Abstract
Orientia tsutsugamushi is the causative agent of scrub typhus. One of the protein antigens of this species, the conserved 47-kDa protein (HtrA), has been shown to induce an antibody response in patients and can provide protective immunity against live challenge by Orientia in mice. Pepscan experiments identified many peptide epitope clusters in different parts of this protein. The majority of the most reactive epitopes are located at the C terminus of the protein (from amino acid 333 to amino acid 430). Protein sequence analysis revealed that the 47-kDa protein contains a trypsin domain and has sequence homology to human serine protease HtrA1 (hHtrA1). As the 47-kDa protein is a potential vaccine candidate and its ability to induce autoimmunity is a concern, the reactivity of scrub typhus patient sera with purified recombinant 47-kDa and hHtrA1 proteins was tested. A significant percentage (>20%) of scrub typhus patient sera reacted strongly with recombinant hHTRA1 and two of the antigenic polypeptide epitopes in hHtrA1. These findings suggest that the safety of the full-length 47-kDa antigen as a vaccine candidate is a significant issue due to its cross-reactivity with a human protein, which may also contribute to autoimmune responses or enhanced pathology in some scrub typhus patients.
Scrub typhus is caused by infection with the obligate intracellular, gram-negative bacterium Orientia tsutsugamushi. It accounts for up to 23% of febrile episodes in areas of the Asia-Pacific region where scrub typhus is endemic, and it can cause up to 33% mortality in untreated patients (4, 5). The symptoms are very broad and may include high fever, pneumonitis, meningitis, rash, and headache. Differentiating scrub typhus from other acute febrile illnesses, such as leptospirosis, murine typhus, and malaria, can be difficult because of similarities in their clinical presentation.
Many microbial pathogens have antigenic epitopes which are shared with host cell components. The antibody responses to such epitopes may cause transient or long-term damage if the antibodies react with host components (17). Many examples have been reported, particularly cases involving intracellular pathogens. Cytoplasmic, nuclear, and platelet autoantibodies are found in the sera of human granulocytic ehrlichiosis patients (25). Induction of antiphospholipid antibodies occurs during spotted fever (19) and Q fever (1, 18) illness. Serine protease 3 (c-ANCA) has been identified as an autoantigen in human neutrophils (6). A mouse monoclonal antibody (MAb) raised against O. tsutsugamushi reacted with a cytokeratin protein, suggesting that an epitope similar to that recognized by this MAb could interact with human antibodies (14).
In general, naturally occurring autoantibodies have been considered to be of little importance in pathogenesis. They occur at low levels in normal individuals (9). Molecular mimicry has been proposed as a pathogenic mechanism for autoimmune disease. This hypothesis is based on experimental evidence of an association of infectious agents with autoimmune disease and observed cross-reactivity of immune reagents with host antigens and microbial determinants (17). Molecular mimicry has been defined as similar structures shared by these two elements. Either linear domains or their conformational structures may be shared. Such epitopes shared by microbial pathogens and host cells may activate autoreactive T cells and B cells during infection (26). The pathogenic antigens which bear epitopes that are identical or very similar to host components can break self-tolerance, resulting in the appearance of autoantibodies. The contribution of these autoantibodies to rickettsial pathogenesis remains largely unknown. In dogs experimentally infected with Ehrlichia canis, antiplatelet antibodies were detected and appeared to contribute to the immune destruction of platelets (22, 13, 23). In patients infected with Rickettsia conorii, the antiphospholipid antibodies may cause membrane damage in endothelial cells (19). Recently, autoantibodies were identified in nonautoimmune individuals during acute bacterial, viral, parasitic, and rickettsial infections (2). The mechanism by which O. tsutsugamushi infection induces the synthesis of self-reactive antibody may be molecular mimicry (14).
In this study, sequence analysis revealed that the 47-kDa protein of O. tsutsugamushi contains a trypsin domain and has significant sequence homology to the human HtrA1 (hHtrA1) protein (Fig. 1). We demonstrated that scrub typhus patients frequently have antibodies that recognize not only the 47-kDa protein (12) but also hHtrA1.
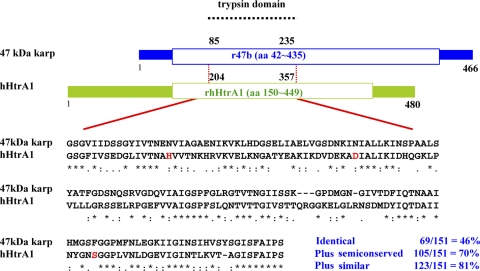
FIG. 1.
The 47-kDa antigen of O. tsutsugamushi has significant sequence homology to hHtrA1 in a 151-amino-acid region. Red type indicates the three amino acids (histidine, aspartic acid, and serine) located in the catalytic site of human HtrA1; asterisks indicate identical amino acids; colons indicate semiconserved amino acids; and periods indicate similar amino acids.
MATERIALS AND METHODS
Bacterial strains and vectors.
Escherichia coli Top10 (Invitrogen, California) cells were used for cloning. The cloned genes were inserted into pET24a and pET28a (Novagen, California) for expression of the 47-kDa protein and hHtrA1, respectively. E. coli BL21(DE3) (Invitrogen, California) was the expression host which expressed proteins under control of the phage T7 lac promoter (21).
Cloning of the gene coding for the 47-kDa protein into the expression vector pET24a.
A pair of primers (47bfB [5′-GGTGGATCCGTAAATAGTTTATCCGATATAGTTG-3′] and 47br [5′-GGTGGTCTCGAGTCTTAGTGTTTTAACAGAAATATCTT-3′]) was designed based on the nucleotide sequence encoding the 47-kDa protein from strain Karp (20). The coding sequence for amino acids 42 to 435 of the 47-kDa protein (r47b) was amplified by PCR using genomic DNA isolated from O. tsutsugamushi strain Karp as the template. The PCR product was digested with BamHI and XhoI and ligated into the expression vector pET24a digested with the same enzymes. Top10 competent cells were transformed with the ligation mixture, and colonies were screened for inserts that had the correct size. The final sequences were confirmed by DNA sequencing of the resulting plasmid, which contained a sequence encoding a His tag at the C terminus of r47b.
Cloning of the gene coding for recombinant hHtrA1 (rhHtrA1).
The codon-optimized gene fragment coding for amino acids 150 to 449 of hHtrA1 was cloned between NdeI and XhoI sites of the pET28a expression vector by BioClone Inc. (California). The encoded protein contained a His tag at the N terminus of the expressed hHtrA1.
Expression and purification of the recombinant 47-kDa and hHtrA1 protein fragments.
E. coli BL21(DE3) cells were transformed with plasmids carrying the correct inserts. The recombinant E. coli colonies with high levels of expression of the r47b or rhHtrA1 protein were grown overnight in Overnight Express medium TB (EMD Biosciences, California) at 37°C with shaking at 200 rpm. Cell pellets from 500-ml cultures were resuspended in 20 ml of buffer A (20 mM Tris-HCl [pH 8.0], 1 mM EDTA, 1 mM dithiothreitol) containing 2% deoxycholic acid. Cells were ruptured by sonication (model VirSonic 475 ultrasonic liquid processor; Virtis Company, New York) five times at setting 3 for 10 s each time, with cooling on ice for 1 min between sonication treatments. Cell extract was centrifuged at 10,000 × g for 30 min in a Thermo centrifuge (model IEC MultiRF). The pellets were resuspended in 2 M urea in buffer A by vortexing, incubated on a shaker at room temperature for an additional 10 min, and centrifuged for 30 min at 10,000 × g. The extraction process was then repeated with 4 M urea and 6 M urea in buffer A. The final pellets were dissolved in 8 M urea in buffer A and loaded onto to a high-pressure liquid chromatography ion-exchange column (DEAE-5PW; TOSOH Bioscience, Pennsylvania) for fractionation. Proteins were eluted over 30 min with a linear gradient of 0.0 to 0.1 M NaCl using buffer B (6 M urea in buffer A) and buffer C (6 M urea and 0.5 M NaCl in buffer A). Fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The purest fractions were pooled and dialyzed against buffer A with 6 M urea and reapplied to the DEAE column as previously described. Protein fractions from the second DEAE run were pooled and dialyzed against Hisbind buffer (20 mM Tris [pH 8.0], 0.5 M NaCl, 10 mM imidazole) containing 8 M urea at room temperature for 2 h and then applied to a 1-ml nickel-nitrilotriacetic acid column equilibrated with Hisbind buffer containing 8 M urea. The recombinant proteins were eluted from the column using a 25 to 400 mM imidazole stepwise gradient in Hisbind buffer containing 8 M urea.
Refolding of the r47b and rhHtrA1 proteins.
Refolding of the r47b and rhHtrA1 proteins in 8 M urea in Hisbind buffer was achieved by sequential dialysis with decreasing amounts of urea in buffer A containing 0.15 M NaCl and finally with buffer A containing 0.15 M NaCl without urea. Before dialysis the protein concentration of the eluates from the nickel column was adjusted to less than 0.5 mg per ml, and then the eluates were dialyzed with 6 M urea in buffer A containing 0.15 M NaCl for 60 min at 4°C. The same procedure was repeated with 4 M, 2 M, and 1 M urea or no urea. Finally, the sample was dialyzed against buffer A containing 0.15 M NaCl.
Synthesis of overlapping peptides for pepscan.
Synthesis of overlapping decapeptides was carried out as described previously (7). Decamers were synthesized on derivatized polyethylene pins arranged in a 96-well microtiter plate format (10, 11). Each sequential peptide overlapped with six amino acids of the preceding peptide. The number used to designate each decamer is the sequence position of the first amino acid of the peptide. A total of 115 peptides were synthesized in order to span the whole open reading frame of the Karp 47-kDa protein. These peptides were tested for the presence of epitopes recognized by human antibodies using a modified enzyme-linked immunosorbent assay (ELISA) (see below).
Synthesis of cross-reactive peptides with a sulfhydryl group at the N terminus.
Two peptides, P216-236 (C-VTNAHVVTNKHRVKVELKNGA) and P285-308 (C-PFSLQNTVTTGIVSTTQRGGKELG), of hHtrA1 corresponding to homologous sequences of the strain Karp 47-kDa protein were synthesized. These peptides were synthesized with an Applied Biosystems 433A peptide synthesizer using Fmoc chemistry. The carboxyl-terminal amino acid was attached to a hydroxymethylphenoxymethyl polystyrene resin. When synthesis was complete, each peptide was cleaved from the resin using gentle agitation in 90% trifluoroacetic acid, 5% thioanisole, 3% ethanedithiol, 2% anisole for 2.5 h. After precipitation and washing, the peptide was dissolved in 40% acetonitrile in water, and the pH was adjusted to between 7 and 8. The peptide solution was frozen and subsequently lyophilized.
ELISA with recombinant proteins.
Microtiter plates (with 96 wells) were coated for 40 h at 4°C with antigens diluted in phosphate-buffered saline (PBS) and subsequently blocked with 10% skim milk in PBS for 1 h. Patient sera diluted 1:50 (for rhHtrA1) or 1:100 (for r47b) in PBS with 5% skim milk were then added to the plate, incubated for 1 h at room temperature, and washed three times with 0.1% Tween 20 in PBS. Peroxidase-conjugated rabbit anti-human immunoglobulin G (IgG) (Santa Cruz Biotechnology, California) that was diluted 500-fold (for rhHtrA1) or 1,000-fold (r47b) was added. After 1 h of incubation at room temperature, the plates were washed as described above before addition of the 2,2′-azinobis(3-ethylbenzthiazoline-6-sulfonate) (ABTS) substrate (Kirkegaard & Perry). Optical densities at 405 nm were measured after 1 h of incubation at room temperature with a Molecular Devices plate reader.
ELISA with synthetic peptides.
A modified ELISA was performed as described previously (7). Peptides attached to polyethylene pins were precoated with 0.5% casein and 0.5% bovine serum albumin in 0.1 M PBS (pH 7.2) for 1 h to prevent nonspecific binding of antibody. The peptides attached to the pins were incubated with 100 μl of an antibody solution (1:100 dilution) overnight at 4°C. The next day the pins were washed four times for 10 min each in PBS containing 0.05% Triton X-100. The bound antibodies were detected with alkaline phosphate-conjugated goat anti-human IgG (Bio-Rad, California). The optical density at 405 nm was measured with a Molecular Devices plate reader. Pins with attached peptides were reused after the bound antibodies were removed by sonication for 1 h at 55°C in 0.1 M sodium phosphate (pH 7.2) containing 1% sodium dodecyl sulfate and 0.1% β-mercaptoethanol. Sonication was followed by two 2-min washes in distilled water heated to 60°C and two 2-min washes in gently boiling methanol. The pin blocks were then air dried for 1 h before use.
ELISA with sulfhydryl-containing cross-reactive peptides.
The synthesized peptides were dissolved in water, the pH was adjusted to pH 8.0 with sodium hydroxide, and the preparations were diluted with PBS to obtain a final concentration of 3 μg/ml. One hundred microliters of a diluted peptide solution was added to each well of a Reacti-Bind maleimide-activated plate (Pierce, Illinois) and incubated at 4°C for 40 h. After the peptides were immobilized on the plate, the procedures described above for ELISA with recombinant proteins were followed.
Prediction servers.
Peptide antigenicity was predicted using the BcePred server. This program combines seven epitope characteristics (hydrophilicity, turns, surface probability, flexibility, polarity, accessibility, and antigenicity) of a protein antigen for prediction of possible epitopes. The tertiary structure was predicted using Cn3D from the NCBI server.
Human sera.
Patient sera from the Pescadore Islands of Taiwan were obtained from a Chinese military garrison stationed there during 1976 and 1977 and were stored at −80°C (3). The clinical diagnosis of scrub typhus was confirmed by demonstration of rising anti-Orientia indirect fluorescent antibody assay titers and by isolation of the agent in many cases. Sera from patients without a history of scrub typhus that were diagnosed with the following diseases were included in the negative control panel (24) (the numbers of sera are indicated in parentheses): bartonellosis (5), cholera (1), leptospirosis (2), malaria (2), rheumatoid arthritis (2), tularemia (6), typhoid (6), antinuclear antibody (21), and other diseases with unknown etiology from the Pescadore Islands (21).
RESULTS
Sequence analysis.
The sequence analysis of the 47-kDa protein by domain research revealed that it contains a trypsin domain similar to that of other proteases. Sequence alignment of the trypsin domains of the 47-kDa protein and the hHtrA1 protein revealed a very high degree of homology (Fig. 1). There is 46% identity (69 of 151 residues) and >70% conservation (105 of 151 residues) at the amino acid level in this domain. The three amino acids (histidine, aspartic acid, and serine) located in the catalytic site of human HtrA1 are indicated in Fig. 1. Although the 47-kDa protein does not have these three conserved amino acids in the corresponding places, N at two corresponding positions is semiconserved. Therefore, the overall folded structure of the 47-kDa protein may be very similar to that of hHtrA1 in this trypsin domain.
Comparison of r47b and rhHtrA1 antigenicities by ELISA.
The refolded purified recombinant 47b and hHtrA1 proteins were used as antigens to detect the presence of specific IgG antibodies in an ELISA. A panel of 480 sera obtained from serial bleeding of 126 individuals with clinically diagnosed scrub typhus and a panel of 82 individual negative control sera (including sera from 66 non-scrub typhus patients) were used. Of the 480 patient sera, 297 sera from 98 patients (77.8%) exhibited interactions with the recombinant 47-kDa protein antigen (Table 1).
TABLE 1.
r47b and rhHtrA1 IgG ELISA reactivities of human sera
| Group | No. of individuals (no. of sera) | No. (%) positive witha:
|
|
|---|---|---|---|
| r47b | rhHtrA1 | ||
| Scrub typhus | 126 (480) | 98 (77.8) | 26 (20.6)b |
| Endemic control | 16 (16) | 0 (0) | 0 (0) |
| Non-scrub typhus | 66 (66) | 1 (1.5) | 2 (3.0) |
The same patient may have had multiple bleedings. Individuals for which at least one sample was positive were considered positive.
The experiment was performed three times. The percentage is the lowest percentage of individuals positive for rhHtrA1; the values for the other two times are 24% and 30% positive individuals.
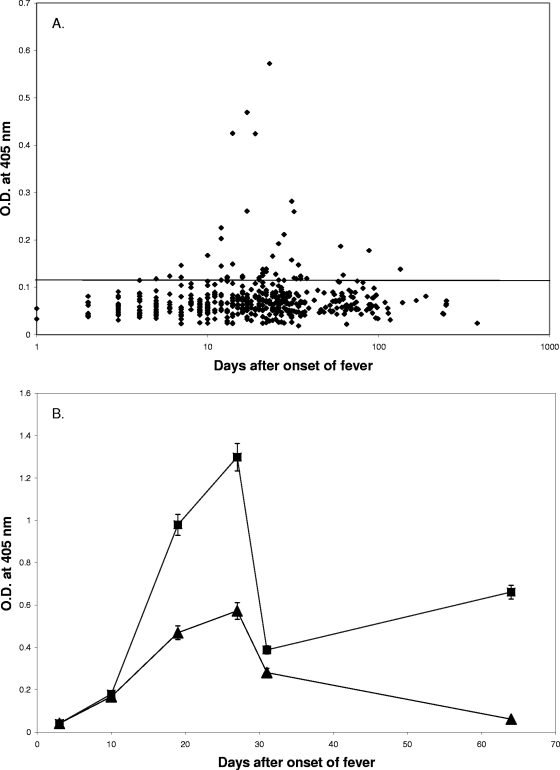
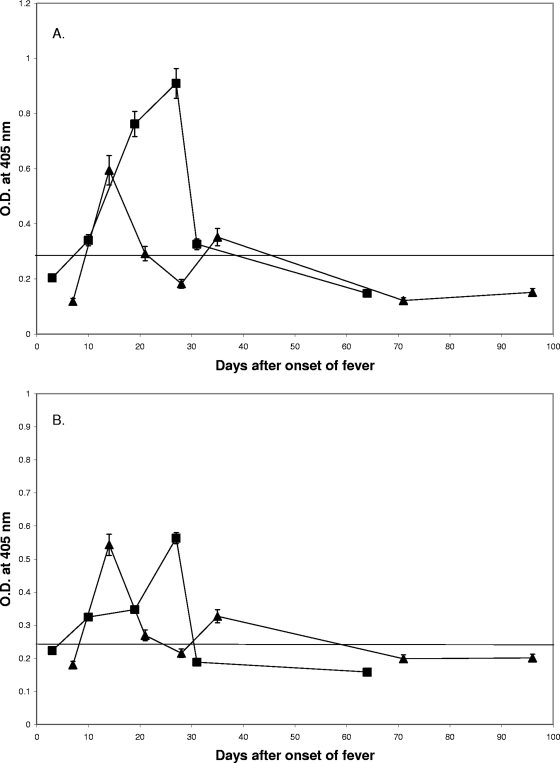
The ELISA results for the 480 scrub typhus patient sera against rhHtrA1 are shown in Fig. 2A. Forty-one sera from 26 patients had levels of IgG antibody against the rhHtrA1 protein greater than the background level. About 21% (26 of 126) of scrub typhus patients produced antibodies against the rhHtrA1 protein, while only 3% (2 of 66) of the non-scrub typhus patients (Table 1) had detectable levels of antibodies in the same ELISA. The difference between the percentage of antibody-positive scrub typhus patients and the percentage of antibody-positive non-scrub typhus patients is statistically significant (P < 0.001). This confirms that infection with O. tsutsugamushi may induce antibodies which cross-react with the hHtrA1 protein. The majority of the sera (92.7% [38 of 41 sera]) which were positive with rhHtrA1 were also positive with the r47b protein. The cross-reactivity with rhHtrA1, like the reactivity with the r47b protein, was transient, but it can last for several months, as some of our serial bleeding data indicated (Fig. 2A and 2B). A typical example is shown in Fig. 2B, as both of the reactivities peaked between day 20 and day 30 after the onset of illness. The antibody level began to rise after day 10, was significantly greater than the background level around week 3, was maximal at week 4, and decreased back to the background level for rhHtrA1.
FIG. 2.
ELISA results for 480 scrub typhus patient sera with rhHtrA1. The cutoff values are 0.117 for rhHtrA1 and 0.158 for r47b (means for 82 negative controls plus 2 standard deviations). (A) Relationship of reactivity (as reflected by the optical density determined by ELISA) to the number of days after the onset of fever. The x axis indicates the number of days after the onset of illness, and the y axis indicates the optical density at 405 nm. (B) Time course of reactivity of patient MAK116 sera collected at days 3, 10, 19, 27, 31, and 64 with the recombinant 47-kDa (▪) and rhHtrA1 (▴) proteins. The symbols indicate the averages of two experiments, and the error bars represent ranges. O.D., optical density.
Identification of antigenic epitopes with pepscan.
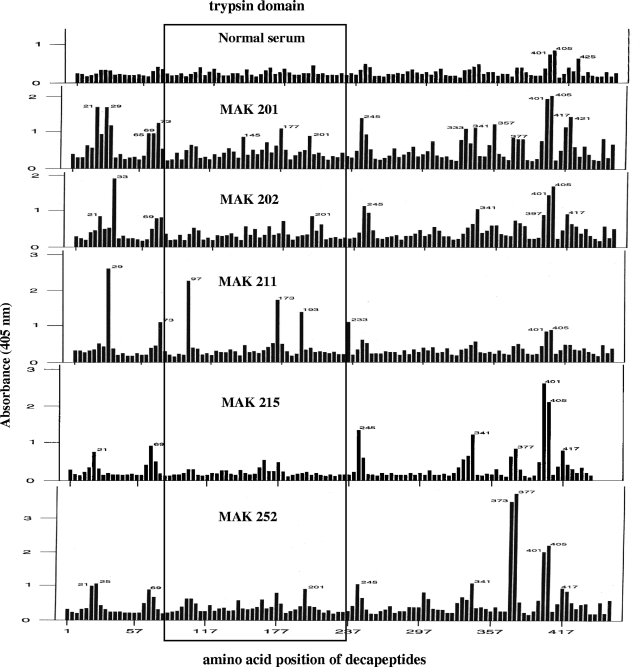
Pepscan with overlapping decapeptides of the 47-kDa protein was tested with five patient sera (selected based on the quantity available) to identify antigenic epitopes (Fig. 3). Many reactive decapeptides are scattered throughout the molecule; however, there are clusters of such decapeptides at the N and C termini of the protein, and some are also located in the trypsin domain which shared sequence homology with hHtrA1 (Fig. 3). Most of the patient sera tested reacted with the clusters in the C-terminal portion (specifically decapeptides 341, 377, 401, 405, and/or 417), and fewer patient sera reacted with the clusters in the N-terminal portion (such as decapeptides 21, 29, and/or 69). A few significant epitope clustering regions (around decapeptides 97 to 101, 145, 165, and/or 173 to 177) were present in the middle portion where there is sequence homology (Fig. 3 and Table 2). Of the 10 decapeptides shown in the Table 2, 1 was recognized by all five patient sera, 6 were recognized by four patient sera, 2 were recognized by three patient sera, and 1 was recognized by only two patient sera. These data demonstrated that there are variations in antibody responses to epitopes among scrub typhus patients.
FIG. 3.
ELISA profiles for five patient sera and one normal serum with overlapping synthetic decapeptides encompassing the 47-kDa antigen of O. tsutsugamushi Karp. The five MAK numbers indicate five different patients. The amino acid position for each decapeptide is the first amino acid. Each successive decapeptide begins four residues after the preceding peptide.
TABLE 2.
Data for 47-kDa antigen epitopes in the trypsin domain identified by five patient sera
| Protein | Position | Sequence homolog | ELISA optical density for:
|
Predicted epitope scorea | |||||
|---|---|---|---|---|---|---|---|---|---|
| Normal serum | Patient 201 | Patient 202 | Patient 211 | Patient 215 | Patient 252 | ||||
| 47 kDa | 97 | VTNENVIAGA | 0.25 | 0.51 | 0.35 | 2.42 | 0.21 | 0.38 | 5.417 |
| hHtrA1 | 216 | VTNAHVVTNK | |||||||
| 47 kDa | 101 | NVIAGAENIK | 0.27 | 0.68 | 0.54 | 0.42 | 0.32 | 0.65 | 4.660 |
| hHtrA1 | 220 | HVVTNKHRVK | |||||||
| 47 kDa | 145 | YATFGDSNQS | 0.30 | 0.84 | 0.64 | 0.47 | 0.29 | 0.63 | 11.391 |
| hHtrA1 | 264 | VLLLGRSSEL | |||||||
| 47 kDa | 153 | QSRVGDQVIA | 0.21 | 0.52 | 0.51 | 0.31 | 0.18 | 0.24 | 7.607 |
| hHtrA1 | 272 | ELRPGEFVVA | |||||||
| 47 kDa | 165 | SPFGLRGTVT | 0.31 | 0.74 | 0.66 | 0.30 | 0.62 | 0.66 | 6.067 |
| hHtrA1 | 284 | SPFSLQNTVT | |||||||
| 47 kDa | 173 | VTNGIISSKG | 0.27 | 0.67 | 0.38 | 1.68 | 0.22 | 0.38 | 8.910 |
| hHtrA1 | 292 | VTTGIVSTTQ | |||||||
| 47 kDa | 177 | IISSKGPDMG | 0.30 | 1.11 | 0.74 | 0.51 | 0.49 | 0.76 | 9.916 |
| hHtrA1 | 296 | IVSTTQRGGK | |||||||
| 47 kDa | 193 | FIQTNAAIHM | 0.24 | 0.47 | 0.35 | 1.41 | 0.19 | 0.22 | 3.358 |
| hHtrA1 | 312 | YIQTDAIINY | |||||||
| 47 kDa | 201 | HMGSFGGPMF | 0.33 | 0.86 | 0.84 | 0.38 | 0.23 | 0.87 | 3.890 |
| hHtrA1 | 320 | NYGNSGGPLV | |||||||
| 47 kDa | 209 | MFNLEGKIIG | 0.26 | 0.44 | 0.71 | 0.25 | 0.20 | 0.38 | 4.419 |
| hHtrA1 | 328 | LVNLDGEVIG | |||||||
Scores for continuous B-cell epitopes were predicted by using the BcePred server.
Design of cross-reactive epitopes and verification by ELISA.
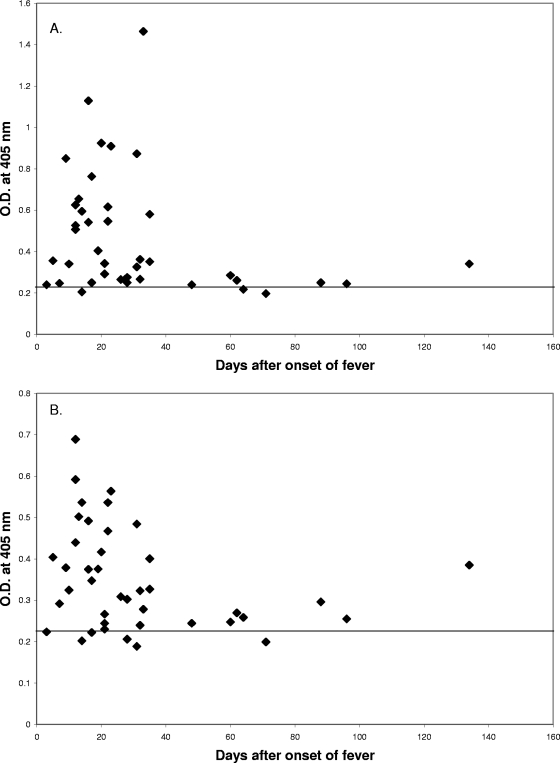
Several potential epitopes in the conserved region (amino acids 85 to 235) in the 47-kDa protein and hHtrA1 were analyzed further for an additional demonstration of cross-reactivity. Table 2 shows (i) the starting position of the peptide, (ii) the amino acid sequence, (iii) the optical density obtained with the modified ELISA, and (iv) the predicted immunogenicity score. Two peptides were selected for synthesis. The selection was based on the following factors: (i) the reactivity of the 47-kDa protein decapeptides with multiple patient sera as determined by pepscan (the decapeptides reacted with at least four of five sera tested), (ii) antigenic prediction (with the BcePred server software as described in Materials and in Methods) based on amino acid sequences of the 47-kDa protein (the higher the value, the higher immunogenicity of the epitope), and (iii) surface location on the 47-kDa antigen based on a tertiary structure prediction showing regions that are located on the surface of the protein. Based on the three factors mentioned above, two longer peptides (P216-236 [VTNAHVVTNKHRVKVELKNGA; 21-mer] and P285-308 [PFSLQNTVTTGIVSTTQRGGKELG; 24-mer]) from hHtrA1 were synthesized to confirm the true reactivities of these epitopes in the trypsin domain. The corresponding decapeptides of the 47-kDa protein were recognized by four of five patient sera tested. These decapeptides were predicted to be surface exposed and had antigenic index values above 4.5 (see Materials and Methods). Of the 41 sera positive with rhHtrA, 38 recognized P216-236, and 36 recognized P285-308. Two sera did not recognize either of these two peptides. Overall, 95% of the sera recognized at least one of these two peptides (Fig. 4). Figure 5A and 5B demonstrate the parallel onset and decline of antibodies that recognized each peptide epitope with sera from two individual patients. This further confirmed the basis of the cross-reactivity of patient sera with the r47b and rhHtrA1 proteins. It also eliminated the possibility that impurities from E. coli that were present in the recombinant protein preparations caused the cross-reactivity that we observed.
FIG. 4.
ELISA results for 41 rhHtrA1-positive patient sera with synthetic peptides. P216-236 (A) and P285-308 (B) were tested with 41 rhHtrA1-positive sera from serial bleeding of 26 scrub typhus patients. The cutoff values for positive results are 0.221 and 0.237 (means of 10 negative controls plus 2 standard deviations) for P216-236 (A) and P285-308 (B), respectively. O.D., optical density.
FIG. 5.
Time course of antibody responses of two patients to hHtrA1 synthetic peptides. P216-236 (A) and P285-308 (B) were tested with sera from serial bleeding of patient MAK116 (▪) collected at days 3, 10, 19, 27, 31, and 64 and patient MAK131 (▴) collected at days 7, 14, 21, 28, 35, 71, and 96. The cutoff values for positive results are 0.221 and 0.237 (means of 10 negative controls plus 2 standard deviations) for P216-236 and P285-308, respectively. The symbols indicate the averages of two experiments, and the error bars represent ranges. O.D., optical density.
DISCUSSION
Sera from scrub typhus patients have been shown to recognize several O. tsutsugamushi antigens, including the major outer membrane 56-kDa protein, which comprises 10 to 15% of the total rickettsial cellular protein content (12, 16), and other minor proteins, such as the 22-kDa, 47-kDa, and 110-kDa proteins. Among these antigens, the 56-kDa, 47-kDa, and 110-kDa antigens have been shown to provide protection against live challenge in a mouse model (W.-M. Ching, C.-C. Chao, T.-C. Chan, J. Jiang, S. Chattopadhay, and A. Richards, unpublished data). Although the antigenically variable 56-kDa outer membrane protein is the major antigen and has been shown to provide protection against homologous challenge, complete cross protection from heterologous challenge with strains having a different 56-kDa protein type has not been obtained. The 47-kDa protein belongs to the HtrA heat shock protein family, which is highly conserved (>97% identity in 25 different strains of O. tsutsugamushi), and provided good homologous protection and cross protection against four heterologous strains in our lethal challenge mouse model (W.-M. Ching et al., unpublished data). It seems to be a good vaccine candidate for broad protection against Orientia infection. Although there are many serine proteases which have the trypsin domain, sequence alignment analysis of these proteins and the 47-kDa protein has not previously revealed a high degree of similarity. We first noticed the high sequence homology between the 47-kDa and hHtrA1 proteins when we constructed an alignment with only the trypsin domain.
The ELISA study with patient sera showed that antibodies interact with both the r47b and rhHtrA1 proteins. Some sera had very strong interactions with rhHtrA1, but the majority of them were very weak, as indicated by the absorbance readings by ELISA. The rise and decline of antibodies against rhHtrA1 was parallel to those of the antibodies against r47b. This work suggested that infection with O. tsutsugamushi induced antibodies which cross-reacted with epitopes on rhHtrA1 in approximately 20% of infected patients. This finding indicates that it is possible that antibodies raised against the 47-kDa protein could also interact with hHtrA1, which raises a major concern regarding the safety of this vaccine candidate.
In our study, 21% (26/126) of scrub typhus patients had antibodies against rhHtrA1. This suggests that the autoantibodies against hHtrA1 may be induced by O. tsutsugamushi infection. Two antigenic epitopes in the trypsin domain of hHtrA1 were identified and interact strongly with scrub typhus patient sera. Patient sera react differently to these two epitopes, similar to what we observed in the pepscan experiment. Unfortunately, we do not have access to the patients' health records and no information about whether the scrub typhus patients with antibodies to rhHtrA1 had more severe illness than the patients without antibodies to rhHtrA1. An immunohistochemistry study showed that hHtrA1 is widely expressed, that there are different levels of expression, and that the level of protein expression is higher in the epithelium of the endometrium and placenta (8, 15). The high degree of cross-reactivity seen in this study suggests that immunization with the full-length 47-kDa protein could potentially produce antibodies that cross-react with host tissues that express hHtrA1. The pepscan analysis with overlapping decapeptides of the 47-kDa protein demonstrated the heterogeneity of the immune responses of scrub typhus patients to the 47-kDa protein, especially in the region homologous to the trypsin domain. Whether deletion of the epitopes which the recombinant 47-kDa antigen shares with hHtrA1 eliminates its vaccine efficacy is unknown, but this might be one way to enhance its safety for human use. Preliminary data for a 12-kDa fragment (amino acids 347 to 466) showed that it provided 20 to 40% protection, which is much less than the protection provided by the full-length candidate, in our mouse model (W.-M. Ching et al., unpublished results). Currently, we are in the process of producing a larger fragment (amino acids 236 to 466, 24 kDa) that can be evaluated for protection in the near future.
Acknowledgments
We thank Michael Flora, Biomedical Instrumentation Center, Uniformed Services University of the Health Sciences, for providing the synthesized cross-reactive peptides. We are also grateful to Barbara Hanson and Paul Graf for their critical reading of the manuscript and helpful comments.
This research was supported by Naval Medical Research Center research work unit 6000.RAD1.J.A0310.
The study protocol was approved by the Naval Medical Research Center Institutional Review Board (case PJT44) in compliance with all applicable federal regulations governing the protection of human subjects.
The views expressed in this paper are those of the authors and do not necessarily reflect the official policy or position of the Department of the Navy, the Department of Defense, the Centers for Disease Control and Prevention, the Department of Health and Human Services, or the U.S. Government. Chien-Chung Chao, Gregory A. Dasch, and Wei-Mei Ching are employees of the U.S. Government.
Editor: A. Camilli
Footnotes
Published ahead of print on 16 March 2009.
REFERENCES
- 1.Aguilar, C., J. L. Ortega, and N. Caro. 2005. Autoimmune type antiphospholipid antibodies in a patient with Q fever. Haematologica 9033-34. [PubMed] [Google Scholar]
- 2.Berlin, T., G. Zandman-Goddard, M. Blank, T. Matthias, S. Pfeiffer, I. Weis, E. Toubi, S. Singh, R. Asherson, A. Fraser, B. Gilburd, T. Sapir, Y. Levy, J. Lukac, B. Rozman, T. Kveder, and Y. Shoenfeld. 2007. Autoantibodies in nonautoimmune individuals during infections. 1108584-593. [DOI] [PubMed] [Google Scholar]
- 3.Bourgeois, A. L., J. G. Olson, R. C. Fang, J. Huang, C. L. Wang, L. Chow, D. Bechthold, D. T. Dennis, J. C. Coolbaugh, and E. Weiss. 1982. Humoral and cellular responses in scrub typhus patients reflecting primary infection and reinfection with Rickettsia tsutsugamushi. Am. J. Trop. Med. Hyg. 31532-540. [DOI] [PubMed] [Google Scholar]
- 4.Brown, G. W., D. M. Robinson, D. L. Huxsoll, T. S. Ng, K. J. Lim, and G. Sannasey. 1976. Scrub typhus: a common cause of illness in indigenous population. Trans. R. Soc. Trop. Med. Hyg. 70444-448. [DOI] [PubMed] [Google Scholar]
- 5.Brown, G. W., J. P. Saunders, S. Singh, D. L. Huxsoll, and A. Shirai. 1978. Single dose deoxycycline therapy for scrub typhus. Trans. R. Soc. Trop. Med. Hyg. 72412-416. [DOI] [PubMed] [Google Scholar]
- 6.Campanelli, D., P. A. Detmers, C. F. Nathan, and J. E. Gabay. 1990. Azurocidin and a homologous serine protease from neutrophils. Differential antimicrobial and proteolytic properties. J. Clin. Investig. 85904-915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ching, W.-M., H. Wang, B. Jan, and G. A. Dasch. 1996. Identification and characterization of epitopes on the 120-kilodalton surface protein antigen of Rickettsia prowazekii with synthetic peptides. Infect. Immun. 641413-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Luca, A., M. De Falco, A. Severino, M. Campioni, D. Santini, F. Baldi, M. G. Paggi, and A. Baldi. 2003. Distribution of the serine protease HtrA1 in normal human tissues. J. Histochem. Cytochem. 511279-1284. [DOI] [PubMed] [Google Scholar]
- 9.Dighiero, G., B. Guilbert, and S. Avrameas. 1982. Naturally occurring antibodies against nine common antigens in human sera. II. High incidence of monoclonal Ig exhibiting antibody activity against actin and tubulin and sharing antibody specificities with natural antibodies. J. Immunol. 1282788-2792. [PubMed] [Google Scholar]
- 10.Geysen, H. M. 1985. Antigen-antibody interactions at the molecular level: adventures in peptide synthesis. Immunol. Today 6364-369. [DOI] [PubMed] [Google Scholar]
- 11.Geysen, H. M., S. J. Rodda, T. J. Mason, G. Tribbick, and P. G. Schoofs. 1987. Strategies for epitope analysis using peptide synthesis. J. Immunol. Methods 102259-274. [DOI] [PubMed] [Google Scholar]
- 12.Hanson, B. 1985. Identification and partial characterization of Rickettsia tsutsugamushi major protein immunogens. Infect. Immun. 50603-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harrus, S., T. Waner, H. Bark, F. Jongejan, and A. W. Cornelissen. 1999. Recent advances in determining the pathogenesis of canine ehrlichiosis. J. Clin. Microbiol. 372745-2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim, M. J., M. K. Kim, and J. S. Kang. 2006. An epitope shared by cellular cytokeratin and Orientia tsutsugamushi. Microb. Pathog. 41125-132. [DOI] [PubMed] [Google Scholar]
- 15.Nie, G., K. Hale, Y. Li, U. Manuelpillai, E. M. Wallace, and L. A. Salamonsen. 2006. Distict expression and localization of serine protease HtrA1 in human endometrium and first-trimester placenta. Dev. Dyn. 2353448-3455. [DOI] [PubMed] [Google Scholar]
- 16.Ohashi, N., A. Tamura, H. Sakurai, and T. Suto. 1988. Immunoblotting analysis of anti-rickettsial antibodies produced in patients of tsutsugamushi disease. Microbiol. Immunol. 321085-1092. [DOI] [PubMed] [Google Scholar]
- 17.Oldstone, M. B. 1998. Molecular mimicry and immune-mediated diseases. FASEB J. 121255-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ordi-Ros, J., A. Selva-O'Callaghan, F. Monegal-Ferran, Y. Monasterio-Aspiri, C. Juste-Sanchez, and M. Vilardell-Tarres. 1994. Prevalence, significance, and specificity of antibodies to phospholipids in Q fever. Clin. Infect. Dis. 18213-218. [DOI] [PubMed] [Google Scholar]
- 19.Sotto, A., M. Berard, D. Bessis, M. Porneuf, J. Jourdan, and M. C. Boffa. 1995. Antiphospholipid antibody production during Mediterranean spotted fever. Autoimmunity 21123-126. [DOI] [PubMed] [Google Scholar]
- 20.Stover, C. K., D. P. Marana, J. M. Carter, B. A. Roe, E. Mardis, and E. V. Oaks. 1990. The 56-kilodalton major protein antigen of Rickettsia tsutsugamushi: molecular cloning and sequence analysis of the sta56 gene and precise identification of a strain-specific epitope. Infect. Immun. 582076-2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Studier, F. W., and B. A. Moffatt. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189113-130. [DOI] [PubMed] [Google Scholar]
- 22.Waner, T., S. Harrus, D. J. Weiss, H. Bark, and A. Keysary. 1995. Demonstration of serum antiplatelet antibodies in experimental acute canine monocytic ehrlichiosis. Vet. Immunol. Immunopathol. 48177-182. [DOI] [PubMed] [Google Scholar]
- 23.Waner, T., I. Leykin, M. Shinitsky, E. Sharabani, H. Buch, A. Keysary, H. Bark, and S. Harrus. 2000. Detection of platelet-bound antibodies in beagle dogs after artificial infection with Ehrlichia canis. Vet. Immunol. Immunopathol. 77145-150. [DOI] [PubMed] [Google Scholar]
- 24.Weddle, J. R., T. C. Chan, K. Thompson, H. Paxton, D. J. Kelly, G. Dasch, and D. Strickman. 1995. Effectiveness of a dot-blot immunoassay of anti-Rickettsia tsutsugamushi antibodies for serologic analysis of scrub typhus. Am. J. Trop. Med. Hyg. 5343-46. [PubMed] [Google Scholar]
- 25.Wong, S. J., and J. A. Thomas. 1998. Cytoplasmic, nuclear, and platelet autoantibodies in human granulocytic ehrlichiosis patients. J. Clin. Microbiol. 361959-1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wucherpfennig, K. W. 2001. Mechanisms for the induction of autoimmunity by infectious agents. J. Clin. Investig. 1081097-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]