Abstract
Capnocytophaga canimorsus is a bacterium of the canine oral flora known since 1976 to cause rare but severe septicemia and peripheral gangrene in patients that have been in contact with a dog. It was recently shown that these bacteria do not elicit an inflammatory response (H. Shin, M. Mally, M. Kuhn, C. Paroz, and G. R. Cornelis, J. Infect. Dis. 195:375-386, 2007). Here, we analyze their sensitivity to the innate immune system. Bacteria from the archetype strain Cc5 were highly resistant to killing by complement. There was little membrane attack complex (MAC) deposition in spite of C3b deposition. Cc5 bacteria were as resistant to phagocytosis by human polymorphonuclear leukocytes (PMNs) as Yersinia enterocolitica MRS40, endowed with an antiphagocytic type III secretion system. We isolated Y1C12, a transposon mutant that is hypersensitive to killing by complement via the antibody-dependent classical pathway. The mutation inactivated a putative glycosyltransferase gene, suggesting that the Y1C12 mutant was affected at the level of a capsular polysaccharide or lipopolysaccharide (LPS) structure. Cc5 appeared to have several polysaccharidic structures, one being altered in Y1C12. The structure missing in Y1C12 could be purified by classical LPS purification procedures and labeled by tritiated palmitate, indicating that it is more likely to be an LPS structure than a capsule. Y1C12 bacteria were also more sensitive to phagocytosis by PMNs than wild-type bacteria. In conclusion, a polysaccharide structure, likely an LPS, protects C. canimorsus from deposition of the complement MAC and from efficient phagocytosis by PMNs.
Since its discovery in 1976, Capnocytophaga canimorsus has been regularly isolated from severe human infections transmitted by dogs or cats (3). The genus Capnocytophaga belongs to the family Flavobacteriaceae and the phylum Bacteroidetes. This phylum is taxonomically remote from the Proteobacteria, which includes most common human pathogens but includes the family Bacteroidaceae, which contains some major commensals of the mammalian intestinal system such as Bacteroides thetaiotaomicron and Bacteroides fragilis (6). The phylum Bacteroidetes includes Porphyromonas gingivalis, a common agent of periodontal disease in humans (33), as well as three animal pathogens: Flavobacterium psychrophilum, the causative agent of cold-water disease in salmonid fish (7); Ornithobacterium rhinotracheale, causing respiratory disease in poultry (29); and Riemerella anatipestifer, the agent of “duckling disease” in waterfowl and turkeys (30, 35). The genus Capnocytophaga includes a variety of commensal bacteria that can be found in the oral flora of humans and animals. Seven Capnocytophaga species are found in humans, while C. canimorsus and Capnocytophaga cynodegmi are found in dogs and cats (4, 9).
Although C. canimorsus has been reported to be present in up to 25.5% of dogs (1, 2, 41), there is only one report in the literature of a dog suffering from a C. canimorsus infection, and it was not lethal (24). In contrast, more than 160 cases of severe human infections have now been reported (15, 36), and most cases are not reported any more. The number of cases of human infections is estimated to be around 1 per 1 million inhabitants per year (26). A pet rabbit infection after a dog bite has also been mentioned in the literature (39). Generally, infections result in fulminant septicemia, peripheral gangrene, or meningitis, with a mortality rate as high as 30%. Splenectomy, alcohol abuse, and immunosuppression have been associated with a number of cases, but more than 40% of the patients had no obvious risk factor (19, 36). Severe infections generally do not occur after dramatic bite injuries, which are preventively treated with antibiotics, but rather after apparently benign bites, scratches, or even licks.
In spite of the fact that these infections are now very well known to clinicians throughout the world, very few studies have addressed the molecular mechanisms of C. canimorsus pathogenesis. Recently, we showed that C. canimorsus cells exhibit robust growth when they are in direct contact with mammalian cells including phagocytes. This property depends on a surface-exposed sialidase allowing C. canimorsus to utilize internal amino sugars of glycan chains from host cell glycoproteins. Although sialidase most likely evolved to sustain commensalism, by releasing carbohydrates from mucosal surfaces, it also contributes to bacterial persistence in a murine infection model. Even more, there is evidence that in the mouse, C. canimorsus cells feed on polymorphonuclear leukocytes (PMNs) by deglycosylating host glycans (21). This observation implies that C. canimorsus cells escape the primary lines of defense. In agreement with this idea, we also showed that C. canimorsus cells are not detected by the usual sentinels of the innate immune system. Macrophages infected with live or heat-killed (HK) bacteria from C. canimorsus strain Cc5, a strain isolated from a patient with fatal septicemia (31), do not release tumor necrosis factor, interleukin-1α (IL-1α), IL-6, IL-8, gamma interferon, macrophage inflammatory protein 1b, and nitric oxide. This absence of a proinflammatory response is characterized by the inability of Toll-like receptor 4 to respond to Cc5 (31). Moreover, live, but not HK, Cc5 cells block the release of tumor necrosis factor and nitric oxide induced by HK Yersinia enterocolitica cells. In addition, live Cc5 cells downregulate the expression of Toll-like receptor 4 and dephosphorylate p38 mitogen-activated protein kinase (31). C. canimorsus cells also resist phagocytosis by a murine macrophage cell line, and some strains, like Cc5, even block the killing of unrelated preys like Escherichia coli by macrophages (23).
Following this investigation of the interaction between C. canimorsus and the innate immune system, we address here the question of the susceptibility to killing by human complement and PMNs. We show that C. canimorsus cells are resistant to complement killing as well as to PMN-mediated phagocytosis and killing. Finally, by isolating and characterizing a highly sensitive mutant, we show that these properties depend on a polysaccharidic structure, likely a lipopolysaccharide (LPS).
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The following bacterial strains were used: C. canimorsus strains Cc5, Cc11, and Cc12 (ATCC type strain), all isolated from human infections as described previously (31); E. coli Top10 (Invitrogen); Yersinia enterocolitica MRS40 (34); Salmonella enterica serovar Typhimurium strain SL1344 aroA (14); and S. enterica SL1344 aroA rfaG (a gift of D. Bumann). C. canimorsus bacteria were routinely grown on heart infusion agar (Difco) supplemented with 5% sheep blood (Oxoid) for 2 days at 37°C in the presence of 5% CO2. For the genetic screen, Cc5 mutant bacteria were grown in 100 μl heart infusion broth (Difco) supplemented with 10% (vol/vol) fetal bovine serum (Invitrogen) in 96-well plates for 48 h without shaking in a 37°C incubator with 5% CO2. Y. enterocolitica cells were grown and type III secretion was induced as described elsewhere previously (5, 18). S. enterica cultures were started (optical density at 600 nm [OD600] of 0.2) from a culture grown overnight and were grown for 2 h at 37°C. Selective agents were added at the following concentrations: ampicillin at 100 μg/ml (E. coli pCF1 and E. coli pCF3), cefoxitin at 10 μg/ml (C. canimorsus cY1C12), erythromycin at 10 μg/ml (C. canimorsus Y1C12), gentamicin at 20 μg/ml (all C. canimorsus strains), kanamycin at 20 μg/ml (S. enterica rfaG strain), nalidixic acid at 3.5 μg/ml (Y. enterocolitica MRS40), and streptomycin at 10 μg/ml (E. coli and wild-type [wt] S. enterica).
Antisera.
Anti-C3c-fluorescein isothiocyanate (FITC) was obtained from Dako, and anti-human factor H (fH) was obtained from Calbiochem. Anti-C4b-binding protein (C4BP) was a kind gift from Anna Blom (Lund University, Lund, Sweden). Anti-human immunoglobulin G (IgG) (Fc specific)-FITC was obtained from Sigma. Anti-dog IgA-FITC was obtained from Bethyl Laboratories. Mouse anti-human C5B-9 antibody (Ab) was obtained from Dako, and anti-mouse IgG-FITC Ab was obtained from Southern Biotech. Anti-goat IgG-FITC was obtained from Santa Cruz, and anti-rabbit IgG-FITC was obtained from Southern Biotech. Polyclonal serum against Cc5 was generated from rabbits by immunization with HK Cc5 (Laboratoire d'Hormonologie, Marloie, Belgium). Polyclonal serum against Y. enterocolitica LPS O:9 was generated from rabbit by immunization with lysate from total Y. enterocolitica E40 pYV− bacteria after 30 min of boiling. The Y1C12-absorbed serum was prepared by incubating an excess amount of Y1C12 mutant bacteria (harvested from blood plates and washed in phosphate-buffered saline [PBS]) with anti-Cc5 serum at 4°C for 4 h. Bacteria were removed by centrifugation and filtration through a 0.2-μm filter.
Detection of human Abs at the bacterial surface.
Bacteria from blood agar plates were washed with PBS and adjusted to an OD600 of 0.1 in PBS. Five hundred microliters of bacteria (2.5 × 107 CFU) was pelleted by centrifugation (6,000 × g for 2 min) and resuspended in 500 μl PBS supplemented with 10% heat-inactivated (HI) normal human serum (NHS). After 30 min of incubation at 37°C, bacteria were washed twice with PBS. Anti-human IgG (Fc specific)-FITC Ab was added to the pellet at a 1:500 dilution and incubated for 15 min at room temperature (RT) in the dark. Bacteria were washed twice with PBS, and the final pellet was resuspended in 500 μl PBS prior to fluorescence-activated cell sorter (FACS) analysis.
Sensitivity to killing by serum.
Bacteria were harvested by gently scraping colonies off the agar surface, washed, and resuspended in PBS to an OD600 of 0.4. NHS from healthy volunteers was pooled, aliquoted, and stored at −80°C. Animal sera were purchased from Innovative Research. Serum was HI at 55°C for 30 min. A total of 1 × 107 bacteria were incubated in 10% NHS in PBS at 37°C with 5% CO2. Serial dilutions were plated onto blood plates, and viable colonies were counted after 24 h (S. enterica serovar Typhimurium and E. coli) or 48 h (C. canimorsus) of incubation. When indicated, NHS was incubated with 10 mM MgCl2 and 10 mM EGTA for 10 min at 37°C before usage. Abs were depleted from NHS by incubation with GammaBind Sepharose (GE Healthcare) for 3 h at 4°C.
Detection of C3b, MAC, fH, and C4BP binding by flow cytometry analysis.
Bacteria from blood agar plates were washed with PBS and adjusted to an OD600 of 1 in PBS. For the detection of C3b, 90 μl of bacterial suspension (4.5 × 107 CFU) was incubated with 10 μl C7-depleted human serum (Sigma) for 30 min at 37°C. After incubation, bacteria were pelleted by centrifugation (6,000 × g for 1 min) and washed once with PBS. Anti-C3c-FITC was added to the pellet at a 1:200 dilution and incubated for 30 min at RT in the dark. For detection of MAC, 90 μl of bacterial suspension (4.5 × 107 CFU) was incubated with 1 μl NHS and 9 μl PBS for 15 min at 37°C. After incubation, bacteria were pelleted by centrifugation (6,000 × g for 1 min) and washed once with PBS. Anti-C5B-9 Ab was added to the pellet at a 1:200 dilution and incubated at RT for 30 min. After washing with PBS, anti-mouse IgG-FITC-conjugated Ab was added to the pellet at a 1:200 dilution and incubated for 15 min at RT in the dark. For the detection of fH and C4BP, 90 μl of bacteria (4.5 × 107 CFU) was incubated with 10 μl HI NHS for 60 min at 37°C. After incubation, bacteria were pelleted by centrifugation (6,000 × g for 1 min) and washed once with PBS. Anti-fH Ab or anti-C4BP Ab was added to the pellet at a 1:200 dilution and incubated for 30 min at RT. After washing with PBS, anti-goat IgG-FITC-conjugated Ab or anti-rabbit IgG-FITC-conjugated Ab was added to the pellet at a 1:200 dilution and incubated for 15 min at RT in the dark. For all detections, bacteria were washed twice in PBS, and the final pellet was resuspended in 1 ml of PBS prior to FACS analysis.
Detection of fH and C4BP deposition by immunoblotting.
Bacteria from blood agar plates were washed once with PBS-0.05% Tween and adjusted to an OD600 of 14. One hundred microliters of bacterial suspension (2.8 × 108 CFU) was incubated with 100 μl HI NHS at 37°C for 1 h. Bacteria were washed extensively with PBS-0.05% Tween. Bound proteins were eluted using 100 μl 1 M glycine (pH 2.2) for 20 min at RT. Eluted proteins were neutralized and analyzed by immunoblotting.
Immunofluorescence.
Falcon culture slides (Becton Dickinson) were coated with poly-d-lysine. Bacteria from blood agar plates were adjusted to an OD600 of 1 in PBS. Slides were washed four times with PBS. One hundred microliters of bacterial suspension, corresponding to 5 × 107 CFU, was added to the slides and incubated for 1 h at 37°C. Slides were washed four times with PBS before being fixed with 3% paraformaldehyde for 15 min at RT. Bacteria were labeled with anti-Cc5 or Y1C12-absorbed anti-Cc5 antiserum at the appropriate dilution for 30 min at 4°C. Slides were washed four times in PBS. FITC-conjugated secondary Ab was added, and slides were incubated at 37°C for 20 min. Slides were washed four times with PBS, mounted with antifade reagent (Vector Laboratories), and analyzed by use of a Leica Dmire2 microscope. Pictures were taken with a digital camera (Hamamatsu Photonics) and OpenLab software (version 3.1.2).
Isolation of human PMNs.
Human PMNs were isolated from healthy volunteers using the dextran-Percoll protocol, adapted with modifications described previously (16). Contaminating erythrocytes were removed by hypotonic lysis with “aqua ad iniectabilia” (Bichsel).
In vitro phagocytosis and killing assay.
Freshly isolated human PMNs were resuspended at 1 × 107 ml−1 in D-PBS (Gibco) supplemented with 10% Ab-depleted HI NHS (Scipac Ltd.) and infected with bacteria at the indicated multiplicity of infection (MOI). Bacteria were incubated at 37°C either in the presence of PMNs or without cells as the reference sample. When specified, bacteria were opsonized for 30 min at 37°C either with 10% HI NHS (as a source of Abs and spontaneously formed C3b) or with 10% C7-depleted human serum (as a source of C3b; Sigma) before infection. Samples were analyzed 120 min after infection. To differentiate between phagocytosis and killing, an aliquot was incubated with “aqua ad iniectabilia” (Bichsel) for 1 min in order to lyse the PMNs. Aliquots from untreated and lysed samples were then plated at different dilutions to count the surviving bacteria. Samples from bacteria incubated without cells were plated in parallel as a reference. Counts from lysed samples gave the total number of surviving bacteria, whereas counts from untreated cell samples gave the numbers of nonphagocytosed, nonadhering, extracellular bacteria. Numbers of phagocytosed and killed bacteria were calculated by subtracting the counts of extracellular or surviving bacteria, respectively, from the counts of the bacteria grown without cells (reference sample). Values are given as percentages of control values.
Tn mutant screen for serum bactericidal assay.
Random Tn4351 mutants were generated as described previously (20).Transposon (Tn) mutant bacteria were grown in 96-well plates (see above) and subsequently diluted in a new plate to an OD590 of 0.025. Fifty microliters of bacterial suspension (6.25 × 105 CFU) was incubated with either 10% NHS or HI NHS as a control in a 100-μl total volume. After 3 h of incubation, a microliter-range aliquot was spotted onto blood agar using a metal stamper and incubated for 48 h. The insertion site of the Tn was mapped by arbitrary PCR as described previously (21). The Tn integration site for mutant Y1C12 was confirmed by using primers 5′-GGACATTGTCTCTCTTTCC-3′ and 5′-CGAGCGTCCAGAGGCAATG-3′, complementary to the glycosyltransferase (gtf) sequence.
Construction of complementation plasmid.
Full-length gtf was amplified with primers 5′-GGAATTCTCTATGCCCAATG TGTCGG-3′ and 5′-ATGGATCCCAAAACCCGAAACTCCTG-3′ and inserted into the EcoRI/BamHI sites (underlined) of vector pUC19, resulting in pCF1. Full-length gtf was amplified from pCF1 using primers 5′-AATTCCATGGGAAAAGTACTTATAGTAACAC-3′ and 5′-GCTCTAGAAATTTTTTTAAATTAAGTATTCG-3′ inserted into the corresponding sites of E. coli-C. canimorsus shuttle vector pMM47 (20) using NcoI and XbaI (underlined), leading to the insertion of a glycine at position 2 and a C-terminal six-histidine tag (pCF3). All constructs were sequenced with an ABI sequencer.
Outer membrane proteins were isolated by a sarcosyl extraction method described previously (21).
Preparation of proteinase K-resistant samples.
Bacteria were washed with PBS and adjusted to an OD600 of 1.5 in PBS. Five hundred microliters of bacterial suspension (3.75 × 108 CFU) was pelleted (20,000 × g for 1 min), and bacteria were resuspended in 100 μl loading buffer (1% sodium dodecyl sulfate [SDS], 10% glycerol, 50 mM dithiothreitol, 0.02% bromophenol blue, 45 mM Tris (pH 6.8). Samples were boiled at 99°C for 10 min and then incubated at 37°C overnight after proteinase K (Roche) had been added to a final concentration of 50 μg/ml. After incubation, samples were boiled again for 10 min at 99°C, and proteinase K was added to a final concentration of 100 μg/ml. Samples were then incubated at 55°C for 3 h, boiled again for 5 min at 99°C, and loaded onto a 15% SDS-polyacrylamide gel electrophoresis (PAGE) gel or 16% Tricine-SDS-PAGE gel. Samples were analyzed by immunoblotting or silver-periodic acid staining (37), respectively.
LPS isolation.
wt and Y1C12 mutant bacteria were grown on 500 blood agar plates, harvested by scraping, and treated with 0.2% phenol. Bacterial pellets were washed with 1 liter each of ethanol, acetone, and diethyl ether with stirring (RT for 60 min). After centrifugation, cells were dried in air, giving 11.2 g (wt) and 30.8 g (Y1C12) of biomass. The isolation of LPS was achieved after phenol-water extraction (40), whereby the LPS was identified in the phenol phase, which was then extracted by the phenol-chloroform-petroleum ether method (11), giving 70 mg (wt) and 200 mg (Y1C12) LPS, respectively.
In vivo radiolabeling with [3H]palmitate and fluorography.
Bacteria were inoculated into a culture of HeLa epithelial cells (ATCC CCL-2) in complete Dulbecco's modified Eagle's medium at 37°C with 5% CO2 at an MOI of 20. Sixteen hours postinfection, [9,10-3H]palmitic acid (48 Ci/mmol; Perkin-Elmer Life Sciences) was added to a final concentration of 50 μCi/ml, and incubation was continued for 8 h, by which time the bacterial culture had reached approximately 108 bacteria/ml, as described elsewhere previously (21). Supernatants of 2 by 1 ml were collected without detaching epithelial cells from the wells. Bacteria were then collected by centrifugation, and pellets were combined from 2 ml and stored at −20°C until processing. Total cell extracts were obtained by solubilizing a pellet for 3 min at 85°C in 60 μl of loading buffer (1% SDS, 10% glycerol, 50 mM dithiothreitol, 0.02% bromophenol blue, 45 mM Tris [pH 6.8]). For LPS analysis, total cell extracts were digested with 50 μg/ml of proteinase K for 2 h at 60°C and heat treated for 5 min at 85°C. Samples of total cell extracts or proteinase K digests (20 μl/lane) were separated by Tricine-SDS-PAGE using 16% polyacrylamide (28). Gels were fixed in 25:65:10 isopropanol:water:acetic acid overnight and subsequently soaked for 30 min in Amplify (Amersham). Gels were vacuum dried and exposed to SuperRX autoradiography film (Fuji) for 18 h at −70°C.
Statistical analyses.
For all experiments, means and standard deviations were calculated. For phagocytosis assays and FACS analyses, statistical significance was evaluated using a two-tailed, unpaired Student's t test. Differences were considered significant when the P value was <0.05 (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
Nucleotide sequence accession number.
The sequence of the Y1C12 locus was deposited in the GenBank database under accession number FJ214098.
RESULTS
C. canimorsus cells are resistant to killing by complement.
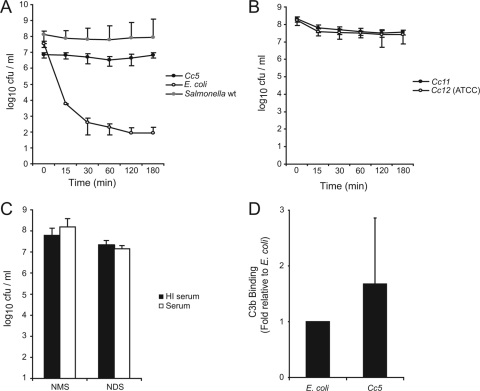
We first tested the sensitivity of C. canimorsus to complement-mediated killing. Cc5 cells were incubated for various periods of time with 10% NHS and plated, and the survival rate was determined by viable counting. For comparison, E. coli Top 10 cells were used as serum-sensitive bacteria, and S. enterica serovar Typhimurium SL1344 cells were used as serum-resistant bacteria. Within 30 min, 10% NHS reduced the number of live E. coli cells by 5 log10 units, but it reduced the number of live Cc5 cells by only 0.1 log10 units and the number of S. enterica serovar Typhimurium cells by 0.5 log10 units (Fig. 1A). After 3 h of incubation, Cc5 counts were even higher than in the inoculum, while E. coli counts went down by almost 6 log10 units and S. enterica serovar Typhimurium counts were reduced by 0.7 log10 units (Fig. 1A). E. coli cells survived incubation with HI NHS (data not shown), demonstrating that killing was indeed complement mediated. Two other C. canimorsus strains isolated from human infections, Cc11 and Cc12 (ATCC type strain), were then tested and found to be as resistant as Cc5 (Fig. 1B), indicating that complement resistance is a common property of C. canimorsus strains.
FIG. 1.
C. canimorsus cells are resistant to complement-mediated killing. (A) Total CFU present after incubation of Cc5 (black circles), E. coli (white circles), and wt S. enterica serovar Typhimurium (gray circles) in 10% NHS for 0, 15, 30, 60, 120, and 180 min at 37°C. (B) Survival of Cc11 (black circles) and Cc12 (white circles) in 10% NHS for 0, 15, 30, 60, 120, and 180 min at 37°C. (C) Total CFU present after incubation of Cc5 in 10% HI (black) or active (white) normal mouse serum (NMS) or normal dog serum (NDS) for 180 min at 37°C. (D) Binding of C3b by Cc5 or E. coli after 30 min incubation with HI NHS. FACS analysis was performed using anti-C3c polyclonal antiserum to quantify the level of C3b on the bacterial surface. The values are the means of three independent experiments ± standard deviations (bars).
Sera obtained from different species of mammals are known to possess different levels of bactericidal activity against gram-negative bacteria (13, 22). Since C. canimorsus is a dog commensal and since mice were shown to be quite resistant to experimental infection by C. canimorsus (21), we tested the sensitivity of Cc5 to normal dog serum and normal mouse serum. As shown in Fig. 1C, it appeared that Cc5 also resists killing by complement from dog and mouse.
We then tested whether this serum resistance was due to the recruitment of negative regulators that are known to interfere with C3b deposition. We first performed a serum absorption experiment in which whole bacteria were incubated with HI NHS, followed by elution of bound serum proteins at a low pH and analysis of eluates by immunoblotting. By this method, some fH, but no C4BP, was found to be recruited at the surface of Cc5, while none was recruited at the surface of E. coli (data not shown). We then tried to confirm this observation by direct flow cytometry analysis with an FITC-conjugated secondary Ab directed against anti-fH or anti-C4BP Abs. However, neither fH nor C4BP could be detected at the bacterial surface by this approach (data not shown). This suggests that the fH deposition observed by the first approach might not be sufficient to prevent C3b deposition.
We next analyzed whether there was C3b deposition. Bacteria were incubated for 30 min with C7-depleted NHS, and deposited C3b was detected by flow cytometry with an FITC-conjugated anti-C3c Ab. There was indeed a significant deposition of C3b on Cc5 bacteria as well as on E. coli bacteria (Fig. 1D). Hence, the resistance of C. canimorsus to complement killing was not the result of a lack of C3b deposition but was likely the result of a lack of MAC insertion. Indeed, very little, if any, C5B-9 was detected at the surface of Cc5 bacteria by flow cytometry (mean fluorescence intensity increased by about 1.3-fold compared to background fluorescence).
Cc5 bacteria resist phagocytosis and killing by PMNs.
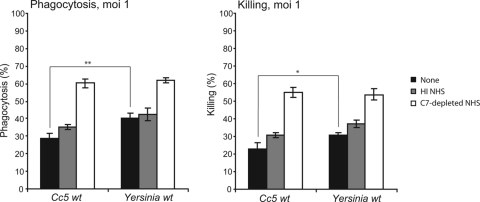
We next examined the interaction between C. canimorsus and human PMNs. We used wt Y. enterocolitica as a control for phagocytosis-resistant bacteria (12, 43). In the absence of opsonization and at an MOI of 1, the levels of phagocytosis of Cc5 and wt Y. enterocolitica reached about 30% and 40%, respectively. About 20% of Cc5 and 30% of Y. enterocolitica bacteria were killed by PMNs (Fig. 2). At an MOI of 50, Cc5 was completely resistant to phagocytosis and killing by PMNs (both <5%), whereas 28% of wt Y. enterocolitica cells were phagocytosed, and 17% were killed (data not shown).
FIG. 2.
Cc5 cells are resistant to PMN-mediated killing. Levels of phagocytosis and killing were determined 120 min after infection of human PMNs. PMNs were infected with either Cc5 or wt Y. enterocolitica at an MOI of 1. Infection with nonopsonized (black), HI NHS-opsonized (gray), or C7-depleted NHS-opsonized bacteria (white) are shown. Results are the means of at least three independent experiments ± standard deviations (bars). The statistical significance of the difference between Cc5 and wt Y. enterocolitica (unopsonized) is given, with * indicating a P value of <0.05 and ** indicating a P value of <0.01, using a two-tailed unpaired Student's t test.
Preopsonization with C7-depleted NHS increased the level of phagocytosis and killing of Cc5 and wt Y. enterocolitica by about twofold at an MOI of 1. To test the opsonizing effect independent of C3b, we compared the effect of HI-NHS (Abs and spontaneously preformed C3b) to the effect of C7-depleted NHS (activated C3b plus Abs). As shown in Fig. 2, HI NHS increased phagocytosis and killing of Cc5 by 5%, suggesting that there could be anti-C. canimorsus Abs in the pool of NHS. We monitored the presence of such Abs by FACS analysis using anti-human IgG-FITC and found that, indeed, the opsonization of Cc5 by HI NHS led to a sixfold increase in the mean fluorescence intensity (data not shown). In comparison, preopsonization of Y. enterocolitica with C7-depleted human serum led to ∼60% phagocytosis and 55% killing of the bacteria at an MOI of 1. At an MOI of 50, Cc5 preopsonization with C7-depleted NHS led to 20% phagocytosis, while Y. enterocolitica was phagocytosed at 60% (data not shown).
In conclusion, Cc5 bacteria were as resistant as or even more resistant to phagocytosis and killing by human PMNs than Y. enterocolitica endowed with the anti-phagocytic type III secretion system.
Isolation of a Cc5 mutant that is sensitive to killing by complement and PMNs.
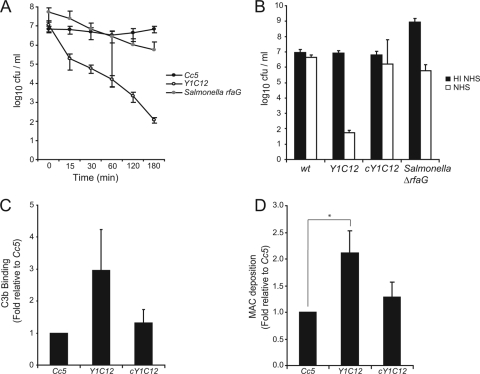
By screening a library of Tn4351 mutants of Cc5, we isolated a clone (designated Y1C12) whose survival rate in NHS was severely decreased compared to that of the wt. As shown in Fig. 3A, 10% NHS reduced the viable counts of Y1C12 by more than 2 log10 units already within 30 min and by almost 5 log10 units after 3 h of incubation. Y1C12 mutant bacteria survived incubation with HI NHS, meaning that the observed serum sensitivity involved complement-dependent killing (Fig. 3B). The Y1C12 mutant was even more sensitive to killing by complement than the rfaG mutant of S. enterica serovar Typhimurium deficient in O-chain formation of LPS (17). Deposition of C3b onto Y1C12 mutant bacteria was not significantly increased compared to that of the wt (Fig. 3C), reinforcing our above-described observation that resistance of the wt to complement does not result from a lack of C3b deposition. However, as expected, there was significantly more MAC formed at the surface of Y1C12 bacteria. Already after 15 min of incubation with NHS, there was twofold more MAC inserted onto the mutant bacteria than onto wt Cc5. Since the already-lysed bacteria are not detected by FACS analysis, this difference is probably even underestimated. In conclusion, mutant Y1C12 bacteria are killed by complement because they allow more MAC deposition onto their surface (Fig. 3D).
FIG. 3.
Characterization of the serum-sensitive Tn mutant of Cc5. (A) Total CFU present after incubation of Cc5 (black circles), Y1C12 (white circles), or an S. enterica serovar Typhimurium rfaG mutant (gray circles) in 10% NHS for 0, 15, 30, 60, 120, and 180 min at 37°C. (B) Total CFU present after incubation of Cc5, Y1C12, cY1C12, or an S. enterica serovar Typhimurium rfaG mutant in 10% HI NHS (black) or 10% NHS (white) for 180 min at 37°C. (C) Binding of C3b by Cc5, Y1C12, or cY1C12 after 30 min incubation with C7-depleted NHS. FACS analysis was performed using anti-C3c polyclonal antiserum to quantify the level of C3b on the bacterial surface. (D) MAC deposition on Cc5, Y1C12, or cY1C12 after 15 min of incubation with NHS (longer incubation leads to significant lysis). FACS analysis was performed using mouse anti-C5B-9 primary and anti-mouse-FITC secondary Abs to quantify the level of MAC insertion into the bacterial surface. The statistical significance of the difference in MAC insertion between Y1C12 and wt C. canimorsus is given, with * indicating a P value of <0.05 using a two-tailed unpaired Student's t test. Results are the means of at least three independent experiments ± standard deviations (bars). cY1C12, Y1C12 mutant bacteria complemented with the gtf gene in trans.
Killing of the serum-sensitive Y1C12 mutant involves the Ab-dependent classical pathway.
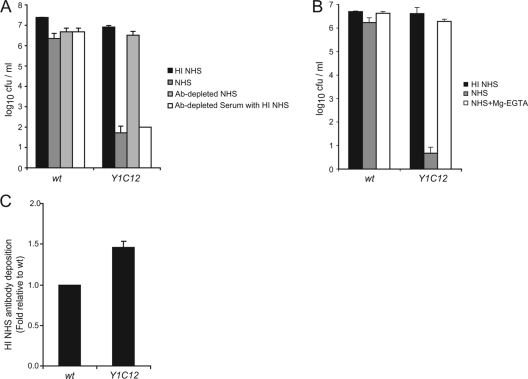
Since the NHS pool contained Abs recognizing Cc5, and as these Abs favored phagocytosis, we investigated the role of these Abs in the killing of Y1C12 mutant bacteria by complement in NHS. There was no significant difference in the survival rates of wt bacteria in 10% NHS, HI NHS, Ab-depleted NHS, and Ab-depleted NHS supplemented with HI NHS (as a source of Abs) (Fig. 4A). In contrast, Y1C12 mutant bacteria survived in HI NHS and Ab-depleted NHS but were readily killed by both NHS and Ab-depleted NHS supplemented with HI NHS (Fig. 4A). These results thus indicate that the killing of Y1C12 bacteria by complement is dependent on the presence of specific Abs recognizing C. canimorsus.
FIG. 4.
Killing of Y1C12 occurs via an IgG-dependent classical pathway. (A) Total CFU present after incubation of wt or Y1C12 C. canimorsus in 10% HI NHS (black), NHS (dark gray), Ab-depleted NHS (light gray), or Ab-depleted NHS supplemented with HI NHS as a source of Abs (white) for 180 min at 37°C. (B) Total CFU present after incubation of wt or Y1C12 C. canimorsus in 10% HI NHS (black), NHS (gray), or Mg-EGTA-treated NHS (white) for 180 min at 37°C. (C) Binding of human serum Abs to wt, Y1C12, or cY1C12 C. canimorsus after 30 min incubation with HI NHS. The level of Ab deposition on bacteria was quantified by FACS analysis using anti-human IgG-FITC Ab. Results are the means of at least three independent experiments ± standard deviations (bars).
To further dissect which complement pathway is responsible for killing Y1C12 mutant bacteria, NHS was treated with Mg-EGTA, which blocks the classical and the lectin pathways (8). After 180 min of incubation with 10% Mg-EGTA-treated NHS, the number of viable bacteria was reduced only by less than 0.5 log10 units, in comparison to 6 log10 units in untreated NHS (Fig. 4B). Thus, it appears that the killing of Y1C12 bacteria occurs primarily by the Ab-dependent classical pathway. However, as already seen for C3b deposition, the deposition of human serum Abs onto Y1C12 mutant bacteria was increased only by 1.5-fold (Fig. 4C), in good agreement with the hypothesis that the increased sensitivity is due essentially to differences in the sensitivity to MAC deposition.
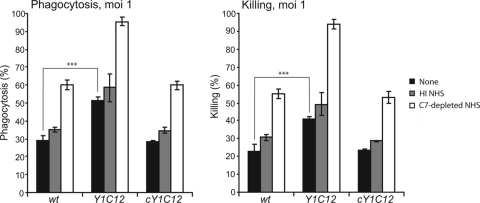
Serum-sensitive Y1C12 mutant bacteria have an increased sensitivity to phagocytosis by PMNs.
At an MOI of 1, the levels of PMN-mediated phagocytosis and killing of nonopsonized Y1C12 reached ∼50% and ∼40%, respectively (Fig. 5), compared to ∼30% and ∼20% for wt Cc5. Preopsonization of the Y1C12 mutant bacteria with C7-depleted NHS increased the level of phagocytosis and killing by PMNs by 40%. When Y1C12 mutant bacteria were preopsonized with HI NHS, phagocytosis increased by only ∼10%, indicating that Abs present in NHS were responsible for only a slight increase in levels of phagocytosis and killing of Y1C12. In conclusion, Y1C12 mutant bacteria were approximately twice as sensitive to phagocytosis and killing by PMNs as the wt.
FIG. 5.
Tn mutant Y1C12 cells exhibit increased sensitivity to phagocytosis and killing by PMNs. Phagocytosis and killing were determined 120 min after infection of human PMNs. PMNs were infected with wt, Y1C12, or cY1C12 C. canimorsus at an MOI of 1. Infections with nonopsonized (black), HI NHS-opsonized (gray), or C7-depleted NHS-opsonized bacteria (white) are shown. Results are the means of at least three independent experiments ± standard deviations (bars), including statistical significance between phagocytosis and killing of the wt and Y1C12 (unopsonized), with *** indicating a P value of <0.001 using a two-tailed, unpaired Student's t test.
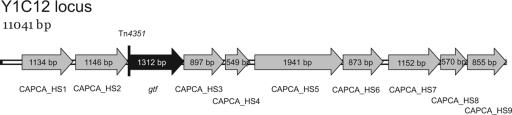
The Y1C12 mutant is affected in a glycosyltransferase.
The transposon integration site in Y1C12 was found to be located after the first 26 bp of a gene encoding a putative glycosyltransferase (gtf). A locus of 11 kb was sequenced, and BLAST search analysis revealed that eight of nine surrounding genes are predicted to be involved in sugar biosynthesis and that one was predicted to be a prokaryote-type ATPase (Fig. 6 and Table 1).
FIG. 6.
Y1C12 is affected in a gene encoding a putative glycosyltransferase. Shown is the genetic locus of Y1C12 including its upstream and downstream genes (GenBank accession number FJ214098).
TABLE 1.
Sequence analysis of the Y1C12 genomic locus
| Locus tag or gene product | Best-hit BLAST accession no., description (species)a | Score | E value | InterProScan accession no. (description)b |
|---|---|---|---|---|
| CAPCA_HS1 | YP_001296180.1, FnlB protein involved in UDP-l-FucpNAc biosynthesis (a nucleotide sugar precursor for antigen-O biosynthesis) (Flavobacterium psychrophilum JIP02/86) | 554 | 4e−156 | IPR000888 (dTDP-4-dehydrorhamnose 3,5-epimerase related), IPR016040 [NAD(P) binding] |
| CAPCA_HS2 | YP_001296179.1, FnlC protein involved in UDP-l-FucpNAc biosynthesis (a nucleotide sugar precursor for antigen-O biosynthesis), UDP-2-acetamino-2,6-dideoxy-l-talose 2-epimerase (Flavobacterium psychrophilum JIP02/86) | 707 | 0.0 | IPR003331 (UDP-N-acetylglucosamine 2-epimerase) |
| Gtf | ZP_03015063.1, hypothetical protein BACINT_02652 (Bacteroides intestinalis DSM 17393) | 441 | 7e−122 | IPR001296 (glycosyltransferase, group 1) |
| CAPCA_HS3 | YP_001192692.1, NAD-dependent epimerase/dehydratase (Flavobacterium johnsoniae UW101) | 315 | 4e−84 | IPR001509 (NAD-dependent epimerase/dehydratase), IPR016040 [NAD(P) binding] |
| CAPCA_HS4 | ZP_01043213.1, sugar transferase (Idiomarina baltica OS145) | 265 | 1e−69 | IPR003362 (bacterial sugar transferase) |
| CAPCA_HS5 | YP_862017.1, capsular polysaccharide biosynthesis protein (Gramella forsetii KT0803) | 623 | 2e−176 | IPR003869 (polysaccharide biosynthesis protein CapD), IPR016040 [NAD(P) binding] |
| CAPCA_HS6 | YP_001302048.1, putative glucose-1-phosphate thymidyltransferase (Parabacteroides distasonis ATCC 8503) | 508 | 1e−142 | IPR005907 (glucose-1-phosphate thymidylyltransferase, long form), IPR005835 (nucleotidyl transferase) |
| CAPCA_HS7 | ZP_02034053.1, hypothetical protein PARMER_04094 (Parabacteroides merdae ATCC 43184) | 384 | 6e−105 | IPR011579 (prokaryotic ATPase) |
| CAPCA_HS8 | NP_810251.1, dTDP-4-dehydrorhamnose-3,5-epimerase (Bacteroides thetaiotaomicron VPI-5482) | 281 | 2e−74 | IPR000888 (dTDP-4-dehydrorhamnose-3,5-epimerase related), IPR014710 (RmlC-like jelly roll fold) |
| CAPCA_HS9 | YP_001299685.1, dTDP-4-dehydrorhamnose reductase (Bacteroides vulgatus ATCC 8482) | 335 | 2e−90 | IPR005913 (dTDP-4-dehydrorhamnose reductase), IPR016040 [NAD(P) binding] |
Nonredundant database as of August 2008.
InterPro accession number and description as of August 2008.
First, we wanted to confirm that the serum sensitivity of the Y1C12 mutant is due to a disruption in the hypothetical gtf gene and not due to polar effects of the transposon. The gtf gene was therefore cloned into a C. canimorsus expression shuttle vector (20), and the resulting plasmid, pCF3, was introduced into Y1C12 in trans. The complemented Y1C12 (designated cY1C12) bacteria exhibited a phenotype similar to that of wt bacteria with regard to C3b deposition, resistance to complement, and resistance to PMN-mediated killing (Fig. 3B and C, 4, and 5). Hence, the serum sensitivity of Y1C12 resulted from the disruption of the gtf gene.
The Y1C12 mutant is affected in the biosynthesis of a lipopolysaccharidic structure.
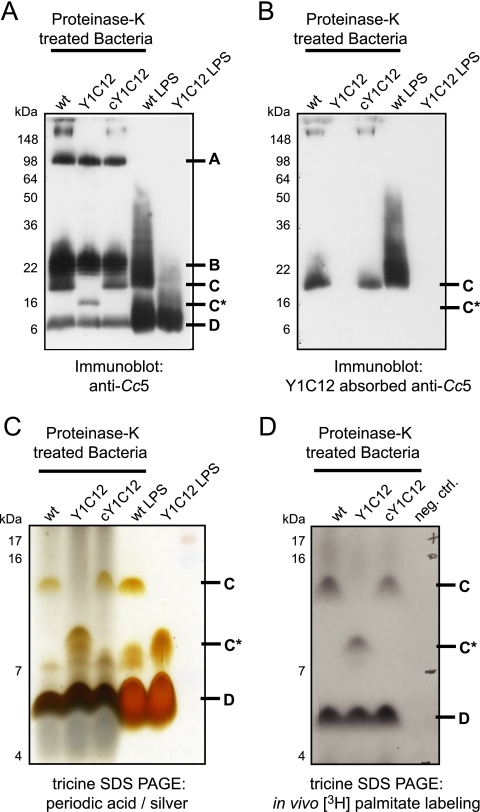
Glycosyltransferases are enzymes that are involved in the formation of polysaccharide structures such as capsules or O antigens of LPS. To gain better insights into possible alterations of the bacterial surface on Y1C12 bacteria, we first absorbed the Cc5 antiserum with a large excess of intact Y1C12 in order to generate an antiserum recognizing the structure affected by the gtf mutation specifically. The efficacy of the serum absorption was controlled by immunofluorescence: while crude anti-Cc5 antiserum recognized wt, Y1C12, and cY1C12 bacteria, the Y1C12-absorbed anti-Cc5 antiserum recognized only wt and cY1C12 bacteria (data not shown). Next, we purified polysaccharide structures by digesting whole bacteria with proteinase K followed by immunoblotting using crude and absorbed anti-Cc5 antiserum (Fig. 7). Immunoblot analysis with anti-Cc5 antiserum of the proteinase K-treated samples from the wt revealed four major bands, labeled A, B, C, and D in Fig. 7A. Bands A, B, and D were present in the proteinase K-treated samples from Y1C12 bacteria, but band C was not, which was replaced by a band migrating faster (C*) (Fig. 7A), suggesting that band C corresponds to the structure altered in Y1C12. In good agreement with this, only band C was visualized in samples from wt and cY1C12 bacteria when the Y1C12-absorbed anti-Cc5 antiserum was used (Fig. 7B). We conclude from this result that a surface structure (band C) is altered in Y1C12 bacteria.
FIG. 7.
Y1C12 has an altered lipidated polysaccharide structure. (A) Immunoblotting analysis of proteinase K-treated Cc5, Y1C12, and cY1C12 bacteria and of LPS isolated from Cc5 and Y1C12 using anti-Cc5. (B) Immunoblotting analysis as described above (A) using Y1C12-absorbed anti-Cc5 antiserum. (C) Silver-periodic acid staining of Tricine gel electrophoresis of the same samples as above (A and B). (D) Proteinase K-resistant structures of wt Cc5, Y1C12, and cY1C12 bacteria that had been labeled in vivo using [9,10-3H]palmitic acid.
To determine the nature of this structure, we specifically extracted and purified the LPS from wt and Y1C12 bacteria and analyzed it by SDS-PAGE followed by either immunoblotting (Fig. 7A) or periodic acid-silver staining (Fig. 7C). In the periodic acid-silver-stained gel, the LPS preparation from wt bacteria appeared as three bands, with two migrating like bands C and D in the proteinase K-treated samples. In contrast, the LPS preparation from Y1C12 bacteria lacked the C band but exhibited a band migrating like band C*. When analyzed by immunoblotting with complete crude anti-Cc5 serum, the LPS from the wt appeared to contain bands B, C, and D, while the LPS from Y1C12 showed only band D clearly (Fig. 7A). When analyzed by immunoblotting using the Y1C12-absorbed serum, band C appeared in the LPS preparation of wt bacteria but not in the LPS preparation of Y1C12 (Fig. 7B). These results suggested that the polysaccharide structure (band C) altered by the gtf mutation is likely to be an LPS, although it does not exhibit the typical ladder pattern by electrophoresis. In contrast, structure B, which does migrate as a ladder, is not present in the LPS preparation and, hence, probably does not represent an LPS structure.
To check whether this structure has a lipid anchor, we incorporated [9,10-3H]palmitic acid into wt or Y1C12 bacteria. To do this, we cultured bacteria in the presence of HeLa epithelial cells because this is the best way to obtain robust growth in a synthetic medium (21). Labeled bacteria were treated with proteinase K and analyzed by Tricine-SDS-PAGE followed by autoradiography (Fig. 7D). A band migrating like band C appeared in wt Cc5 bacteria (Fig. 7A) but not in Y1C12 mutant bacteria, where a band migrating more slowly appeared (C*). These data suggest that the structure altered in Y1C12 has a lipid anchor, reinforcing the idea that it represents a form of LPS. Structure B did not appear to be palmitoylated and, hence, probably does not correspond to LPS in spite of its migration profile. It could represent a capsule. Structure D was also lipidated, suggesting that Cc5 could have two different LPS structures (structures C and D) besides other polysaccharidic structures (structures A and B).
DISCUSSION
In an effort to understand the pathogenesis of human C. canimorsus infections, we investigated here the sensitivity of C. canimorsus to killing by complement and phagocytosis by human PMNs. The archetypal C. canimorsus strain Cc5 and two other strains isolated from human infections turned out to be highly resistant to killing by the human, dog, and mouse complement systems. C3b deposition was not prevented, but nevertheless, there was very little MAC assembly. This suggested that some surface structure prevents MAC assembly. This hypothesis was reinforced by the fact that a mutant that was hypersensitive to complement killing turned out to undergo significantly more MAC deposition than the wt. The mutation turned out to affect a glycosyltransferase, hinting that a polysaccharide structure is responsible for complement resistance. In good agreement with this prediction, the mutant turned out to be affected in the glycosidic moiety of a lipidated structure. However, this analysis revealed the presence of more than one polysaccharide structure. Since there was no obvious change in the patterns of outer membrane proteins themselves (data not shown), we thus conclude that a polysaccharide structure, likely LPS, prevents the formation or insertion of the MAC and makes C. canimorsus resistant to complement killing. Although the critical structure is lipidated, one cannot formally exclude that it could be a capsule rather than the LPS. Indeed, Neisseria meningitidis polysialic acid capsules have been shown to have a lipid anchor (38). More work is thus required to characterize the surface structure of C. canimorsus that is responsible for complement resistance. Polysaccharide surface structures are well known for protecting gram-negative bacteria, including the closely related P. gingivalis (32), from complement killing, either by steric hindrance of the MAC or by binding fH, eventually through decoration with sialic acid residues (10, 27). According to our data, the first mechanism is the most likely here, but the presence of sialic acid on a surface structure cannot be definitely excluded so far. We detected some fH binding on wt Cc5, but the hypersensitive mutant could still bind fH, reinforcing the idea that fH binding is not key to complement resistance. The hypersensitive mutant was found to be killed by the classical or lectin pathway rather than by the alternative pathway, and the pools of NHS used as a complement source were found to contain Abs recognizing Cc5. This observation is not surprising since pools of human serum have been reported to contain Abs recognizing Capnocytophaga sp. belonging to normal human oral flora (42), and we confirmed that the pools of HI NHS used in this study contained Abs recognizing Capnocytophaga ochracea, Capnocytophaga gingivalis, and C. cynodegmi (data not shown).
C. canimorsus also turned out to be quite resistant to phagocytosis and killing by human PMNs. The complement-sensitive gtf mutant bacteria were more sensitive to phagocytosis and killing by human PMNs than wt Cc5 bacteria, indicating that the same polysaccharide structure protects from complement killing and from phagocytosis. The resistance to phagocytosis was more pronounced at a high MOI, and this suggests that a factor other than the polysaccharide might contribute to this resistance. In conclusion, by virtue of a particular polysaccharidic structure, C. canimorsus can resist the assaults from the innate immune system. This resistance, combined with the low proinflammatory and even immunosuppressive properties described previously (31) as well as a capacity to feed in vivo from host glycans (21), explains easily that C. canimorsus may cause extremely severe infections. However, in view of these virulence properties and the high prevalence of C. canimorsus in dogs, one must question why human infections are so rare. One possibility would be that the C. canimorsus strains colonizing most dogs would not be endowed with all these virulence functions or would have only some of them. Dog strains are presently being collected and analyzed to address this hypothesis. Besides this, a thorough immunological investigation of patients who survived a C. canimorsus infection would be of the utmost interest but is difficult to organize given the low prevalence of infections. These two approaches may shed some light on this problem, but one should keep in mind that the same haunting question applies to many pathogens causing severe diseases, like Neisseria meningitidis, to cite only one (25).
Acknowledgments
We thank Regine Landmann and Naja Jann for their help and advice concerning the isolation and handling of PMNs and Anna Blom for the gift of the anti-C4BP serum.
This work was supported by the Swiss National Science Foundation (grant 32-65393.01). We declare that no competing interests exist.
Editor: J. B. Bliska
Footnotes
Published ahead of print on 23 March 2009.
REFERENCES
- 1.Bailie, W. E., E. C. Stowe, and A. M. Schmitt. 1978. Aerobic bacterial flora of oral and nasal fluids of canines with reference to bacteria associated with bites. J. Clin. Microbiol. 7223-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blanche, P., E. Bloch, and D. Sicard. 1998. Capnocytophaga canimorsus in the oral flora of dogs and cats. J. Infect. 36134. [DOI] [PubMed] [Google Scholar]
- 3.Bobo, R. A., and E. J. Newton. 1976. A previously undescribed gram-negative bacillus causing septicemia and meningitis. Am. J. Clin. Pathol. 65564-569. [DOI] [PubMed] [Google Scholar]
- 4.Brenner, D. J., D. G. Hollis, G. R. Fanning, and R. E. Weaver. 1989. Capnocytophaga canimorsus sp. nov. (formerly CDC group DF-2), a cause of septicemia following dog bite, and C. cynodegmi sp. nov., a cause of localized wound infection following dog bite. J. Clin. Microbiol. 27231-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cornelis, G., J. C. Vanootegem, and C. Sluiters. 1987. Transcription of the yop regulon from Y. enterocolitica requires trans acting pYV and chromosomal genes. Microb. Pathog. 2367-379. [DOI] [PubMed] [Google Scholar]
- 6.Coyne, M. J., and L. E. Comstock. 2008. Niche-specific features of the intestinal Bacteroidales. J. Bacteriol. 190736-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duchaud, E., M. Boussaha, V. Loux, J. F. Bernardet, C. Michel, B. Kerouault, S. Mondot, P. Nicolas, R. Bossy, C. Caron, P. Bessieres, J. F. Gibrat, S. Claverol, F. Dumetz, M. Le Henaff, and A. Benmansour. 2007. Complete genome sequence of the fish pathogen Flavobacterium psychrophilum. Nat. Biotechnol. 25763-769. [DOI] [PubMed] [Google Scholar]
- 8.Fine, D. P., S. R. Marney, Jr., D. G. Colley, J. S. Sergent, and R. M. Des Prez. 1972. C3 shunt activation in human serum chelated with EGTA. J. Immunol. 109807-809. [PubMed] [Google Scholar]
- 9.Frandsen, E. V., K. Poulsen, E. Kononen, and M. Kilian. 2008. Diversity of Capnocytophaga species in children and description of Capnocytophaga leadbetteri sp. nov. and Capnocytophaga genospecies AHN8471. Int. J. Syst. Evol. Microbiol. 58324-336. [DOI] [PubMed] [Google Scholar]
- 10.Frank, M. M., K. Joiner, and C. Hammer. 1987. The function of antibody and complement in the lysis of bacteria. Rev. Infect. Dis. 9(Suppl. 5)S537-S545. [DOI] [PubMed] [Google Scholar]
- 11.Galanos, C., O. Luderitz, and O. Westphal. 1969. A new method for the extraction of R lipopolysaccharides. Eur. J. Biochem. 9245-249. [DOI] [PubMed] [Google Scholar]
- 12.Grosdent, N., I. Maridonneau-Parini, M. P. Sory, and G. R. Cornelis. 2002. Role of Yops and adhesins in resistance of Yersinia enterocolitica to phagocytosis. Infect. Immun. 704165-4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hanski, C., M. Naumann, A. Grutzkau, G. Pluschke, B. Friedrich, H. Hahn, and E. O. Riecken. 1991. Humoral and cellular defense against intestinal murine infection with Yersinia enterocolitica. Infect. Immun. 591106-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoiseth, S. K., and B. A. Stocker. 1981. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature 291238-239. [DOI] [PubMed] [Google Scholar]
- 15.Janda, J. M., M. H. Graves, D. Lindquist, and W. S. Probert. 2006. Diagnosing Capnocytophaga canimorsus infections. Emerg. Infect. Dis. 12340-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jepsen, L. V., and T. Skottun. 1982. A rapid one-step method for the isolation of human granulocytes from whole blood. Scand. J. Clin. Lab. Investig. 42235-238. [PubMed] [Google Scholar]
- 17.Kadam, S. K., A. Rehemtulla, and K. E. Sanderson. 1985. Cloning of rfaG, B, I, and J genes for glycosyltransferase enzymes for synthesis of the lipopolysaccharide core of Salmonella typhimurium. J. Bacteriol. 161277-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Letzelter, M., I. Sorg, L. J. Mota, S. Meyer, J. Stalder, M. Feldman, M. Kuhn, I. Callebaut, and G. R. Cornelis. 2006. The discovery of SycO highlights a new function for type III secretion effector chaperones. EMBO J. 253223-3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lion, C., F. Escande, and J. C. Burdin. 1996. Capnocytophaga canimorsus infections in human: review of the literature and cases report. Eur. J. Epidemiol. 12521-533. [DOI] [PubMed] [Google Scholar]
- 20.Mally, M., and G. R. Cornelis. 2008. Genetic tools for studying Capnocytophaga canimorsus. Appl. Environ. Microbiol. 746369-6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mally, M., H. Shin, C. Paroz, R. Landmann, and G. R. Cornelis. 2008. Capnocytophaga canimorsus: a human pathogen feeding at the surface of epithelial cells and phagocytes. PLoS Pathog. 4e1000164. doi: 10.1371/journal.ppat.1000164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marcus, S., D. W. Esplin, and D. M. Donaldson. 1954. Lack of bactericidal effect of mouse serum on a number of common microorganisms. Science 119877. [DOI] [PubMed] [Google Scholar]
- 23.Meyer, S., H. Shin, and G. R. Cornelis. 2008. Capnocytophaga canimorsus resists phagocytosis by macrophages and pages the ability of macrophages to kill other bacteria. Immunobiology 213805-814. [DOI] [PubMed] [Google Scholar]
- 24.Meyers, B., J. P. Schoeman, A. Goddard, and J. Picard. 2008. The bacteriology and antimicrobial susceptibility of infected and non-infected dog bite wounds: fifty cases. Vet. Microbiol. 127360-368. [DOI] [PubMed] [Google Scholar]
- 25.Parkhill, J., M. Achtman, K. D. James, S. D. Bentley, C. Churcher, S. R. Klee, G. Morelli, D. Basham, D. Brown, T. Chillingworth, R. M. Davies, P. Davis, K. Devlin, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, S. Leather, S. Moule, K. Mungall, M. A. Quail, M. A. Rajandream, K. M. Rutherford, M. Simmonds, J. Skelton, S. Whitehead, B. G. Spratt, and B. G. Barrell. 2000. Complete DNA sequence of a serogroup A strain of Neisseria meningitidis Z2491. Nature 404502-506. [DOI] [PubMed] [Google Scholar]
- 26.Pers, C., B. Gahrn-Hansen, and W. Frederiksen. 1996. Capnocytophaga canimorsus septicemia in Denmark, 1982-1995: review of 39 cases. Clin. Infect. Dis. 2371-75. [DOI] [PubMed] [Google Scholar]
- 27.Rautemaa, R., and S. Meri. 1999. Complement-resistance mechanisms of bacteria. Microbes Infect. 1785-794. [DOI] [PubMed] [Google Scholar]
- 28.Schagger, H. 2006. Tricine-SDS-PAGE. Nat. Protoc. 116-22. [DOI] [PubMed] [Google Scholar]
- 29.Schuijffel, D. F., P. C. van Empel, A. M. Pennings, J. P. van Putten, and P. J. Nuijten. 2005. Successful selection of cross-protective vaccine candidates for Ornithobacterium rhinotracheale infection. Infect. Immun. 736812-6821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Segers, P., W. Mannheim, M. Vancanneyt, K. De Brandt, K. H. Hinz, K. Kersters, and P. Vandamme. 1993. Riemerella anatipestifer gen. nov., comb. nov., the causative agent of septicemia anserum exsudativa, and its phylogenetic affiliation within the Flavobacterium-Cytophaga rRNA homology group. Int. J. Syst. Bacteriol. 43768-776. [DOI] [PubMed] [Google Scholar]
- 31.Shin, H., M. Mally, M. Kuhn, C. Paroz, and G. R. Cornelis. 2007. Escape from immune surveillance by Capnocytophaga canimorsus. J. Infect. Dis. 195375-386. [DOI] [PubMed] [Google Scholar]
- 32.Slaney, J. M., A. Gallagher, J. Aduse-Opoku, K. Pell, and M. A. Curtis. 2006. Mechanisms of resistance of Porphyromonas gingivalis to killing by serum complement. Infect. Immun. 745352-5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Socransky, S. S., and A. D. Haffajee. 2005. Periodontal microbial ecology. Periodontol. 2000 38135-187. [DOI] [PubMed] [Google Scholar]
- 34.Sory, M. P., and G. R. Cornelis. 1994. Translocation of a hybrid YopE-adenylate cyclase from Yersinia enterocolitica into HeLa cells. Mol. Microbiol. 14583-594. [DOI] [PubMed] [Google Scholar]
- 35.Subramaniam, S., B. Huang, H. Loh, J. Kwang, H. M. Tan, K. L. Chua, and J. Frey. 2000. Characterization of a predominant immunogenic outer membrane protein of Riemerella anatipestifer. Clin. Diagn. Lab. Immunol. 7168-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tierney, D. M., L. P. Strauss, and J. L. Sanchez. 2006. Capnocytophaga canimorsus mycotic abdominal aortic aneurysm: why the mailman is afraid of dogs. J. Clin. Microbiol. 44649-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsai, C. M., and C. E. Frasch. 1982. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal. Biochem. 119115-119. [DOI] [PubMed] [Google Scholar]
- 38.Tzeng, Y. L., A. K. Datta, C. A. Strole, M. A. Lobritz, R. W. Carlson, and D. S. Stephens. 2005. Translocation and surface expression of lipidated serogroup B capsular polysaccharide in Neisseria meningitidis. Infect. Immun. 731491-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Duijkeren, E., C. van Mourik, M. Broekhuizen, M. Leuven, W. Gaastra, and D. Houwers. 2006. First documented Capnocytophaga canimorsus infection in a species other than humans. Vet. Microbiol. 118148-150. [DOI] [PubMed] [Google Scholar]
- 40.Westphal, O., and K. Jann. 1965. Bacterial lipopolysaccharides. Extraction with phenol-water and further applications of the procedure. Methods Carbohydr. Chem. 583-91. [Google Scholar]
- 41.Westwell, A. J., K. Kerr, M. B. Spencer, and D. N. Hutchinson. 1989. DF-2 infection. BMJ 298116-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilson, M. E., R. Burstein, J. T. Jonak-Urbanczyk, and R. J. Genco. 1985. Sensitivity of Capnocytophaga species to bactericidal properties of human serum. Infect. Immun. 50123-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Woestyn, S., A. Allaoui, P. Wattiau, and G. R. Cornelis. 1994. YscN, the putative energizer of the Yersinia Yop secretion machinery. J. Bacteriol. 1761561-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]