of bacterial adherence at the time of the initiation of flow were comparable for all strains tested. Furthermore, biofilm formation at the 6-h time point was equivalent among all strains based on Comstat analyses (Fig. 2A and B). This suggests that adherence to the HBE cells was not a factor in the differences in biofilm development between the hrpA mutant and the wild type. The second possibility correlates with the observation of HrpA involvement in escape from invaded tissue (46). If only wild-type bacteria can escape the tissue culture cells, then the wild-type organisms could provide an additional population of bacteria to contribute to the biofilm after escape while the hrpA mutant would be deficient. The third possibility is that HrpA has a role in the maintenance of the biofilm structure. The 48-h biofilms grown on collagen demonstrated that HrpA was involved in biofilm development because of the biofilm defect in the absence of cells (Fig. 5). FHA previously was implicated in multiple functions in B. pertussis, including binding to self and host ligands (31, 35, 55). This example of multiple roles for FHA demonstrates the possibility of HrpA having multiple domains for interaction with different cellular or bacterial ligands. It also is possible that the different processed forms of HrpA have different roles.
Based on our results, we hypothesize that HrpA is active mainly in its 180-kDa processed form and its presence is enhanced in anaerobic environments, as suggested by the real-time RT-PCR data, which demonstrate the upregulation of the hrpA transcript (Table 1), and the Western blot data, which demonstrates the enhanced processing of HrpA (Fig. 1A). This corresponds to the data of a biofilm defect in the HrpA mutant. Biofilms are known to develop oxygen gradients as they grow (54). This suggests that microaerophilic or anaerobic environments develop within larger biofilms, where the hrpA transcript could be upregulated and the production of the 180-kDa form of HrpA could be enhanced. Our data suggest that HrpA has a role in maintaining the biofilm structure, possibly through bacteria-bacteria interaction. We hypothesize that mutant meningococci lacking HrpA form smaller biofilms with smaller oxygen gradients, since the anaerobically active form of the protein is absent in the mutant strain and therefore not able to hold the larger biofilm together.
We are unsure of the reason why HrpA remains surface bound in our study, while studies from other groups found HrpA in the supernatant (42, 48). It may be because the previous studies grew their cultures to ODs that most likely represented stationary-phase organisms, while ours were still in the log phase of growth. Since Neisseria organisms bleb and undergo autolysis, these studies could be detecting HrpA from lysed organisms or HrpA bound to membrane fragments that are too small to pellet in the low-speed centrifugation that is used to pellet the bacteria (33, 40). Alternatively, N. meningitidis may actively release bound HrpA at stationary phase, and we cannot exclude the possibility that our strains are unable to release HrpA from the surface. One candidate to aid in the release of HrpA from the meningococcal surface is NalP. This protein has 30% homology to the SphB1 protein that is involved in the final maturation of FHA in B. pertussis (9). The nalP gene is likely phase variable in N. meningitidis due to a stretch of ∼10 cytosine residues in the coding sequence that participate in slip-stranded mispairing to turn the gene on and off (47, 50). However, the sequence analysis of strain NMB demonstrated nalP is phase on (data not shown).
Studies of the two-partner secretion system of N. meningitidis have led to interesting discoveries regarding the pathogenesis of epithelial cell layer infection models. The first is that HrpA is important in intracellular escape (46). The mechanism by which this occurs is not understood. The second is that HrpA has a role in biofilm formation on epithelia, as demonstrated in this study. Biofilms may be important for N. meningitidis during nasopharyngeal colonization. The genes encoding the HrpA proteins are found in virtually all strains of N. meningitidis (42, 48). Since the HrpA paralog FHA is a component of the acellular B. pertussis vaccine, future research should determine if HrpA undergoes antigenic variation, which could limit the usefulness of HrpA in a broadly reactive meningococcal vaccine (12). In addition, investigations of the cellular or self receptors accounting for HrpA protein involvement in bacteria-cell or bacteria-bacteria interactions should be undertaken.
Supplementary Material
Acknowledgments
This work was supported by AI045728 and AI007511.
We thank Yu-Hui Chang for assistance in statistical analysis.
Editor: J. N. Weiser
Footnotes
Published ahead of print on 16 March 2009.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Abramson, T., H. Kedem, and D. A. Relman. 2001. Proinflammatory and proapoptotic activities associated with Bordetella pertussis filamentous hemagglutinin. Infect. Immun. 692650-2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balder, R., J. Hassel, S. Lipski, and E. R. Lafontaine. 2007. Moraxella catarrhalis strain O35E expresses two filamentous hemagglutinin-like proteins that mediate adherence to human epithelial cells. Infect. Immun. 752765-2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Capecchi, B., J. Adu-Bobie, F. Di Marcello, L. Ciucchi, V. Masignani, A. Taddei, R. Rappuoli, M. Pizza, and B. Arico. 2005. Neisseria meningitidis NadA is a new invasin which promotes bacterial adhesion to and penetration into human epithelial cells. Mol. Microbiol. 55687-698. [DOI] [PubMed] [Google Scholar]
- 4.Cartwright, K. A., J. M. Stuart, D. M. Jones, and N. D. Noah. 1987. The Stonehouse survey: nasopharyngeal carriage of meningococci and Neisseria lactamica. Epidemiol. Infect. 99591-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colombi, D., M. L. Oliveira, I. B. Campos, V. Monedero, G. Perez-Martinez, and P. L. Ho. 2006. Haemagglutination induced by Bordetella pertussis filamentous haemagglutinin adhesin (FHA) is inhibited by antibodies produced against FHA(430-873) fragment expressed in Lactobacillus casei. Curr. Microbiol. 53462-466. [DOI] [PubMed] [Google Scholar]
- 6.Cotter, P. A., M. H. Yuk, S. Mattoo, B. J. Akerley, J. Boschwitz, D. A. Relman, and J. F. Miller. 1998. Filamentous hemagglutinin of Bordetella bronchiseptica is required for efficient establishment of tracheal colonization. Infect. Immun. 665921-5929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coutte, L., S. Alonso, N. Reveneau, E. Willery, B. Quatannens, C. Locht, and F. Jacob-Dubuisson. 2003. Role of adhesin release for mucosal colonization by a bacterial pathogen. J. Exp. Med. 197735-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coutte, L., R. Antoine, H. Drobecq, C. Locht, and F. Jacob-Dubuisson. 2001. Subtilisin-like autotransporter serves as maturation protease in a bacterial secretion pathway. EMBO J. 205040-5048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coutte, L., E. Willery, R. Antoine, H. Drobecq, C. Locht, and F. Jacob-Dubuisson. 2003. Surface anchoring of bacterial subtilisin important for maturation function. Mol. Microbiol. 49529-539. [DOI] [PubMed] [Google Scholar]
- 10.Delisse-Gathoye, A. M., C. Locht, F. Jacob, M. Raaschou-Nielsen, I. Heron, J. L. Ruelle, M. de Wilde, and T. Cabezon. 1990. Cloning, partial sequence, expression, and antigenic analysis of the filamentous hemagglutinin gene of Bordetella pertussis. Infect. Immun. 582895-2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edwards, J. L., and M. A. Apicella. 2005. I-domain-containing integrins serve as pilus receptors for Neisseria gonorrhoeae adherence to human epithelial cells. Cell. Microbiol. 71197-1211. [DOI] [PubMed] [Google Scholar]
- 12.Fishbein, D. B., K. R. Broder, L. Markowitz, and N. Messonnier. 2008. New, and some not-so-new, vaccines for adolescents and diseases they prevent. Pediatrics 121S5-14. [DOI] [PubMed] [Google Scholar]
- 13.Fleckenstein, J. M., K. Roy, J. F. Fischer, and M. Burkitt. 2006. Identification of a two-partner secretion locus of enterotoxigenic Escherichia coli. Infect. Immun. 742245-2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greiner, L. L., J. L. Edwards, J. Shao, C. Rabinak, D. Entz, and M. A. Apicella. 2005. Biofilm formation by Neisseria gonorrhoeae. Infect. Immun. 731964-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grifantini, R., E. Bartolini, A. Muzzi, M. Draghi, E. Frigimelica, J. Berger, F. Randazzo, and G. Grandi. 2002. Gene expression profile in Neisseria meningitidis and Neisseria lactamica upon host-cell contact: from basic research to vaccine development. Ann. N. Y. Acad. Sci. 975202-216. [DOI] [PubMed] [Google Scholar]
- 16.Grifantini, R., S. Sebastian, E. Frigimelica, M. Draghi, E. Bartolini, A. Muzzi, R. Rappuoli, G. Grandi, and C. A. Genco. 2003. Identification of iron-activated and -repressed Fur-dependent genes by transcriptome analysis of Neisseria meningitidis group B. Proc. Natl. Acad. Sci. USA 1009542-9547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guédin, S., E. Willery, J. Tommassen, E. Fort, H. Drobecq, C. Locht, and F. Jacob-Dubuisson. 2000. Novel topological features of FhaC, the outer membrane transporter involved in the secretion of the Bordetella pertussis filamentous hemagglutinin. J. Biol. Chem. 27530202-30210. [DOI] [PubMed] [Google Scholar]
- 18.Hagen, T. A., and C. N. Cornelissen. 2006. Neisseria gonorrhoeae requires expression of TonB and the putative transporter TdfF to replicate within cervical epithelial cells. Mol. Microbiol. 621144-1157. [DOI] [PubMed] [Google Scholar]
- 19.Hannah, J. H., M. F. Renauld, G. Locht, and C. M. J. Brennan. 1994. Sulfated glycoconjugate receptors for the Bordetella pertussis adhesin filamentous hemagglutinin (FHA) and mapping of the heparin-binding domain on FHA. Infect. Immun. 625010-5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henderson, I. R., F. Navarro-Garcia, M. Desvaux, R. C. Fernandez, and D. Ala'Aldeen. 2004. Type V protein secretion pathway: the autotransporter story. Microbiol. Mol. Biol. Rev. 68692-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heydorn, A., A. T. Nielsen, M. Hentzer, C. Sternberg, M. Givskov, B. K. Ersboll, and S. Molin. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 1462395-2407. [DOI] [PubMed] [Google Scholar]
- 22.Hodak, H., B. Clantin, E. Willery, V. Villeret, C. Locht, and F. Jacob-Dubuisson. 2006. Secretion signal of the filamentous haemagglutinin, a model two-partner secretion substrate. Mol. Microbiol. 61368-382. [DOI] [PubMed] [Google Scholar]
- 23.Irie, Y., S. Mattoo, and M. H. Yuk. 2004. The Bvg virulence control system regulates biofilm formation in Bordetella bronchiseptica. J. Bacteriol. 1865692-5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ishibashi, Y., D. A. Relman, and A. Nishikawa. 2001. Invasion of human respiratory epithelial cells by Bordetella pertussis: possible role for a filamentous hemagglutinin Arg-Gly-Asp sequence and α5β1 integrin. Microb. Pathog. 30279-288. [DOI] [PubMed] [Google Scholar]
- 25.Ishibashi, Y., K. Yoshimura, A. Nishikawa, S. Claus, C. Laudanna, and D. A. Relman. 2002. Role of phosphatidylinositol 3-kinase in the binding of Bordetella pertussis to human monocytes. Cell Microbiol. 4825-833. [DOI] [PubMed] [Google Scholar]
- 26.Jorgensen, J., S. Crawford, and K. Fiebelkorn. 2005. Susceptibility of Neisseria meningitidis to 16 antimicrobial agents and characterization of resistance mechanisms affecting some agents. J. Clin. Microbiol. 433162-3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kajava, A. V., and A. C. Steven. 2006. The turn of the screw: variations of the abundant beta-solenoid motif in passenger domains of Type V secretory proteins. J. Struct. Biol. 155306-315. [DOI] [PubMed] [Google Scholar]
- 28.Lappann, M., J. A. J. Haagensen, H. Claus, U. Vogel, and S. Molin. 2006. Meningococcal biofilm formation: structure, development and phenotypes in a standardized continuous flow system. Mol. Microbiol. 621292-1309. [DOI] [PubMed] [Google Scholar]
- 29.Lim, K. H., C. E. Jones, R. N. van den Hoven, J. L. Edwards, M. L. Falsetta, M. A. Apicella, M. P. Jennings, and A. G. McEwan. 2008. Metal binding specificity of the MntABC permease of Neisseria gonorrhoeae and its influence on bacterial growth and interaction with cervical epithelial cells. Infect. Immun. 763569-3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mazar, J., and P. A. Cotter. 2006. Topology and maturation of filamentous haemagglutinin suggest a new model for two-partner secretion. Mol. Microbiol. 62641-654. [DOI] [PubMed] [Google Scholar]
- 31.Menozzi, F. D., P. E. Boucher, G. Riveau, C. Gantiez, and C. Locht. 1994. Surface-associated filamentous hemagglutinin induces autoagglutination of Bordetella pertussis. Infect. Immun. 624261-4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mobberley-Schuman, P. S., and A. A. Weiss. 2005. Influence of CR3 (CD11b/CD18) expression on phagocytosis of Bordetella pertussis by human neutrophils. Infect. Immun. 737317-7323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Namork, E., and P. Brandtzaeg. 2002. Fatal meningococcal septicaemia with “blebbing” meningococcus. Lancet 3601741. [DOI] [PubMed] [Google Scholar]
- 34.Neil, R. B., J. Shao, and M. A. Apicella. 2009. Biofilm formation on human airway epithelia by encapsulated Neisseria meningitidis serogroup B. Microbes Infect. 11281-287. [DOI] [PubMed] [Google Scholar]
- 35.Perez Vidakovics, M. L., Y. Lamberti, W. L. van der Pol, O. Yantorno, and M. E. Rodriguez. 2006. Adenylate cyclase influences filamentous haemagglutinin-mediated attachment of Bordetella pertussis to epithelial alveolar cells. FEMS Immunol. Med. Microbiol. 48140-147. [DOI] [PubMed] [Google Scholar]
- 36.Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Post, D. M., M. R. Ketterer, N. J. Phillips, B. W. Gibson, and M. A. Apicella. 2003. The msbB mutant of Neisseria meningitidis strain NMB has a defect in lipooligosaccharide assembly and transport to the outer membrane. Infect. Immun. 71647-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prasad, S. M., Y. Yin, E. Rodzinski, E. I. Tuomanen, and H. R. Masure. 1993. Identification of a carbohydrate recognition domain in filamentous hemagglutinin from Bordetella pertussis. Infect. Immun. 612780-2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rock, J. D., M. R. Mahnane, M. F. Anjum, J. G. Shaw, R. C. Read, and J. W. Moir. 2005. The pathogen Neisseria meningitidis requires oxygen, but supplements growth by denitrification. Nitrite, nitric oxide and oxygen control respiratory flux at genetic and metabolic levels. Mol. Microbiol. 58800-809. [DOI] [PubMed] [Google Scholar]
- 40.Rosenstein, N. E., B. A. Perkins, D. S. Stephens, T. Popovic, and J. M. Hughes. 2001. Meningococcal disease. N. Engl. J. Med. 3441378-1388. [DOI] [PubMed] [Google Scholar]
- 41.Scarselli, M., D. Serruto, P. Montanari, B. Capecchi, J. Adu-Bobie, D. Veggi, R. Rappuoli, M. Pizza, and B. Arico. 2006. Neisseria meningitidis NhhA is a multifunctional trimeric autotransporter adhesin. Mol. Microbiol. 61631-644. [DOI] [PubMed] [Google Scholar]
- 42.Schmitt, C., D. Turner, M. Boesl, M. Abele, M. Frosch, and O. Kurzai. 2007. A functional two-partner secretion system contributes to adhesion of Neisseria meningitidis to epithelial cells. J. Bacteriol. 1897968-7976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seib, K. L., H. J. Wu, Y. N. Srikhanta, J. L. Edwards, M. L. Falsetta, A. J. Hamilton, T. L. Maguire, S. M. Grimmond, M. A. Apicella, A. G. McEwan, and M. P. Jennings. 2007. Characterization of the OxyR regulon of Neisseria gonorrhoeae. Mol. Microbiol. 6354-68. [DOI] [PubMed] [Google Scholar]
- 44.Shultz, T., J. Tapsall, P. White, C. Ryan, D. Lyras, J. Rood, E. Binotto, and C. Richardson. 2003. Chloramphenicol-resistant Neisseria meningitidis containing catP isolated in Australia. J. Antimicrob. Chemother. 52856-859. [DOI] [PubMed] [Google Scholar]
- 45.Stabler, R. A., G. L. Marsden, A. A. Witney, Y. Li, S. D. Bentley, C. M. Tang, and J. Hinds. 2005. Identification of pathogen-specific genes through microarray analysis of pathogenic and commensal Neisseria species. Microbiology 1512907-2922. [DOI] [PubMed] [Google Scholar]
- 46.Tala, A., C. Progida, M. De Stefano, L. Cogli, M. R. Spinosa, C. Bucci, and P. Alifano. 2008. The HrpB-HrpA two-partner secretion system is essential for intracellular survival of Neisseria meningitidis. Cell Microbiol. 102461-2482. [DOI] [PubMed] [Google Scholar]
- 47.Turner, D. P., K. G. Wooldridge, and D. A. Ala'Aldeen. 2002. Autotransported serine protease A of Neisseria meningitidis: an immunogenic, surface-exposed outer membrane, and secreted protein. Infect. Immun. 704447-4461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Ulsen, P., L. Rutten, M. Feller, J. Tommassen, and A. van der Ende. 2008. Two-partner secretion systems of Neisseria meningitidis associated with invasive clonal complexes. Infect. Immun. 764649-4658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van Ulsen, P., and J. Tommassen. 2006. Protein secretion and secreted proteins in pathogenic Neisseriaceae. FEMS Microbiol. Rev. 30292-319. [DOI] [PubMed] [Google Scholar]
- 50.van Ulsen, P., L. van Alphen, J. ten Hove, F. Fransen, P. van der Ley, and J. Tommassen. 2003. A neisserial autotransporter NalP modulating the processing of other autotransporters. Mol. Microbiol. 501017-1030. [DOI] [PubMed] [Google Scholar]
- 51.Virji, M., K. Makepeace, I. R. Peak, D. J. Ferguson, M. P. Jennings, and E. R. Moxon. 1995. Opc- and pilus-dependent interactions of meningococci with human endothelial cells: molecular mechanisms and modulation by surface polysaccharides. Mol. Microbiol. 18741-754. [DOI] [PubMed] [Google Scholar]
- 52.Virji, M., M. K., Ferguson, D. J., Achtman, M., and E. R. Moxon. 1993. Meningococcal Opa and Opc proteins: their role in colonization and invasion of human epithelial and endothelial cells. Mol. Microbiol. 10499-510. [DOI] [PubMed] [Google Scholar]
- 53.Yasukawa, K., P. Martin, C. R. Tinsley, and X. Nassif. 2006. Pilus-mediated adhesion of Neisseria meningitidis is negatively controlled by the pilus-retraction machinery. Mol. Microbiol. 59579-589. [DOI] [PubMed] [Google Scholar]
- 54.Yoon, S. S., R. F. Hennigan, G. M. Hilliard, U. A. Ochsner, K. Parvatiyar, M. C. Kamani, H. L. Allen, T. R. DeKievit, P. R. Gardner, U. Schwab, J. J. Rowe, B. H. Iglewski, T. R. McDermott, R. P. Mason, D. J. Wozniak, R. E. Hancock, M. R. Parsek, T. L. Noah, R. C. Boucher, and D. J. Hassett. 2002. Pseudomonas aeruginosa anaerobic respiration in biofilms: relationships to cystic fibrosis pathogenesis. Dev. Cell 3593-603. [DOI] [PubMed] [Google Scholar]
- 55.Zaretzky, F. R., M. C. Gray, and E. L. Hewlett. 2002. Mechanism of association of adenylate cyclase toxin with the surface of Bordetella pertussis: a role for toxin-filamentous haemagglutinin interaction. Mol. Microbiol. 451589-1598. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.