The genus Yersinia consists of 15 species (www.bacterio.cict.fr/xz/yersinia.html), and only three of them, Y. pestis, Y. pseudotuberculosis, and Y. enterocolitica, are pathogenic to mammals, including humans. The Y. pseudotuberculosis-Y. pestis evolutionary linkage diverged from Y. enterocolitica between 41 and 186 million years ago, while Y. pestis diverged from Y. pseudotuberculosis within the last 1,500 to 20,000 years (1, 65). In accordance with this evolutionary cascade, wide genetic diversity exists between Y. pseudotuberculosis and Y. enterocolitica, while very close genetic similarity is found between Y. pseudotuberculosis and Y. pestis. Y. pseudotuberculosis causes only nonfatal gastrointestinal disease in mammalian hosts, including humans, and the disease is transmitted by the food-borne route. Y. pestis causes plague, which is one of the most deadly diseases (47). Three pandemics of plague have been recorded in human history and have claimed hundreds of thousands of lives (47). Plague is a typical enzootic disease (an infection of the animal population[s] in one or more confined natural foci without the need for external inputs), and epidemics of rodent plague are restricted in various enzootic plague foci especially in Asia, the Americas, and Africa (80). Compared to its progenitor Y. pseudotuberculosis, Y. pestis utilizes a radically different mechanism of transmission in rodent reservoirs that relies primarily upon biting by flea vectors. This review deals with how genetic changes (gene inactivation, loss, and acquisition) and remodeling of gene regulation encourage Y. pestis to switch from an enteric lifestyle to a mammalian blood-borne lifestyle that relies on vector-borne transmission.
PROGRESSION OF PLAGUE INFECTION
Rodents and humans acquire Y. pestis by the bite of an infected flea, contact with infected tissues, or inhalation of respiratory droplets or aerosols, with manifestations of bubonic, septicemic, and pneumonic plague (47). After the flea bite, there is an initial subcutaneous and intradermal colonization, and then the bacteria migrate into the regional lymph nodes and inflammation, cellulitis, and occasionally large carbuncles develop around the bubo (bubonic plague) (60). Without timely effective treatment, the bacteria will rapidly escape from containment in the lymph node and spread systemically through the blood to various organs, causing fatal sepsis (septicemic plague) (82). An intracellular growth of Y. pestis in macrophages at early stages of infection is thought to allow this pathogen to proliferate and to synthesize virulence determinants, enabling the releasing bacteria to acquire the ability to eliminate the host immune response (39). In addition, secondary pneumonic plague could result from hematogenous spread from the bubo to the lung, presenting in patients as severe bronchopneumonia, cavitation, or consolidation with production of bloody or purulent sputum (82). Primary pneumonic plague could be caused directly by the inhalation of infectious droplets or aerosols, with symptoms including acute pneumonia, intra-alveolar hemorrhage and edema, profound lobular exudation, fibrin deposition, and bacillary aggregation (33). Both primary and secondary pneumonic plagues are highly contagious for close contacts by airborne transmission.
ECOLOGICAL AND EPIDEMIOLOGICAL DIFFERENCES BETWEEN Y. PESTIS AND Y. PSEUDOTUBERCULOSIS
Animals, food, and the abiotic environment are Y. pseudotuberculosis reservoirs from which epizootic and human infection may arise, and the disease is mild and transmitted by the food-borne route. In humans, typical symptoms include fever and right-side abdominal pain. In rare cases the disease may cause skin complaints (erythema nodosum), joint stiffness and pain (reactive arthritis), or spread of bacteria to the blood (bacteremia).
Due to acute and systemic infection, the mortality rate of plague reaches 70 to 100% without treatment depending on routes of infection. Y. pestis has a limited ability to live in the environment, although there is evidence that Y. pestis can live in soil for up to 30 weeks (3). Maintenance of plague in enzootic plague foci is almost absolutely dependent upon cyclic transmission between fleas and mammals (80). Blocked fleas are important for transmission of plague (24). Blockage of fleas (heavy proliferation of bacteria in the adhesive biofilms in the proventriculus) makes them feel hungry and repeatedly attempt to feed, and the plague bacilli will be pumped into the host body during these futile feeding attempts (25). The development of heavy bacteremia (bacterial concentration reaching at least 106 CFU/ml) in hosts is necessary to reliably infect the fleas (38). Such a high level of bacteremia raises the risk of hosts' rapid death. Nevertheless, once some fleas achieve feeding prior to the host's death, they will seek alternative hosts, thereby increasing the likelihood of transmission to other individuals from the hosts (15).
Generally, it takes about 2 weeks for blockage to develop, which is not sufficient to explain the high rate of spread that typifies plague epidemics. Various species of rodent fleas are immediately infectious after biting a septicemic host and transmit bacteria efficiently for at least 4 days postinfection, and accordingly the mode of “early-phase transmission” by unblocked fleas has been proposed (14). Early-phase transmission helps explain not only the rapid spread that typifies plague epidemics but also previous inconsistencies between the rates of pathogen spread expected from the blocked fleas. It was suggested that mechanical transmission by unblocked fleas is significant during epidemics that represent the periods when Y. pestis can spread rapidly across landscapes but that transmission by blocked fleas is important primarily during interepizootic transmission (15). In addition, a combination of early-phase transmission and blocking probably helps to explain the observed high mortality rates of susceptible host populations, including humans during the Black Death (74).
The rodent reservoirs, the flea vectors, and Y. pestis constitute a well-balanced biocommunity in the plague foci. Y. pestis possesses potential to attack humans, and human infection usually occurs with the transmission of the pathogen from rodents. Although cases of human plague can be well controlled by timely antibiotic administration, plague still remains a significant concern for public health because it can be transmitted from person to person through respiratory droplets and used for bioterrorism and biological warfare (68).
Y. PESTIS VIRULENCE DETERMINANTS SHARED BY Y. PSEUDOTUBERCULOSIS
Y. pestis has developed specialized strategies for virulence in hosts and transmission by fleas (Table 1), and many of these determinants are harbored in the genome of Y. pseudotuberculosis.
TABLE 1.
Virulence determinants or functions involved in molecular evolution of Y. pestis
| Function and protein name | Gene identifier(s) | Description | Status in organism:
|
Reference(s) | Labela | |
|---|---|---|---|---|---|---|
| Y. pseudotuberculosis | Y. pestis | |||||
| Flea transmission | ||||||
| GmhA | YPO3243 | Enzymes for biofilm formation | Present | Present | 11 | |
| SpeA and SpeC | YPO0929 and YPO1201 | Enzymes for biofilm formation | Present | Present | 46 | |
| YrbH | YPO3577 | Enzymes for biofilm formation | Present | Present | 71 | |
| Hms | YPO1951 to YPO1954 | Enzymes for biofilm formation | Present | Inhibited | 28 | |
| NghA | YPO2632 | Biofilm-inhibiting glycosyl hydrolase | Present | Inactivated | 16 | β |
| RcsA | YPO2449 | Transcriptional repressor of biofilm formation | Present | Inactivated | 69 | β |
| Ymt | YPMT1.74 | Phospholipase D required for survival in flea | Absent | Present | 26 | α |
| Colonization and dissemination | ||||||
| Inv | YPO1793 | Invasin | Present | Inactivated | 63 | β |
| YadA | YPCD1.87c | Adhesin and invasin | Present | Inactivated | 45 | β |
| Ail | YPO2905 | Adhesin and invasin | Present | Present | 31 | |
| YadBC | YPO1387 to YPO1388 | Invasin | Present | Present | 20 | |
| YapC | YPO2796 | Autotransporter adhesin | Present | Present | 18 | |
| YapE | YPO3984 | Autotransporter adhesin | Present | Present | 35 | |
| Pla | YPPCP1.07 | Plasminogen activator promoting bacterial in vivo dissemination | Absent | Present | 34, 61 | α |
| Intracellular growth | ||||||
| RipA | YPO1926 | Putative acetyl coenzyme A transferase | Present | Present | 54 | |
| Ugd | YPO2174 | LPS modification | Present | Present | 22 | |
| MgtCB | YPO1660 to YPO1661 | Magnesium uptake | Present | Present | 22 | |
| Yfe | YPO2439 to YPO2442 | ABC-type iron transporter | Present | Present | 48 | |
| FeoBA | YPO0132 to YPO0133 | Ferrous iron transporter | Present | Present | 48 | |
| Elimination of host immune response | ||||||
| YadA | YPCD1.87c | Serum resistance | Present | Inactivated | 45 | β |
| Ail | YPO2905 | Serum resistance | Present | Present | 31 | |
| pH 6 antigen | YPO1301 to YPO1305 | Resistance to phagocytosis | Present | Present | 27 | |
| T3SS | Elimination of innate immune cells | Present | Present | 56 | ||
| Tc proteins | Toxicity to mammalian cells | Present | Present | 23 | ||
| F1 capsule | YPMT1.81c to YPMT1.84 | Resistance to phagocytosis | Absent | Present | 13 | α |
| O-antigen genes | Lack of O antigen is essential for Pla function | Present | Inactivated | 32 | β | |
| Iron uptake | ||||||
| Yfe | YPO2439 to YPO2442 | ABC-type iron transporters | Present | Present | 5 | |
| High-pathogenicity island | YPO1906 to YPO1916 | Siderophore yersiniabactin-based iron acquisition system | Present | Present | 5 | |
| Virulence-required regulators | ||||||
| VirF | YPCD1.49 | Positive regulator of T3SS | Present | Present | 19 | |
| CRP | YPO0175 | Positive regulator of Pla | Present | Present | 79 | |
| PhoP-PhoQ | YPO1633 to YPO1634 | Positive regulator of Ugd and MgtCB | Present | Present | 44 | |
| RovA | YPO2374 | Positive regulator of pH 6 antigen | Present | Present | 8 | |
| Other virulence determinants | ||||||
| Dam | YPO0154 | DNA adenine methylation | Present | Present | 58 | |
| HtrA | YPO3382 | Serine protease | Present | Present | 78 | |
| Lpp | YPO2394 | Braun lipoprotein | Present | Present | 62 | |
| YpfΦ | YPO2272 to YPO2281 | Filamentous prophage | Absent | Present | 12 | α |
| Y. pestis-specific chromosomal regions | ||||||
| YPO0387 to YPO0396 | Hypothetical proteins | Absent | Present | 76 | α | |
| YPO2087 to YPO2093 | Prophage proteins | Absent | Present | 76 | α | |
| Y. pseudotuberculosis-specific chromosome regions | ||||||
| LpxL | YPTB2046 | Loss of LpxL enables Y. pestis to evade LPS-induced inflammation | Present | Absent | 41 | γ |
| R1 | YPTB0872 to YPTB0878 | Methionine salvage pathway required for virulence of Y. pseudotuberculosis | Present | Absent | 51 | γ |
| ORF3 and ORF4 | YPTB1495 and YPTB3368 | Hypothetical proteins essential for viability of Y. pseudotuberculosis | Present | Absent | 51 | γ |
| ORF2 | YPTB1058 | Putative pseudouridylate synthase necessary for optimal growth of Y. pseudotuberculosis | Present | Absent | 51 | γ |
| R3 | YPTB2193 to YPTB2201 | Hypothetical proteins necessary for optimal growth of Y. pseudotuberculosis | Present | Absent | 51 | γ |
| Genes linked to human-virulence attenuation of biovar Microtus strains | ||||||
| AspA | YPO0348 | Inactivated in typical Y. pestis rather than human-avirulent biovar Microtus or Pestoides strains | Present | Present | 6, 75 | |
| YPMT1.43c | Inactivated in biovar Microtus strains rather than typical Y. pestis | Present | Present | 83 | ||
| YPO1956 | Inactivated in biovar Microtus strains rather than typical Y. pestis | Present | Present | 83 | ||
| YPO1973 | Inactivated in biovar Microtus strains rather than typical Y. pestis | Present | Present | 83 | ||
| YPO2258 | Inactivated in biovar Microtus strains rather than typical Y. pestis | Present | Present | 83 | ||
| YPO2729 | Inactivated in biovar Microtus strains rather than typical Y. pestis | Present | Present | 83 | ||
| YPO2731 | Inactivated in biovar Microtus strains rather than typical Y. pestis | Present | Present | 83 | ||
| YPO3049 | Inactivated in biovar Microtus strains rather than typical Y. pestis | Present | Present | 83 | ||
| YPO1986 to YPO1987 | Absent in biovar Microtus strains rather than typical Y. pestis | Present | Present | 83 | ||
| YPO2096 to YPO2135 | Absent in biovar Microtus strains rather than typical Y. pestis | Present | Present | 83 | ||
| YPO2469 | Absent in biovar Microtus strains rather than typical Y. pestis | Present | Present | 83 | ||
| YPO2487 to YPO2489 | Absent in biovar Microtus strains rather than typical Y. pestis | Present | Present | 83 | ||
| YPO3046 to YPO3047 | Absent in biovar Microtus strains rather than typical Y. pestis | Present | Present | 83 | ||
α, acquired by Y. pestis through lateral gene transfer; β, conserved in Y. pseudotuberculosis but inactivated in Y. pestis; γ, conserved in Y. pseudotuberculosis but absent from Y. pestis.
COLONIZATION AND DISSEMINATION
The major adhesin and invasin, YadA and Inv, respectively, specific for gastrointestinal infection are inactivated in Y. pestis (see below), but this pathogen still has additional proteins (Ail [31], YadBC [20], and YapE [35]) that account for bacterial colonization and dissemination during infection.
INTRACELLULAR GROWTH
The ability to replicate in macrophages is conserved in Y. pestis and Y. pseudotuberculosis (53). RipABC (54), MgtCB (22), Ugd (22), Yfe (48), and Feo (48) have been shown to be required for the replication of Y. pestis in macrophages. Both MgtCB and Ugd are positively regulated by the PhoP-PhoQ two-component system (37) that is important for survival under conditions of macrophage-induced stress and virulence in Y. pestis (44).
ELIMINATION OF HOST IMMUNE RESPONSE
The plasmid pCD1-borne type III secretion system (T3SS) is composed of a secretion machinery, a set of translocation proteins, a control system, and six Yop effector proteins (56). Through the T3SS, pathogenic yersiniae inject effectors into the cytosol of eukaryotic cells when docking at the surface of host cells, and the injected Yops mediate suppression of phagocytosis and the inflammatory reaction (56). Y. pestis utilizes T3SS to selectively destroy innate immune cells that represent the first line of host defense, thereby preventing adaptive responses and precipitating the fatal outcome of plague (40). Y. pestis still employs pH 6 antigen fimbriae to function as an antiphagocytic factor independent of Yops (27).
The Tc genes were first identified in the insect pathogen and encode a protein complex toxic to insects. Tc proteins in Y. pseudotuberculosis and Y. pestis are not insecticidal toxins but have evolved toxicity to mammalian cells (23).
Heavy proliferation of Y. pestis in the bloodstream is essential for its transmission by fleas. Resistance to complement-mediated lysis (serum resistance) is required for bacterial survival in mammalian blood. The Ail protein (4) and lipopolysaccharide (LPS) (50) promote serum resistance, which appears to be a conserved mechanism in pathogenic yersiniae.
IRON UPTAKE
In mammals, iron is bound to Fe3+-binding proteins and hemoproteins, and thus free iron is too rare to sustain bacterial growth. Iron acquisition is critical for the survival of pathogenic bacteria during infection. A wide array of iron acquisition systems have been characterized or annotated for Y. pestis (21), and at least two (Ybt and Yfe) of them were proven to be required for full virulence (5). Ybt, also known as the high-pathogenicity island (59), is essential to iron acquisition at the site of the flea bite and in the lymphatic system, while Yfe is likely used in the later stages of the disease, i.e., blood-borne systemic dissemination (5).
LATERAL ACQUISITION OF NOVEL VIRULENCE DETERMINANTS BY Y. PESTIS
Lateral gene transfer directly introduces foreign DNA elements into the host genome, which will effectively alter the pathogenic characters of bacterial species (42). Y. pestis has acquired two unique virulence plasmids, pPCP1 and pMT1, through lateral gene transfer. pPCP1 encodes plasminogen activator (Pla), while pMT1 encodes murine toxin (Ymt) and F1 capsule (Table 1).
Pla is essential for bubonic and primary pneumonic plague (but not primary and secondary septicemic forms), since it specifically promotes Y. pestis dissemination from peripheral infection routes (34, 61). At 37°C but not 26°C, Y. pestis expresses a capsule-like antigen, called F1 antigen. F1 provides Y. pestis the ability to block phagocytosis by a mechanism different from those of T3SS and pH 6 antigen (13). Ymt does not play a role in mouse infection (57) but shows phospholipase D activity and is required for survival of Y. pestis in fleas (26). It was thought that intracellular phospholipase D activity appeared to protect Y. pestis from a cytotoxic digestion product of plasma in the flea gut (26).
Unexpectedly, only two chromosomal regions seem to be specific to Y. pestis (76) (Table 1). They are located in two different genomic islands probably acquired through lateral gene transfer (45). These two Y. pestis-specific chromosomal regions deserve more attention to investigation of their roles in virulence and/or transmission by fleas. However, it has been argued that analysis of more bacterial strains could further reduce the number of Y. pestis-specific chromosomal genes, perhaps to zero (7, 45).
DECAY OF REDUNDANT OR DELETERIOUS FUNCTIONS IN Y. PESTIS
About 13% of Y. pseudotuberculosis genes no longer function (inactivated or absent) in Y. pestis CO92 (45). Genome decay (gene loss and inactivation) appears to be closely linked to flea-borne transmission and increased virulence of Y. pestis (Table 1).
GENE INACTIVATION
yadA and inv encode the major adhesin and invasin, respectively, in Y. pseudotuberculosis and enable this enteropathogen to specifically adhere to surfaces of host intestines and invade lining epithelial cells. Both of them are inactivated in Y. pestis (30). Urease plays its role in using urea as a source of nitrogen. Production of urease by the ure operon is necessary for oral transmission of Y. pseudotuberculosis, but it is inactivated in Y. pestis due to the mutation causing a premature stop codon in ureD (33). Since Y. pestis spends its life almost exclusively in a flea-host-flea cycle, the organism can lose with impunity the function of urease needed for survival in natural environments.
Y. pestis expresses rough LPS lacking the O antigen, due to the inactivation of several genes in the O-antigen gene cluster (52). Y. pseudotuberculosis produces a rough LPS at 37°C but not at 26°C, and a variable number of LPS genes are seen to be defective when various biovars of Y. pestis are compared (64). Expression of rough LPS is essential for Pla activity and virulence in Y. pestis (32). A pathogenic advantage of rough LPS in Y. pestis is that it enables efficient Pla-mediated bacterial dissemination to cause systemic disease.
For the blocked fleas, Y. pestis synthesizes an attached biofilm in the flea proventriculus and in its midgut posteriorly (25). Three distinct operons, hmsHFRS, hmsT, and hmsP, are involved in the synthesis of bacterial extracellular matrix, which is the primary component of Yersinia biofilm (28). Y. pseudotuberculosis contains all of these hms genes, which are 99% identical to the Y. pestis homologues (9). In addition to Hms, several other proteins (GmhA, SpeAC, and YrbH) involved in biofilm formation by Y. pestis are harbored in Y. pseudotuberculosis (Table 1).
Only a small number of Y. pseudotuberculosis strains are able to form biofilm on Caenorhabditis elegans, and none of them has the ability to form adhesive biofilms in fleas (17). The transcriptional regulator RcsA (69) and the glycosyl hydrolase NghA (16) have been shown to inhibit Yersinia biofilm formation, but both of them are functional in Y. pseudotuberculosis but inactivated in Y. pestis (16, 69). Expression of functional RcsA or NghA in Y. pestis strongly represses biofilm formation and abolishes flea blockage (16). Therefore, Y. pestis has evolved the changes in regulatory functions on biofilm development to ensure stable biofilm formation in the flea proventriculus and to result in efficient arthropod-borne transmission.
GENE LOSS
Hexa-acylated LPS observed in many gram-negative pathogens is able to activate TLR4 signaling and to further stimulate the host innate immune response (55). The acyltransferase LpxL is required for addition of the secondary acyl chains to the tetra-acylated precursor (55). The lpxL gene is absent from Y. pestis, leading this pathogen to produce tetra-acylated LPS that inhibits TLR4 activation, which allows this pathogen to evade the protective inflammatory response and establish fatal infection (41).
Five additional Y. pseudotuberculosis-specific chromosomal loci (R1, R3, ORF2, ORF3, and ORF4) required for its survival, optimal growth, or virulence are absent from Y. pestis (51) (Table 1). ORF3 and ORF4, with unknown function, are essential for the viability of Y. pseudotuberculosis, while ORF2 (a putative pseudouridylate synthase involved in RNA stability) and R3 (a genomic region composed mostly of genes of unknown functions) are necessary for its optimal growth in a chemically defined medium (51). Deletion of R1 (a genomic region responsible for the methionine salvage pathway) alters the mutant's virulence, suggesting that the availability of free methionine is severely restricted in vivo (51).
REMODELING OF GENE REGULATION
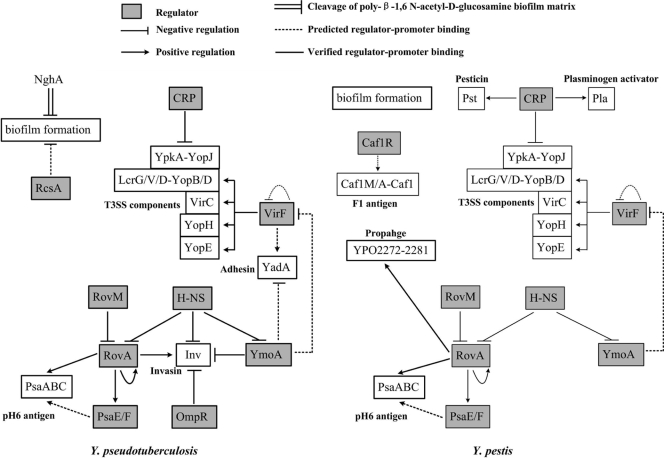
Virulence determinants are tightly and coordinately regulated during infection. Virulence-related regulators can sense host signals, e.g., changes in temperature, and then differentially regulate not only virulence genes but other large sets of genes required for adaptation to the host niche (84). A number of virulence-required regulators have been characterized in Y. pestis and Y. pseudotuberculosis (Table 1), indicating that remodeling of gene regulation contributes to the indicated differences between these two pathogens (Fig. 1).
FIG. 1.
Remodeling of gene regulation in Y. pestis and Y. pseudotuberculosis. Transcription regulators listed here bind to cis-acting DNA sequences within the promoters of their target genes and either activate or repress transcription initiation of these targets. Some regulators (e.g., CRP) can function as either activators or repressors according to the target promoters. Shown also is autoregulation of some regulators (e.g., RovA).
INTEGRATION OF LATERALLY ACQUIRED VIRULENCE GENES
The global transcriptional regulators cyclic AMP receptor protein (CRP) (49, 79) and RovA (8, 73) are conserved and required for virulence in the three pathogenic Yersinia species. In addition, the Y. pestis CRP is 98.6% identical to the Escherichia coli one with the same length, and CRPs from these two bacteria share an identical consensus box sequence (TGTGA-N6-TCACA) that represents the conserved signals for CRP recognition of promoter DNA (79). Through regulator-promoter DNA interaction in Y. pestis, CRP activates two laterally acquired plasmid genes, pla and pst (30, 79), while RovA upregulates a genomic region, YPO2272 to YPO2281 (8). The prophage YPO2272 to YPO2281 is acquired by the Y. pestis ancestor, and its genome forms an unstable episome in biovars Antiqua and Medievalis whereas it is stably integrated in biovar Orientalis (12, 36). The acquisition of this prophage does not correlate with flea transmission but contributes to virulence in mice (12). These “newly” acquired virulence genes have evolved to integrate themselves into the “ancestral” Yersinia regulatory cascade. The plague pathogen integrates laterally acquired genes to coordinate virulence factor expression within global gene regulatory networks to maintain homeostasis through the infectious life cycle.
ADDITION OF LATERALLY ACQUIRED VIRULENCE GENE AND ITS OWN SPECIFIC REGULATOR
The pMT1-borne F1 antigen locus is transcribed as two operons (caf1M-caf1A-caf1 and caf1R) with opposite directions. Caf1R is a positive transcriptional activator responsible for the regulation of capsule formation (29). Thus, the newly acquired caf genes constitute a self-controlling regulatory cascade. According to our microarray expression data, CRP activated caf1R whereas it repressed the caf1M-caf1A-caf1 operon (79), and the nucleoid-associated protein Fis stimulates all the caf genes (unpublished data). Whether these two caf operons are controlled by the host's regulators still needs to be elucidated.
DECAY OF REDUNDANT OR DELETERIOUS REGULATORS/TARGETS
As mentioned above, Y. pestis has evolved the genetic inactivation of inhibitory functions (RcsA or NghA) on biofilm formation, which greatly contributes to efficient flea-borne transmission. RovA and VirF positively control inv and yadA, respectively, while reductive inactivation of these two targets has occurred in Y. pestis (see above). However, the upstream regulators RovA and VirF still function to regulate other virulence genes in Y. pestis, and both of them are required for virulence of Y. pestis (8, 19). The established target of the activator RovA is PsaABC (pH 6 antigen) (8), while VirF stimulates several components (YpkA-YopJ, LcrG/V/D-YopB/D, VirC, YopH, and YopE) of T3SS (77), which appears to be conserved between Y. pestis and Y. pseudotuberculosis.
PROBABLE DARWINIAN EMERGENCE OF PLAGUE
Y. pseudotuberculosis harbors a set of functional or structural determinants including adhesion/invasin, T3SS, pH 6 antigen, Tc proteins, iron uptake systems, and enzymes for biofilm formation. Thereby, it has the potential to attack mammals to cause systemic infection and to be transmitted by fleas. At a certain stage of history, the change of natural environment might have led to the dramatic increase of population size or to behaviors of a certain rodent, probably the woodchuck (70). This change in animal reservoir might trigger the speciation of Y. pestis from Y. pseudotuberculosis as a directional positive selection (Darwinian selection). Y. pseudotuberculosis is found widely in the environment and is a common cause of animal infections. The bacteria can invade individual rodents suffering from cold, hunger, or illness due to drastic in-species competition or a harsh environment and then are sucked into the bodies of fleas through flea biting.
Y. pseudotuberculosis or ancestral Y. pestis shares a niche with other organisms in rodents and fleas, which might allow the random occurrence of lateral gene transfer. At the same time, gene loss or inactivation occurs randomly as well. Beneficial events of genetic variations would be stabilized by vertical inheritance under Darwinian selection. The positive and directional selection promotes the acquisition of novel virulence determinants as well as the decay of redundant or deleterious functions, which would stimulate the emergence of Y. pestis. In addition, remodeling of gene regulation enables the coordinated regulation of existent and newly acquired virulence markers. The Darwinian selective advantages contribute to the demonstrated differences between Y. pestis and Y. pseudotuberculosis and favor an ordered buildup of specific combinations of virulence determinants enabling the establishment and transmission of Y. pestis as a new clone from Y. pseudotuberculosis.
ADAPTIVE INTRASPECIFIC MICROEVOLUTION AND DIVERSIFICATION OF Y. PESTIS
Y. pestis has been historically divided into three biovars, namely, Antiqua, Medievalis, and Orientalis, and they are thought to be linked with the first to third plague pandemics, respectively. Each biovar has unique genes, a different profile of inactivated genes, and a distinct genome structure according to the relevant sequenced genomes. In addition to the above three classical biovars, another distinct group of strains, called biovar Microtus (83), are avirulent in primates and some large rodent species and are thought to be the intermediate evolutionary clade between Y. pseudotuberculosis and Y. pestis (36, 72, 81). Compared to other types of Y. pestis, biovar Microtus strains have a unique genomic profile of gene loss and pseudogene distribution (36, 72, 81, 83). The specific loss of genes or gene functions documented for this group of strains is thought to be responsible for the human attenuation of these strains, providing candidates for further hypothesis-driven investigations of virulence microevolution of Y. pestis.
Y. pestis strains in North and South America are clonally derived from a biovar Orientalis strain due to the third pandemic of human plague in the early 20th century and thus genetically restricted (2). In China, Mongolia, and the former Soviet Union, there are large areas of enzootic plague foci containing genetically diverse strains (2). Y. pestis isolates from these plague foci can be classified into many different genotypes (also known as genomovars) (2, 10, 36, 72, 81). Accumulation of genetic variations promotes the diversification of Y. pestis, while distribution of Y. pestis genotypes is plague focus specific (36, 81). The parallel expansion of plague foci as well as the directional diversification of Y. pestis within these foci is likely subject to the action of the complex of interactions between the environment, the hosts, and the pathogen (81). For a specific plague focus, Y. pestis genotypes can be assigned into major and minor genomovars (36). Strains of major genomovars represent the dominant populations in a specific plague focus, and they are generally isolated from the main reservoirs/vectors and distributed throughout the focus. In contrast, the strains of minor ones account for a very small portion of the tested stains from the specific plague focus and are distributed sporadically in a focus or along the border of neighboring foci. Notably, major and minor genomovars make sense in combination with the concept of a specific plague focus. The major genomovar in one plague focus might be the minor one in the others.
In summary, adaptive microevolution likely promotes diversification of Y. pestis (different major and minor genomovars) within enzootic foci; major genotypes would play a crucial role in the maintenance of plague in enzootic foci, whereas minor ones (likely representing evolutionary dead ends) would make little contribution to the well-balanced interactions between environment, hosts, and Y. pestis (36, 81). This speculation still needs further evidence to show phenotypic effects of gene loss/gain during microevolution of Y. pestis.
DARWINIAN ADAPTIVE EVOLUTION VERSUS NEUTRAL GENETIC DRIFT
The driving forces of molecular evolution involve two aspects, namely, “genetic drift” and “natural [or Darwinian] selection.” Genetic drift promotes the accumulation of neutral, random changes in a gene pool, and thus it affects only genotypic frequencies within a population and has no phenotypic causes. Darwinian selection makes alleles more or less widespread in a population due to their effects on fitness advantages; therefore, it influences both phenotype and genotype components in a population. Darwinian selection impels the creation of adaptations, while genetic drift does not. Classical Darwinian evolution refers to the specific selection and fixation of small allelic variants (point mutations) that confer positive evolutionary advantage. It is becoming more and more evident that accumulation of large genomic changes (gene loss/acquisition) can alter the phenotypes in “quantum leaps” during the evolution of bacterial pathogenesis (43). Herein, selection and fixation of inactivation, loss, and acquisition of functional genes are all thought to be under the framework of Darwinian adaptive evolution.
Although evolved from the mildly pathogenic bacterium Y. pseudotuberculosis, Y. pestis is a highly virulent pathogen and has switched from an enteric lifestyle to a mammalian blood-borne one. For it to become a more virulent pathogen, at least three adaptive evolution mechanisms were involved to gain more pathogenic phenotypes: (i) the horizontal acquisition of genes encoding specific virulence determinants (“gain-of-function” mechanism), (ii) an appropriate functional inactivation or loss of preexisting genetic materials (“loss-of-function” mechanism), and (iii) laterally acquired virulence genes either with or without their own specific regulators, which evolve to being integrated into the host's regulatory cascades to coordinate expression of virulence factors within global gene regulatory networks for maintaining homeostasis through the infectious life cycle (“regulation-remodeling” mechanism). Genetic variations occur randomly, yet only those beneficial to mammalian blood-borne infection or vector-borne transmission of Y. pestis would be stabilized by vertical inheritance under Darwinian selection. Darwinian adaptive evolution (rather than neutral genetic drift) would induce Y. pestis to evolve from Y. pseudotuberculosis to a newly emerged pathogen that is not only able to parasitize insects in part of its life cycle but also highly virulent to rodents and humans, causing pandemics of a systemic and often fatal disease.
PERSPECTIVES
Comparison of available genome sequences of Y. pestis and Y. pseudotuberculosis enables us to find genetic differences in minute detail. Subsequent hypothesis-directed studies have presented a wealth of experimental evidence that promotes our understanding of the positive evolution of virulence during speciation of Y. pestis, whereas phenotypic characterization of intraspecific microevolution of Y. pestis and its link to expansion of enzootic plague foci is lacking.
Tightly regulated virulence genes are important components of the global gene regulation network, which is a three-dimensional architecture involving various regulators and structural genes. Y. pestis has evolved extremely complex signaling and regulatory pathways that are activated during different stages of infection. Global comparative analysis of gene regulation (84) in Y. pestis and Y. pseudotuberculosis would bring a dynamic and complete picture of virulence and host adaption in Y. pestis.
The insertion sequence (IS) elements constitute about 3% of the whole genome in Y. pestis (67). Due to IS-mediated homologous recombination, the genome structures of different Y. pestis strains are markedly different, although their genome contents and sequences show high similarity (67). There are no reported data for the IS-mediated recombination event that is experimentally linked to the virulence evolution of Y. pestis.
An evolutionary “source-sink” model was recently proposed to detect genetic adaptation of bacterial pathogens, with which the evolution of bacterial pathogens can be seen from the angle of continuous switching between permanent (source) and transient (sink) habitats (66). Virulence habitats are marginal sink habitats for some pathogens, and virulence-enhancing genetic adaptation is mostly transient in nature. The adaptation to harsh, or “sink,” environments is the supply of beneficial mutations via migration from a “source” population. Experimental evidence to support the source-sink model for Y. pestis needs to be characterized and would provide a conceptual framework for understanding the population dynamics and molecular mechanisms of virulence evolution.
Acknowledgments
Related work in our laboratory has been supported by grants from the National Science Foundation of China for Distinguished Young Scholars (30525025), the State Key Development Program for Basic Research of China (2009CB522604 and 2009CB522601), the National Natural Science Foundation of China (30771179 and 30600353), and the Beijing Natural Science Foundation (5072040).
Editor: J. B. Kaper
Footnotes
Published ahead of print on 16 March 2009.
REFERENCES
- 1.Achtman, M., K. Zurth, G. Morelli, G. Torrea, A. Guiyoule, and E. Carniel. 1999. Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. Proc. Natl. Acad. Sci. USA 9614043-14048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anisimov, A. P., L. E. Lindler, and G. B. Pier. 2004. Intraspecific diversity of Yersinia pestis. Clin. Microbiol. Rev. 17434-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ayyadurai, S., L. Houhamdi, H. Lepidi, C. Nappez, D. Raoult, and M. Drancourt. 2008. Long-term persistence of virulent Yersinia pestis in soil. Microbiology 1542865-2871. [DOI] [PubMed] [Google Scholar]
- 4.Bartra, S. S., K. L. Styer, D. M. O'Bryant, M. L. Nilles, B. J. Hinnebusch, A. Aballay, and G. V. Plano. 2008. Resistance of Yersinia pestis to complement-dependent killing is mediated by the Ail outer membrane protein. Infect. Immun. 76612-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bearden, S. W., and R. D. Perry. 1999. The Yfe system of Yersinia pestis transports iron and manganese and is required for full virulence of plague. Mol. Microbiol. 32403-414. [DOI] [PubMed] [Google Scholar]
- 6.Bearden, S. W., C. Sexton, J. Pare, J. M. Fowler, C. G. Arvidson, L. Yerman, R. E. Viola, and R. R. Brubaker. 2009. Attenuated enzootic (pestoides) isolates of Yersinia pestis express active aspartase. Microbiology 155198-209. [DOI] [PubMed] [Google Scholar]
- 7.Brubaker, R. R. 2005. Influence of Na+, dicarboxylic amino acids, and pH in modulating the low-calcium response of Yersinia pestis. Infect. Immun. 734743-4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cathelyn, J. S., S. D. Crosby, W. W. Lathem, W. E. Goldman, and V. L. Miller. 2006. RovA, a global regulator of Yersinia pestis, specifically required for bubonic plague. Proc. Natl. Acad. Sci. USA 10313514-13519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chain, P. S., E. Carniel, F. W. Larimer, J. Lamerdin, P. O. Stoutland, W. M. Regala, A. M. Georgescu, L. M. Vergez, M. L. Land, V. L. Motin, R. R. Brubaker, J. Fowler, J. Hinnebusch, M. Marceau, C. Medigue, M. Simonet, V. Chenal-Francisque, B. Souza, D. Dacheux, J. M. Elliott, A. Derbise, L. J. Hauser, and E. Garcia. 2004. Insights into the evolution of Yersinia pestis through whole-genome comparison with Yersinia pseudotuberculosis. Proc. Natl. Acad. Sci. USA 10113826-13831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cui, Y., Y. Li, O. Gorge, M. E. Platonov, Y. Yan, Z. Guo, C. Pourcel, S. V. Dentovskaya, S. V. Balakhonov, X. Wang, Y. Song, A. P. Anisimov, G. Vergnaud, and R. Yang. 2008. Insight into microevolution of Yersinia pestis by clustered regularly interspaced short palindromic repeats. PLoS ONE. 3e2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Darby, C., S. L. Ananth, L. Tan, and B. J. Hinnebusch. 2005. Identification of gmhA, a Yersinia pestis gene required for flea blockage, by using a Caenorhabditis elegans biofilm system. Infect. Immun. 737236-7242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Derbise, A., V. Chenal-Francisque, F. Pouillot, C. Fayolle, M. C. Prevost, C. Medigue, B. J. Hinnebusch, and E. Carniel. 2007. A horizontally acquired filamentous phage contributes to the pathogenicity of the plague bacillus. Mol. Microbiol. 631145-1157. [DOI] [PubMed] [Google Scholar]
- 13.Du, Y., R. Rosqvist, and A. Forsberg. 2002. Role of fraction 1 antigen of Yersinia pestis in inhibition of phagocytosis. Infect. Immun. 701453-1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eisen, R. J., S. W. Bearden, A. P. Wilder, J. A. Montenieri, M. F. Antolin, and K. L. Gage. 2006. Early-phase transmission of Yersinia pestis by unblocked fleas as a mechanism explaining rapidly spreading plague epizootics. Proc. Natl. Acad. Sci. USA 10315380-15385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eisen, R. J., and K. L. Gage. 2008. Adaptive strategies of Yersinia pestis to persist during inter-epizootic and epizootic periods. Vet. Res. 401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Erickson, D. L., C. O. Jarrett, J. A. Callison, E. R. Fischer, and B. J. Hinnebusch. 2008. Loss of a biofilm-inhibiting glycosyl hydrolase during the emergence of Yersinia pestis. J. Bacteriol. 1908163-8170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Erickson, D. L., C. O. Jarrett, B. W. Wren, and B. J. Hinnebusch. 2006. Serotype differences and lack of biofilm formation characterize Yersinia pseudotuberculosis infection of the Xenopsylla cheopis flea vector of Yersinia pestis. J. Bacteriol. 1881113-1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Felek, S., M. B. Lawrenz, and E. S. Krukonis. 2008. The Yersinia pestis autotransporter YapC mediates host cell binding, autoaggregation and biofilm formation. Microbiology 1541802-1812. [DOI] [PubMed] [Google Scholar]
- 19.Flashner, Y., E. Mamroud, A. Tidhar, R. Ber, M. Aftalion, D. Gur, S. Lazar, A. Zvi, T. Bino, N. Ariel, B. Velan, A. Shafferman, and S. Cohen. 2004. Generation of Yersinia pestis attenuated strains by signature-tagged mutagenesis in search of novel vaccine candidates. Infect. Immun. 72908-915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Forman, S., C. R. Wulff, T. Myers-Morales, C. Cowan, R. D. Perry, and S. C. Straley. 2008. yadBC of Yersinia pestis, a new virulence determinant for bubonic plague. Infect. Immun. 76578-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao, H., D. Zhou, Y. Li, Z. Guo, Y. Han, Y. Song, J. Zhai, Z. Du, X. Wang, J. Lu, and R. Yang. 2008. The iron-responsive Fur regulon in Yersinia pestis. J. Bacteriol. 1903063-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grabenstein, J. P., H. S. Fukuto, L. E. Palmer, and J. B. Bliska. 2006. Characterization of phagosome trafficking and identification of PhoP-regulated genes important for survival of Yersinia pestis in macrophages. Infect. Immun. 743727-3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hares, M. C., S. J. Hinchliffe, P. C. Strong, I. Eleftherianos, A. J. Dowling, R. H. Ffrench-Constant, and N. Waterfield. 2008. The Yersinia pseudotuberculosis and Yersinia pestis toxin complex is active against cultured mammalian cells. Microbiology 1543503-3517. [DOI] [PubMed] [Google Scholar]
- 24.Hinnebusch, B. J. 2003. Transmission factors: Yersinia pestis genes required to infect the flea vector of plague. Adv. Exp. Med. Biol. 52955-62. [DOI] [PubMed] [Google Scholar]
- 25.Hinnebusch, B. J., and D. L. Erickson. 2008. Yersinia pestis biofilm in the flea vector and its role in the transmission of plague. Curr. Top. Microbiol. Immunol. 322229-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hinnebusch, B. J., A. E. Rudolph, P. Cherepanov, J. E. Dixon, T. G. Schwan, and A. Forsberg. 2002. Role of Yersinia murine toxin in survival of Yersinia pestis in the midgut of the flea vector. Science 296733-735. [DOI] [PubMed] [Google Scholar]
- 27.Huang, X. Z., and L. E. Lindler. 2004. The pH 6 antigen is an antiphagocytic factor produced by Yersinia pestis independent of Yersinia outer proteins and capsule antigen. Infect. Immun. 727212-7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jarrett, C. O., E. Deak, K. E. Isherwood, P. C. Oyston, E. R. Fischer, A. R. Whitney, S. D. Kobayashi, F. R. DeLeo, and B. J. Hinnebusch. 2004. Transmission of Yersinia pestis from an infectious biofilm in the flea vector. J. Infect. Dis. 190783-792. [DOI] [PubMed] [Google Scholar]
- 29.Karlyshev, A. V., E. E. Galyov, V. M. Abramov, and V. P. Zav'yalov. 1992. Caf1R gene and its role in the regulation of capsule formation of Y. pestis. FEBS Lett. 30537-40. [DOI] [PubMed] [Google Scholar]
- 30.Kim, T. J., S. Chauhan, V. L. Motin, E. B. Goh, M. M. Igo, and G. M. Young. 2007. Direct transcriptional control of the plasminogen activator gene of Yersinia pestis by the cyclic AMP receptor protein. J. Bacteriol. 1898890-8900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kolodziejek, A. M., D. J. Sinclair, K. S. Seo, D. R. Schnider, C. F. Deobald, H. N. Rohde, A. K. Viall, S. S. Minnich, C. J. Hovde, S. A. Minnich, and G. A. Bohach. 2007. Phenotypic characterization of OmpX, an Ail homologue of Yersinia pestis KIM. Microbiology 1532941-2951. [DOI] [PubMed] [Google Scholar]
- 32.Kukkonen, M., M. Suomalainen, P. Kyllonen, K. Lahteenmaki, H. Lang, R. Virkola, I. M. Helander, O. Holst, and T. K. Korhonen. 2004. Lack of O-antigen is essential for plasminogen activation by Yersinia pestis and Salmonella enterica. Mol. Microbiol. 51215-225. [DOI] [PubMed] [Google Scholar]
- 33.Lathem, W. W., S. D. Crosby, V. L. Miller, and W. E. Goldman. 2005. Progression of primary pneumonic plague: a mouse model of infection, pathology, and bacterial transcriptional activity. Proc. Natl. Acad. Sci. USA 10217786-17791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lathem, W. W., P. A. Price, V. L. Miller, and W. E. Goldman. 2007. A plasminogen-activating protease specifically controls the development of primary pneumonic plague. Science 315509-513. [DOI] [PubMed] [Google Scholar]
- 35.Lawrenz, M. B., J. D. Lenz, and V. L. Miller. 2009. A novel autotransporter adhesin is required for efficient colonization during bubonic plague. Infect. Immun. 77317-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li, Y., E. Dai, Y. Cui, M. Li, Y. Zhang, M. Wu, D. Zhou, Z. Guo, X. Dai, B. Cui, Z. Qi, Z. Wang, H. Wang, X. Dong, Z. Song, J. Zhai, Y. Song, and R. Yang. 2008. Different region analysis for genotyping Yersinia pestis isolates from China. PLoS ONE. 3e2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li, Y. L., H. Gao, L. Qin, B. Li, Y. P. Han, Z. B. Guo, Y. J. Song, J. H. Zhai, Z. M. Du, X. Y. Wang, D. S. Zhou, and R. F. Yang. 2008. Identification and characterization of PhoP regulon members in Yersinia pestis biovar Microtus. BMC Genomics 9143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lorange, E. A., B. L. Race, F. Sebbane, and B. J. Hinnebusch. 2005. Poor vector competence of fleas and the evolution of hypervirulence in Yersinia pestis. J. Infect. Dis. 1911907-1912. [DOI] [PubMed] [Google Scholar]
- 39.Lukaszewski, R. A., D. J. Kenny, R. Taylor, D. G. Rees, M. G. Hartley, and P. C. Oyston. 2005. Pathogenesis of Yersinia pestis infection in BALB/c mice: effects on host macrophages and neutrophils. Infect. Immun. 737142-7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marketon, M. M., R. W. DePaolo, K. L. DeBord, B. Jabri, and O. Schneewind. 2005. Plague bacteria target immune cells during infection. Science 3091739-1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Montminy, S. W., N. Khan, S. McGrath, M. J. Walkowicz, F. Sharp, J. E. Conlon, K. Fukase, S. Kusumoto, C. Sweet, K. Miyake, S. Akira, R. J. Cotter, J. D. Goguen, and E. Lien. 2006. Virulence factors of Yersinia pestis are overcome by a strong lipopolysaccharide response. Nat. Immunol. 71066-1073. [DOI] [PubMed] [Google Scholar]
- 42.Ochman, H., J. G. Lawrence, and E. A. Groisman. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405299-304. [DOI] [PubMed] [Google Scholar]
- 43.Ochman, H., and N. A. Moran. 2001. Genes lost and genes found: evolution of bacterial pathogenesis and symbiosis. Science 2921096-1099. [DOI] [PubMed] [Google Scholar]
- 44.Oyston, P. C., N. Dorrell, K. Williams, S. R. Li, M. Green, R. W. Titball, and B. W. Wren. 2000. The response regulator PhoP is important for survival under conditions of macrophage-induced stress and virulence in Yersinia pestis. Infect. Immun. 683419-3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parkhill, J., B. W. Wren, N. R. Thomson, R. W. Titball, M. T. Holden, M. B. Prentice, M. Sebaihia, K. D. James, C. Churcher, K. L. Mungall, S. Baker, D. Basham, S. D. Bentley, K. Brooks, A. M. Cerdeno-Tarraga, T. Chillingworth, A. Cronin, R. M. Davies, P. Davis, G. Dougan, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Leather, S. Moule, P. C. Oyston, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Genome sequence of Yersinia pestis, the causative agent of plague. Nature 413523-527. [DOI] [PubMed] [Google Scholar]
- 46.Patel, C. N., B. W. Wortham, J. L. Lines, J. D. Fetherston, R. D. Perry, and M. A. Oliveira. 2006. Polyamines are essential for the formation of plague biofilm. J. Bacteriol. 1882355-2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Perry, R. D., and J. D. Fetherston. 1997. Yersinia pestis—etiologic agent of plague. Clin. Microbiol. Rev. 1035-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Perry, R. D., I. Mier, Jr., and J. D. Fetherston. 2007. Roles of the Yfe and Feo transporters of Yersinia pestis in iron uptake and intracellular growth. Biometals 20699-703. [DOI] [PubMed] [Google Scholar]
- 49.Petersen, S., and G. M. Young. 2002. Essential role for cyclic AMP and its receptor protein in Yersinia enterocolitica virulence. Infect. Immun. 703665-3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Porat, R., W. R. McCabe, and R. R. Brubaker. 1995. Lipopolysaccharide associated resistance to killing of yersiniae by complement. J. Endotoxin Res. 291-97. [Google Scholar]
- 51.Pouillot, F., C. Fayolle, and E. Carniel. 2008. Characterization of chromosomal regions conserved in Yersinia pseudotuberculosis and lost by Yersinia pestis. Infect. Immun. 764592-4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Prior, J. L., J. Parkhill, P. G. Hitchen, K. L. Mungall, K. Stevens, H. R. Morris, A. J. Reason, P. C. Oyston, A. Dell, B. W. Wren, and R. W. Titball. 2001. The failure of different strains of Yersinia pestis to produce lipopolysaccharide O-antigen under different growth conditions is due to mutations in the O-antigen gene cluster. FEMS Microbiol. Lett. 197229-233. [DOI] [PubMed] [Google Scholar]
- 53.Pujol, C., and J. B. Bliska. 2003. The ability to replicate in macrophages is conserved between Yersinia pestis and Yersinia pseudotuberculosis. Infect. Immun. 715892-5899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pujol, C., J. P. Grabenstein, R. D. Perry, and J. B. Bliska. 2005. Replication of Yersinia pestis in interferon gamma-activated macrophages requires ripA, a gene encoded in the pigmentation locus. Proc. Natl. Acad. Sci. USA 10212909-12914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Raetz, C. R., and C. Whitfield. 2002. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71635-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ramamurthi, K. S., and O. Schneewind. 2002. Type iii protein secretion in yersinia species. Annu. Rev. Cell Dev. Biol. 18107-133. [DOI] [PubMed] [Google Scholar]
- 57.Rebeil, R., R. K. Ernst, B. B. Gowen, S. I. Miller, and B. J. Hinnebusch. 2004. Variation in lipid A structure in the pathogenic yersiniae. Mol. Microbiol. 521363-1373. [DOI] [PubMed] [Google Scholar]
- 58.Robinson, V. L., P. C. Oyston, and R. W. Titball. 2005. A dam mutant of Yersinia pestis is attenuated and induces protection against plague. FEMS Microbiol. Lett. 252251-256. [DOI] [PubMed] [Google Scholar]
- 59.Schubert, S., A. Rakin, and J. Heesemann. 2004. The Yersinia high-pathogenicity island (HPI): evolutionary and functional aspects. Int. J. Med. Microbiol. 29483-94. [DOI] [PubMed] [Google Scholar]
- 60.Sebbane, F., D. Gardner, D. Long, B. B. Gowen, and B. J. Hinnebusch. 2005. Kinetics of disease progression and host response in a rat model of bubonic plague. Am. J. Pathol. 1661427-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sebbane, F., C. O. Jarrett, D. Gardner, D. Long, and B. J. Hinnebusch. 2006. Role of the Yersinia pestis plasminogen activator in the incidence of distinct septicemic and bubonic forms of flea-borne plague. Proc. Natl. Acad. Sci. USA 1035526-5530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sha, J., S. L. Agar, W. B. Baze, J. P. Olano, A. A. Fadl, T. E. Erova, S. Wang, S. M. Foltz, G. Suarez, V. L. Motin, S. Chauhan, G. R. Klimpel, J. W. Peterson, and A. K. Chopra. 2008. Braun lipoprotein (Lpp) contributes to virulence of yersiniae: potential role of Lpp in inducing bubonic and pneumonic plague. Infect. Immun. 761390-1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Simonet, M., B. Riot, N. Fortineau, and P. Berche. 1996. Invasin production by Yersinia pestis is abolished by insertion of an IS200-like element within the inv gene. Infect. Immun. 64375-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Skurnik, M., and J. A. Bengoechea. 2003. The biosynthesis and biological role of lipopolysaccharide O-antigens of pathogenic Yersiniae. Carbohydr. Res. 3382521-2529. [DOI] [PubMed] [Google Scholar]
- 65.Skurnik, M., A. Peippo, and E. Ervela. 2000. Characterization of the O-antigen gene clusters of Yersinia pseudotuberculosis and the cryptic O-antigen gene cluster of Yersinia pestis shows that the plague bacillus is most closely related to and has evolved from Y. pseudotuberculosis serotype O:1b. Mol. Microbiol. 37316-330. [DOI] [PubMed] [Google Scholar]
- 66.Sokurenko, E. V., R. Gomulkiewicz, and D. E. Dykhuizen. 2006. Source-sink dynamics of virulence evolution. Nat. Rev. Microbiol. 4548-555. [DOI] [PubMed] [Google Scholar]
- 67.Song, Y., Z. Tong, J. Wang, L. Wang, Z. Guo, Y. Han, J. Zhang, D. Pei, D. Zhou, H. Qin, X. Pang, J. Zhai, M. Li, B. Cui, Z. Qi, L. Jin, R. Dai, F. Chen, S. Li, C. Ye, Z. Du, W. Lin, J. Yu, H. Yang, P. Huang, and R. Yang. 2004. Complete genome sequence of Yersinia pestis strain 91001, an isolate avirulent to humans. DNA Res. 11179-197. [DOI] [PubMed] [Google Scholar]
- 68.Stenseth, N. C., B. B. Atshabar, M. Begon, S. R. Belmain, E. Bertherat, E. Carniel, K. L. Gage, H. Leirs, and L. Rahalison. 2008. Plague: past, present, and future. PLoS Med. 5(1)e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sun, Y. C., B. J. Hinnebusch, and C. Darby. 2008. Experimental evidence for negative selection in the evolution of a Yersinia pestis pseudogene. Proc. Natl. Acad. Sci. USA 1058097-8101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Suntsov, V. V., and N. I. Suntsova. 2008. Macro- and microevolution as related to the problem of origin and global expansion of the plague pathogen Yersinia pestis. Izv. Akad. Nauk Ser. Biol. July-August:389-395. (In Russian.) [PubMed]
- 71.Tan, L., and C. Darby. 2006. Yersinia pestis YrbH is a multifunctional protein required for both 3-deoxy-D-manno-oct-2-ulosonic acid biosynthesis and biofilm formation. Mol. Microbiol. 61861-870. [DOI] [PubMed] [Google Scholar]
- 72.Tong, Z., D. Zhou, Y. Song, L. Zhang, D. Pei, Y. Han, X. Pang, M. Li, B. Cui, J. Wang, Z. Guo, Z. Qi, L. Jin, J. Zhai, Z. Du, X. Wang, J. Yu, P. Huang, H. Yang, and R. Yang. 2005. Pseudogene accumulation might promote the adaptive microevolution of Yersinia pestis. J. Med. Microbiol. 54259-268. [DOI] [PubMed] [Google Scholar]
- 73.Tran, H. J., A. K. Heroven, L. Winkler, T. Spreter, B. Beatrix, and P. Dersch. 2005. Analysis of RovA, a transcriptional regulator of Yersinia pseudotuberculosis virulence that acts through antirepression and direct transcriptional activation. J. Biol. Chem. 28042423-42432. [DOI] [PubMed] [Google Scholar]
- 74.Twigg, G. 2003. The Black Death and DNA. Lancet Infect. Dis. 311. [DOI] [PubMed] [Google Scholar]
- 75.Viola, R. E., L. Yerman, J. M. Fowler, C. G. Arvidson, and R. R. Brubaker. 2008. A missense mutation causes aspartase deficiency in Yersinia pestis. Microbiology 1541271-1280. [DOI] [PubMed] [Google Scholar]
- 76.Wang, X., Y. Han, Y. Li, Z. Guo, Y. Song, Y. Tan, Z. Du, A. Rakin, D. Zhou, and R. Yang. 2007. Yersinia genome diversity disclosed by Yersinia pestis genome-wide DNA microarray. Can. J. Microbiol. 531211-1221. [DOI] [PubMed] [Google Scholar]
- 77.Wattiau, P., and G. R. Cornelis. 1994. Identification of DNA sequences recognized by VirF, the transcriptional activator of the Yersinia yop regulon. J. Bacteriol. 1763878-3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Williams, K., P. C. Oyston, N. Dorrell, S. Li, R. W. Titball, and B. W. Wren. 2000. Investigation into the role of the serine protease HtrA in Yersinia pestis pathogenesis. FEMS Microbiol. Lett. 186281-286. [DOI] [PubMed] [Google Scholar]
- 79.Zhan, L., Y. Han, L. Yang, J. Geng, Y. Li, H. Gao, Z. Guo, W. Fan, G. Li, L. Zhang, C. Qin, D. Zhou, and R. Yang. 2008. The cyclic AMP receptor protein, CRP, is required for both virulence and expression of the minimal CRP regulon in Yersinia pestis biovar microtus. Infect. Immun. 765028-5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhou, D., Y. Han, Y. Song, P. Huang, and R. Yang. 2004. Comparative and evolutionary genomics of Yersinia pestis. Microbes Infect. 61226-1234. [DOI] [PubMed] [Google Scholar]
- 81.Zhou, D., Y. Han, Y. Song, Z. Tong, J. Wang, Z. Guo, D. Pei, X. Pang, J. Zhai, M. Li, B. Cui, Z. Qi, L. Jin, R. Dai, Z. Du, J. Bao, X. Zhang, J. Yu, P. Huang, and R. Yang. 2004. DNA microarray analysis of genome dynamics in Yersinia pestis: insights into bacterial genome microevolution and niche adaptation. J. Bacteriol. 1865138-5146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhou, D., Y. Han, and R. Yang. 2006. Molecular and physiological insights into plague transmission, virulence and etiology. Microbes Infect. 8273-284. [DOI] [PubMed] [Google Scholar]
- 83.Zhou, D., Z. Tong, Y. Song, Y. Han, D. Pei, X. Pang, J. Zhai, M. Li, B. Cui, Z. Qi, L. Jin, R. Dai, Z. Du, J. Wang, Z. Guo, J. Wang, P. Huang, and R. Yang. 2004. Genetics of metabolic variations between Yersinia pestis biovars and the proposal of a new biovar, microtus. J. Bacteriol. 1865147-5152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhou, D., and R. Yang. 2006. Global analysis of gene transcription regulation in prokaryotes. Cell. Mol. Life Sci. 632260-2290. [DOI] [PMC free article] [PubMed] [Google Scholar]