Abstract
Lipopolysaccharide (LPS) is a major constituent of the outer membrane and an important virulence factor of Salmonella enterica subspecies 1 serovar Typhimurium (serovar Typhimurium). To evaluate the role of LPS in eliciting intestinal inflammation in streptomycin-treated mice, we constructed an O-antigen-deficient serovar Typhimurium strain through deletion of the wbaP gene. The resulting strain was highly susceptible to human complement activity and the antimicrobial peptide mimic polymyxin B. Furthermore, it showed a severe defect in motility and an attenuated phenotype in a competitive mouse infection experiment, where the ΔwbaP strain (SKI12) was directly compared to wild-type Salmonella. Nevertheless, the ΔwbaP strain (SKI12) efficiently invaded HeLa cells in vitro and elicited acute intestinal inflammation in streptomycin-pretreated mice. Our experiments prove that the presence of complete LPS is not essential for in vitro invasion or for triggering acute colitis.
Salmonella spp. are a common cause of bacterial food-borne infections. Diseases caused by Salmonella spp. range from gastrointestinal symptoms such as fever, diarrhea, abdominal pain, and nausea to severe systemic infections. Salmonella enterica subspecies 1 serovar Typhimurium (serovar Typhimurium) is one of the most frequent enteropathogens, causing large numbers of diarrheal infections worldwide by colonizing the gut and triggering mucosal inflammation (33). The type III secretion system 1 (TTSS-1) and TTSS-2 encoded on Salmonella pathogenicity island 1 (SPI1) and SPI2 on the Salmonella genome are employed by the pathogen for mediating bacterial entry into the gut mucosa (SPI1) as well as the intracellular survival followed by systemic spread of the bacteria (SPI2) (9). Acute enteric serovar Typhimurium infection and the mechanisms leading to intestinal inflammation can be analyzed using a well-defined mouse model for Salmonella colitis: streptomycin-pretreated, naïve mice develop a vigorous local inflammation of the large intestine upon intragastric infection with serovar Typhimurium (3).
Besides the SPI1- and SPI2-encoded TTSSs, serovar Typhimurium requires numerous additional virulence factors for colonizing the host, resisting host immune defense, and finally, triggering disease. One key virulence factor for serovar Typhimurium is lipopolysaccharide (LPS), a major surface component (42). It contributes to the stability of the outer membrane, serves as a permeability barrier, and protects the bacterium against environmental challenges (34). LPS is composed of three domains. The lipid A part, also known as endotoxin, anchors LPS molecules in the outer membrane with its fatty acid chains. It is connected through the inner core consisting of heptoses and Kdo (3-deoxy-d-manno-octulosonic acid), with the outer core containing hexoses and N-acetylhexoses. Linked to the last glucose of the outer core is the polymeric O-antigen region. This region is composed of 16 to >100 repeats of an oligosaccharide structure containing four to six monosaccharides (27).
The endotoxic properties of LPS are mediated by the lipid A moiety, which can be recognized by Toll-like receptor 4 and thus triggers an innate immune response (16, 32). The O antigen, in combination with the inner and outer cores, serves as protection against complement antimicrobial peptides, detergents, and certain antibiotics. Furthermore, the O-antigen region is a key determinant for recognition by the adaptive immune response (40).
A number of studies have established an important role for O-antigen side chains in Salmonella virulence. A signature-tagged mutagenesis screening by Morgan and coworkers proved that mutations in genes for enzymes involved in the biosynthesis of O-antigen side chains attenuated bacteria in their ability to colonize chick and calf intestines (25). Interestingly, a mutant in wbaP, the phosphogalactosyltransferase starting O-antigen biosynthesis, was able to colonize calves but showed an attenuated phenotype in chicks (25). Moreover, screening for Salmonella genes required for long-term systemic infection after intraperitoneal injection showed negative selection for mutants in O-antigen biosynthesis (21). Coinfection experiments by Nevola et al. show that mutants lacking O antigen are still able to colonize the murine intestine but are attenuated in competitive infection experiments (30). Furthermore, a recent in vitro study with Salmonella enterica serovar Typhi showed that O-antigen side chains are not necessary for adhesion to and invasion of epithelial cells. However, mutants lacking the complete outer core are severely attenuated (14). In general, the loss of core structures seems more detrimental than the loss of O-antigen side chains. However, it had remained unclear whether the O-antigen side chains are required for triggering intestinal inflammation.
We wanted to analyze the role of O-antigen side chains in a well-established mouse model for enteric infections (3) and in an in vitro cellular invasion assay (36). Thus, we deleted the gene encoding the phosphogalactosyltransferase WbaP. This enzyme adds phosphogalactose to undecaprenylphosphate, the first step in O-antigen side chain biosynthesis in the cytoplasm of serovar Typhimurium (35, 43, 44). Streptomycin-pretreated mice were orally infected with the wbaP mutant strain (SKI12), and in line with published work, we found that the ΔwbaP mutant strain (SKI12) was significantly attenuated in a competitive infection assay. In spite of this, the wbaP mutant alone was able to trigger acute colitis. This demonstrates that serovar Typhimurium permits substantial manipulation of the O-antigen structure without losing its ability to trigger mucosal inflammation.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
A summary of the bacterial strains used in this study is given in Table 1. Bacteria were grown in Luria-Bertani (LB) medium (10 g/liter Bacto tryptone, 5 g/liter Bacto yeast extract, 5 g/liter NaCl). LB agar plates were supplemented with 1.5% (wt/vol) agar. MacConkey agar plates were prepared according to the manufacturer's instructions (Difco). Antibiotics were used in final concentrations, as follows: ampicillin, 100 μg/ml; kanamycin (Kan), 50 μg/ml; chloramphenicol (Cam), 25 μg/ml; streptomycin (Sm), 50 μg/ml; and tetracycline (Tet), 10 μg/ml. For mouse infection experiments and the in vitro cellular invasion assay, bacteria were grown as described previously (3).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype(s) and/or phenotype(s)a | Source or reference(s) |
|---|---|---|
| Salmonella enterica serovar Typhimurium strains | ||
| SL1344 | Wild type; Strr | 15 |
| SB161 | SL1344 ΔinvG; Strr | 18 |
| SKI12 | SL1344 ΔwbaP; Strr | This study |
| SKI11 | SL1344 ΔwbaP::cat; Strr Camr | This study |
| SKI33 | SL1344 ΔwbaP::pKI9; Strr Tetr | This study |
| M939 | SL1344 ϕsopE::aph; Strr Kanr | 1 |
| M933 | SL1344 ΔinvG; sseD::aphT; fliGHI::Tn10; Strr Kanr Tetr | 38 |
| Escherichia coli strains | ||
| CC118 λpir | Δ(ara-leu) araD ΔlacX74 galE galK phoA20 thi-1 rpsE rpoB argE(Am) recA λpir | 13 |
| Sm10λpir | thi thr leu tonA lacY supE recA::RP4 2-Tc::Mu λpir; Kanr | 23 |
| DH5α | supE44 ΔlacU169 ϕ80dlacZΔM15 hsdR17 recA1 endA1 gyrA96 thi- 1 relA1 | 11 |
| Plasmids | ||
| pGEM-T Easy | bla | Promega |
| pSB377 | tetAB oriR6K | 24 |
| pKI9 | tetAB 3′manBwbaP oriR6K | This study |
| pM975 | bla PssaGgfpmut2 oripMB1 | 12, 36 |
| pKD3 | bla FRT cat FRT PS1 PS2 oriR6K | 7 |
| pKD46 | bla PBADgam bet exo pSC101 oriTS | 7 |
| pCP20 | bla cat cI857 PRflp pSC101 oriTS | 7 |
Str, streptomycin.
Construction of a wbaP deletion in serovar Typhimurium and its complementation.
Construction of the wbaP deletion mutant of serovar Typhimurium wild-type strain SL1344 was carried out as described previously (7). Briefly, primers RfbP H1P1 (CTTAATATGCCTATTTTATTTACATTATGCACGGTCAGAGGGTGAGGATTAAGTGTAGGCTGGAGCTGCTTC) and RfbP H2P2 (GAT TTTACGCAGGCTAAT.TTATACAATTATTATTCAGTACTTCTCGGTAA GCCATATGAATATCCTCCTTAGTTCCTATTCC), annealing to template DNA from plasmid pKD3, which carries a chloramphenicol resistance gene flanked by FLP recognition target (FRT) sites and 40 to 45 additional nucleotides corresponding to regions adjacent to the wbaP gene, were constructed. These sequences were employed to amplify a gene cassette for an in-frame deletion of wbaP, as described previously. The resulting chloramphenicol-resistant strain (wbaP::cat) was termed the serovar Typhimurium ΔwbaP::cat strain (SKI11) and verified by PCR. Removal of the chloramphenicol resistance cassette was achieved by using pCP20, and the resulting strain was termed the serovar Typhimurium ΔwbaP strain (SKI12). The presence of the chromosomal deletion was verified by PCR.
All cloning steps involving the suicide plasmid pSB377 for constructing plasmid pKI9 were carried out using Escherichia coli CC118λpir. All other cloning steps were carried out using E. coli DH5α. wbaP and the region 500 bp upstream belonging to manB were amplified by PCR (primers Fwd_wbaK_BamHI, CAGGATCCCGGAGTTATAGTCGTATTGTCGG, and Rev_wbaP_PstI, AACCTGCAGTTAATACGCACCATCTCGCCG). The PCR fragment was inserted into pGEM-T Easy (Promega) and moved into pSB377 via BamHI and PstI, yielding pKI9. pKI9 was introduced into the serovar Typhimurium ΔwbaP strain (SKI12) by conjugation, and clones carrying the complementing plasmid integrated in the chromosome were selected by plating on LB medium (Sm and Tet), yielding the serovar Typhimurium ΔwbaP::pKI9 strain (SKI33).
Immunoblot analysis and silver staining of glycoconjugates.
The equivalent of 2 OD600 units/ml (where OD600 is the optical density at 600 nm) of exponentially growing cultures from serovar Typhimurium wild-type strain SL1344, the serovar Typhimurium ΔwbaP strain (SKI12), or the serovar Typhimurium ΔwbaP::pKI9 strain (SKI33) was pelleted by centrifugation at 16,000 × g for 2 min, and the supernatant was discarded. Cells were resuspended in 100 μl Lämmli sample buffer (0.065 M Tris-HCl [pH 6.8], 2% [wt/vol] sodium dodecyl sulfate [SDS], 5% [vol/vol] β-mercaptoethanol, 10% [vol/vol] glycerol, 0.05% [wt/vol] bromophenol blue) and lysed for 5 min at 95°C. After being cooled to room temperature, proteinase K (Gibco/Life Technologies) was added (final concentration, 0.4 mg/ml), and samples were incubated for 1 h at 60°C before subjecting equal amounts to 12% SDS-polyacrylamide gel electrophoresis (SDS-PAGE). To detect serovar Typhimurium O antigen, Salmonella O antiserum (group B, factors 1, 4, 5, and 12; Difco) was used. Enterobacterial common antigen (ECA) was detected with the mouse monoclonal antibody 898 against ECA (17). Glycoconjugate bands were detected with goat anti-rabbit immunoglobulin G-horseradish peroxidase or goat anti-mouse immunoglobulin G-horseradish peroxidase conjugates (Santa Cruz) and ECL (Amersham), as recommended by the manufacturer. Silver staining of 12% SDS-PAGE gels was carried out as described previously (41).
Analysis of polymyxin B resistance.
The equivalent of 1 OD600 unit/ml of exponentially growing cultures from serovar Typhimurium wild-type strain SL1344 or the serovar Typhimurium ΔwbaP strain (SKI12) was spun down, resuspended in 150 μl cold sterile 1× phosphate-buffered saline (PBS), and diluted 5 × 106-fold before use. For the assay, 45 μl of the diluted cultures was mixed with 5 μl of polymyxin B (final concentration, 1 μg/ml; Sigma) or 5 μl PBS and incubated for 1 h at 37°C under slight agitation. After addition of 80 μl LB agar, bacteria were plated on LB agar plates containing streptomycin. The survival efficiency was calculated by dividing the number of CFU of the peptide-treated culture by the number of CFU of untreated culture and multiplying by 100. The assay was performed in triplicate for two independent experiments, and data are shown as means ± standard deviations.
Swimming motility assay.
Motility of bacteria was tested on soft agar plates (0.3% [wt/vol] agar, 5 g/liter NaCl, 10 g/liter Bacto tryptone). A total of 1 μl of overnight cultures of the serovar Typhimurium wild type (SL1344), serovar Typhimurium ΔwbaP strain (SKI12), serovar Typhimurium ΔwbaP::pKI9 strain (SKI33), or serovar Typhimurium fliGHI::Tn10 strain (M933) was spotted in the middle of plates, and motility was quantified by measuring the diameter of the halo visible after 4.75 h and 9.5 h of incubation at 37°C. Each experiment was carried out in triplicate on two different occasions, and throughout, data are shown as means ± standard deviations.
Analysis of serum resistance.
Bactericidal activity of the complement was tested essentially as described previously (4). In brief, the serovar Typhimurium ΔwbaP::cat strain (SKI11), M939, a kanamycin-resistant derivative of serovar Typhimurium wild-type strain SL1344 (aph integrated downstream of sopE), and cells from the serovar Typhimurium ΔwbaP::pKI9 strain (SKI33), taken from exponentially growing cultures, were mixed in equal amounts (3 × 108 CFU/ml for M393; 4 × 108 CFU/ml for SKI11 and SKI33) and diluted 5 × 104-fold before use in sterile 1× PBS. This diluted bacterial culture was mixed 1:1 with 20% human serum containing no antibodies against serovar Typhimurium LPS and incubated at 37°C with slight agitation. Aliquots were taken at 0 min, 15 min, and 30 min after mixing, and complement activity was quenched by adding brain heart infusion broth. The aliquots were kept on ice until plating on LB medium (Sm, Kan) selecting for the wild type, LB medium (Sm, Cam) selecting for wbaP::cat, and LB medium (Sm, Tet) to determine the number of ΔwbaP::pKI9 CFU. The same experiment was carried out using serum, where the complement was heat inactivated at 56°C for 30 min. Data are shown as the means of log CFU ± standard deviations.
In vitro invasion assay and automated imaging analysis.
Effects of deletion of O antigen on host cell invasion of bacteria into HeLa cells were tested as described previously (36). Approximately 1.2 × 103 HeLa cells per well were seeded in 96-well plates (half size, μClear; Greiner) and, after overnight growth, infected with serial dilutions of a 4-h subculture of serovar Typhimurium wild-type strain SL1344, the ΔinvG strain (SB161), or the ΔwbaP strain (SKI12) harboring plasmid pM975. Plasmid pM975 encodes green fluorescent protein (GFP) under the control of the ssaG promoter (SPI2), which is activated only by intracellular bacteria. After 26 min of infection, medium was replaced with Dulbecco modified Eagle medium containing gentamicin at 400 μg/ml to kill remaining bacteria that had not invaded the mammalian cells. The mammalian cells were then incubated further for 3 h 30 min at 37°C and subsequently fixed with 4% paraformaldehyde and 4% sucrose in PBS for 20 min. After being stained with DAPI (4′,6′-diamidino-2-phenylindole) at a final concentration of 10 μg/ml and 0.1% Triton X-100 for 7 min at room temperature, cells were washed twice with PBS and left in PBS containing 400 μg/ml gentamicin. Automated microscopy was performed using a cellWoRx microscope (API) with a 10× objective. Intracellular bacteria were quantified using SPI2-dependent GFP fluorescence, and approximately 10,000 cells per well were analyzed. Image analysis was carried out essentially as described previously (5, 36).
Mouse infection experiments.
Salmonella infections were performed in individually ventilated cages at the RCHCI, Zurich, as previously described (39). In brief, C57BL/6 mice (specific pathogen free; colony of the RCHCI, Zurich) were pretreated by gavaging with 20 mg of streptomycin. Twenty-four hours later, the mice were inoculated with 5 × 107 CFU of serovar Typhimurium strain or mixtures of strains, as indicated (coinfection experiment). Bacterial loads (CFU) in fresh fecal pellets and mesenteric lymph node (MLN), spleen, and cecal content were determined by plating on MacConkey agar plates (50 μg/ml streptomycin), as previously described (3). Samples of cecal tissue were cryoembedded, and inflammation was quantified using hematoxylin and eosin (H&E)-stained tissue sections, as described. Cryosections (5 μm, cross-sectional) were stained with H&E. Cecum pathology was evaluated by using a histopathological scoring scheme, as previously described (12, 39).
Animal experiments were approved by the Swiss authorities (license number 201/2007; Kantonales Veterinäramt, Zurich, Switzerland) and performed according to the legal requirements. For coinfection experiments, the competitive indices (CI) were determined according to the following formula: CI = (mutant output/wild-type output)/(mutant input/wild-type input).
Statistical analysis.
Statistical analysis was performed using the exact Mann-Whitney U test (Prism 4.0c). A P value of <0.05 (two tailed) was considered to be statistically significant. In mouse experiments, values were set to the minimal detectable value (10 CFU for MLNs, 20 CFU for the spleen) for samples harboring “no bacteria.”
RESULTS
Analysis of an O-antigen-negative serovar Typhimurium strain: the wbaP mutant (SKI12) is susceptible to complement-mediated killing and polymyxin B.
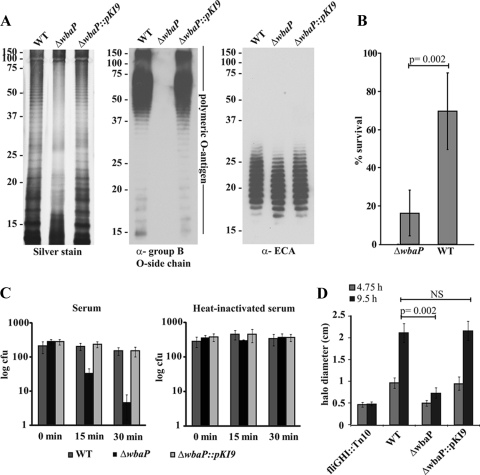
To study the role of O-antigen side chains, we deleted the gene encoding the phosphogalactosyltransferase WbaP in the serovar Typhimurium wild-type strain, as described previously (7). The glycoconjugates of the resulting strain were analyzed, and we verified the lack of O-antigen side chains by silver staining (41) and immunoblotting with Salmonella group B-specific anti-O antiserum (Fig. 1A, left and middle). This revealed the typical LPS ladder pattern of the polymeric O antigen for the wild type and the absence of this pattern in the ΔwbaP strain. O-antigen formation was restored by inserting the wbaP open reading frame (Fig. 1A, left and middle panels). The ΔwbaP strain (SKI12) and the complemented Δwba::pKI9 strain (SKI33) produce ECA in amounts similar to that of the wild-type strain (Fig. 1A, right).
FIG. 1.
The O-antigen-negative serovar Typhimurium ΔwbaP strain (SKI12) is susceptible to complement-mediated killing and polymyxin B. (A) Silver stain (left) and anti-Salmonella group B O-antigen immunoblot (middle) of SDS-PAGE of the serovar Typhimurium wild-type strain (WT), ΔwbaP, or ΔwbaP::pKI9 whole-cell extracts treated with proteinase K. This confirms the lack of polymeric O antigen in the ΔwbaP strain (SKI12), its restoration in the ΔwbaP::pKI9 strain (SKI33), and normal expression of ECA (right). α, anti. (B) Polymyxin B sensitivity was analyzed by incubating the serovar Typhimurium wild-type strain and the ΔwbaP strain (SKI12) for 1 h at 37°C in the presence (final concentration, 1 μg/ml) or absence of polymyxin B. The number of CFU was analyzed by plating, and the percentage of survival was calculated by dividing the number of CFU determined with and without the polymyxin B treatment. The assay was performed in triplicate on two independent occasions; data are shown as means ± standard deviations, and statistical significance was determined using the exact Mann-Whitney U test. (C) Complement-mediated killing of the kanamycin-resistant serovar Typhimurium wild-type strain, M939, the O-antigen-negative ΔwbaP::cat strain (SKI11), and the complemented ΔwbaP::pKI9 mutant (SKI33) was tested by incubating a 1:1:1 dilution mixture of wild-type, ΔwbaP, and ΔwbaP::pKI9 (SKI33) Salmonella strains for the indicated times with 20% human serum or 20% heat-inactivated human serum. Survival was analyzed by plating on differentiating media. (D) Motility assay of the serovar Typhimurium wild-type strain, the ΔwbaP strain (SKI12), and SKI33. Motility of the wild-type and ΔwbaP (SKI12) Salmonella strains was tested on soft agar plates by measuring the halo after 4.75 h and 9.5 h of incubation at 37°C. The assay was performed in triplicate on two independent occasions; data are shown as means ± standard deviations, and statistical significance was determined using the exact Mann-Whitney U test. NS, not statistically significant.
Since LPS is known to protect against host defenses, the resistance of the O-antigen-negative serovar Typhimurium ΔwbaP strain (SKI12) against complement-mediated killing (serum resistance) and polymyxin B was determined. To examine resistance against polymyxin B, the serovar Typhimurium wild type and the ΔwbaP strain (SKI12) were incubated for 1 h with polymyxin B or PBS. Polymyxin B mimics the action of antimicrobial peptides by increasing membrane permeability. In the serovar Typhimurium wild-type strain, 69% (±20%) of the cells survived the polymyxin B treatment, whereas only 16% (±11%) of the ΔwbaP bacteria were viable after the incubation phase (P = 0.002, exact Mann-Whitney U test) (Fig. 1B).
To test serum resistance, serovar Typhimurium wild-type strain M939, the serovar Typhimurium ΔwbaP::cat strain (SKI11), and the serovar Typhimurium ΔwbaP::pKI9 strain (SKI33) were mixed and incubated in 10% human serum lacking antibodies directed against serovar Typhimurium LPS (data not shown). Complement resistance was determined by plating bacteria on media distinguishing the wild-type strain and ΔwbaP strain (SKI12) after the indicated exposure times. Incubation in heat-inactivated human serum served as a negative control. As expected, the serovar Typhimurium wild-type strain was resistant to complement-mediated killing (Fig. 1C, left). In contrast, the ΔwbaP strain (SKI12) was more sensitive. After 15 min of incubation with 20% human serum, bacterial counts of the ΔwbaP strain (SKI12) were reduced by approximately eightfold compared to the counts achieved at the beginning of the incubation period. After 30 min of incubation, the counts for the ΔwbaP strain (SKI12) were sixty times less than those at the beginning of the incubation time. This phenotype could be complemented (the wbaP::pKI9 strain [SKI33]) (Fig. 1C, left). This verified the role of the O side chain in complement resistance.
The serovar Typhimurium ΔwbaP strain (SKI12) shows a severe defect in swimming motility.
Swimming motility is an important virulence factor of serovar Typhimurium. To test if motility is impaired in the O-antigen-negative serovar Typhimurium ΔwbaP strain (SKI12), bacteria were grown for the indicated times on motility agar. As shown in Fig. 1D, the ΔwbaP strain (SKI12) was significantly less motile than the serovar Typhimurium wild-type strain (P = 0.002, exact Mann-Whitney U test) but still showed a higher motility than the nonmotile fliGHI::Tn10 control strain. Motility could be restored to wild-type levels by complementing the wbaP deletion (the wbaP::pKI9 strain [SKI33]) (Fig. 1D).
The serovar Typhimurium ΔwbaP strain (SKI12) invades HeLa cells in vitro.
The TTSS-1 (inv-spa) is a major Salmonella virulence factor required for the induction of membrane ruffling and invasion of nonphagocytic cells. In Shigella flexneri and Pseudomonas aeruginosa, the activity of TTSS is known to be modulated by the length of the O side chain of LPS (2, 45). Therefore, we tested the functionality of TTSS-1 in the O-antigen-deficient ΔwbaP strain (SKI12) using an in vitro invasion assay. HeLa cells were infected at the indicated multiplicities of infection (MOIs) with the serovar Typhimurium wild-type strain, the ΔwbaP strain (SKI12), and the TTSS-1-deficient ΔinvG strain. The bacteria carried plasmid pM975, which leads to ssaG-dependent expression of GFP in the bacterial cytoplasm if the bacteria are inside the HeLa cells (12, 36). The invasion efficiency was determined by automated microscopy and image analysis (see Materials and Methods). The wild-type strain of serovar Typhimurium and the ΔwbaP mutant (SKI12) efficiently invaded HeLa cells (Fig. 2). Considering the role of motility in host cell invasion (20) and the motility defect of the ΔwbaP strain (Fig. 1D), the invasiveness of this mutant was unexpected. We speculate that an enhanced TTSS-1 activity may compensate for the reduced motility of the ΔwbaP strain (see Discussion). In contrast, the ΔinvG mutant was noninvasive (Fig. 2). The number of HeLa cells present per well did not differ between the different experimental groups, indicating that Salmonella infection did not lead to cell death and detachment at the tested MOIs (data not shown). Taken together, our in vitro data show that the serovar Typhimurium ΔwbaP strain (SKI12), although significantly less motile than the wild-type strain, is invasive in a cellular invasion assay.
FIG. 2.
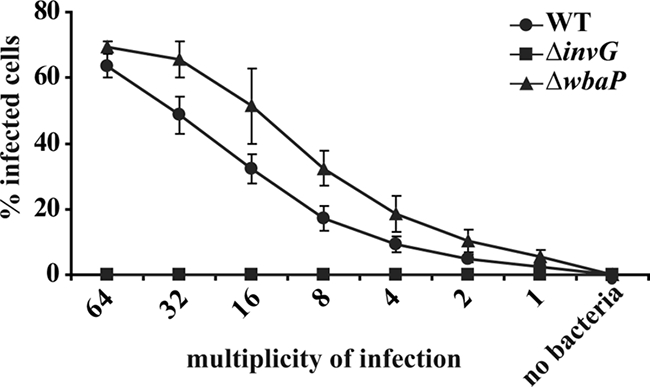
The serovar Typhimurium ΔwbaP strain (SKI12) can invade HeLa cells. HeLa cells were infected for 26 min with the wild type (WT), the invasion-disabled ΔinvG mutant (negative control), or the isogenic O-antigen-deficient ΔwbaP mutant at the indicated MOIs. The invasion efficiency was analyzed by an automated imaging assay (see Materials and Methods). The assay was performed in quadruplicate, and data are presented as medians ± standard deviation. Serovar Typhimurium wild type, filled circles; the ΔinvG mutant, filled squares; the ΔwbaP mutant, filled triangles.
The serovar Typhimurium ΔwbaP strain (SKI12) can induce colitis at day 2 p.i. in streptomycin-treated mice.
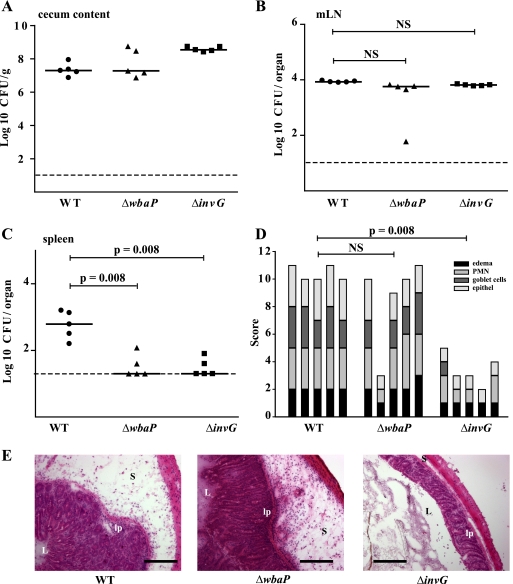
Having found that the ΔwbaP strain (SKI12) is as potent as the wild-type strain in terms of epithelial invasion efficacy in vitro, we investigated its capacity to induce colitis. Three groups of five streptomycin-pretreated C57BL/6 mice were infected intragastrically with 5 × 107 CFU of the serovar Typhimurium wild type, the ΔwbaP strain (SKI12), or a ΔinvG strain. At day 2 postinfection (p.i.), the animals were sacrificed and analyzed for colonization of the cecal lumen, the spleen, and the MLNs by serovar Typhimurium as well as for pathological changes in the cecal tissue. Colonization levels in the cecal lumen and the MLNs by all three strains were not significantly different (Fig. 3A and B). The ΔwbaP strain (SKI12) could be detected at slightly lower levels in the spleen (P ≥ 0.05) (Fig. 3C). This could be due to an impairment of intracellular replication or to complement-mediated lysis of the ΔwbaP strain (SKI12) at systemic sites. Histopathological analysis of the cecum revealed no significant difference in the total cecal pathological score between the ΔwbaP strain (SKI12) and the wild-type strain (Fig. 3D), whereas the SPI1-deficient ΔinvG control strain showed reduced or no cecal inflammation (Fig. 3D and E), as previously published (3). This indicated that the ΔwbaP strain (SKI12) was able to induce a severe cecal inflammation at day 2 p.i. Surprisingly, the lack of the O-antigen side chains did not impair MLN colonization and initiation of colitis in streptomycin-pretreated mice.
FIG. 3.
The serovar Typhimurium ΔwbaP strain (SKI12) is a potent inducer of colitis at day 2 p.i. Three groups of streptomycin-treated wild-type C57BL/6 mice (n = 5 per group) were infected for 2 days with the serovar Typhimurium wild-type strain (WT), ΔwbaP strain (SKI12), or ΔinvG strain (total of 5 × 107 CFU intragastrically). Serovar Typhimurium colonization of cecum, MLNs, and spleen and cecal pathology were examined at day 2 p.i. (A to C) The number of serovar Typhimurium CFU in different organs (cecal content [A], MLNs [B], spleen [C]). Horizontal bars, median; dashed line, minimal detectable level; NS, not statistically significant (P ≥ 0.05). (D) Cecal pathology and its quantification at day 2 p.i. PMN, polymorphonuclear leukocytes. (E) Thin sections (5 μm) of cryoembedded cecal tissues were stained with H&E, as described in Materials and Methods. L, intestinal lumen; lp, lamina propria; S, submucosa. Scale bars, 200 μm.
The serovar Typhimurium ΔwbaP strain (SKI12) is attenuated in a competitive infection assay.
Since the ΔwbaP strain (SKI12) was impaired in motility and resistance to serum and polymyxin B but was still able to induce severe cecal pathologies in streptomycin-treated mice, we further determined the role of LPS in gut colonization. A coinfection experiment with the serovar Typhimurium wild type (M939) and the ΔwbaP strain (SKI11) was performed. Five streptomycin-treated mice were infected with a 1:2 dilution mixture (total of 5 × 107 CFU) intragastrically of the ΔwbaP strain (SKI11) and wild-type strain. The ratio of the two strains (CI; see Materials and Methods) was determined in the feces at days 1 to 4 p.i. A decrease in the ΔwbaP strain counts in relation to those of the wild type was detected (1 log scale per day). This was in line with earlier data (25, 30) and showed that the ΔwbaP strain (SKI11) indeed had a severe competitive defect in comparison to the wild-type serovar Typhimurium strain in the intestinal tract (P > 0.05) (Fig. 4A). Thus, O-antigen side chains clearly are important for competitive growth under the conditions prevailing in the inflamed mouse intestine. Moreover, the CI of the two strains at systemic sites (MLNs, liver, spleen) at day 4 p.i. also demonstrated a significant competitive defect of the serovar Typhimurium ΔwbaP strain (SKI12). Nevertheless, the defect was less pronounced than in the intestine (Fig. 4B). In conclusion, the coinfection experiment revealed that the lack of O antigen decreases the competitive fitness of the serovar Typhimurium strain in the intestinal lumen. Despite this defect, the serovar Typhimurium ΔwbaP strain was still proficient in the induction of colitis (Fig. 3).
FIG. 4.
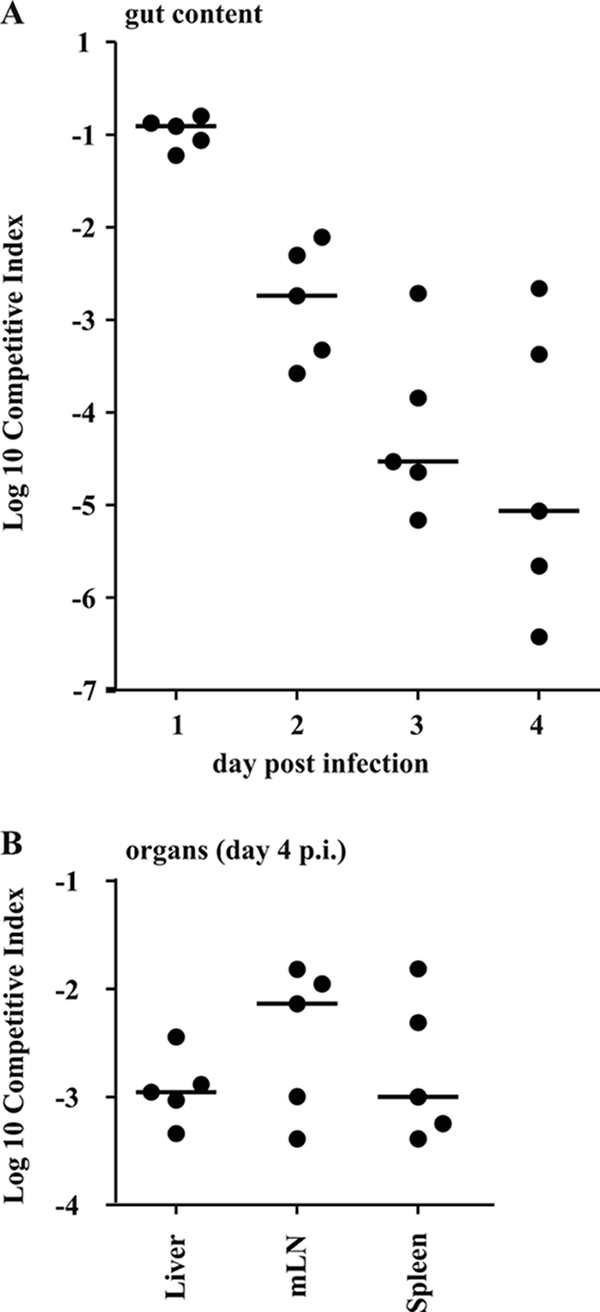
The serovar Typhimurium ΔwbaP strain (SKI12) is attenuated in competitive infection. (A) Streptomycin-pretreated wild-type C57BL/6 mice (n = 5) were infected for 4 days with a 1:2 dilution mixture (total of 5 × 107 CFU intragastrically) of the serovar Typhimurium ΔwbaP strain and wild-type strain SL1344. CI [(mutant output/wild-type output)/(mutant input/wild-type input)] of the serovar Typhimurium ΔwbaP strain (SKI12) and wild-type strain were determined at days 1 to 3 p.i. in feces and at day 4 p.i. in the cecal content (see Materials and Methods) (B) CI in the MLNs, spleens, and livers at day 4 p.i.
DISCUSSION
LPS is involved in cell adhesion, organ colonization, and resistance against antimicrobial effector molecules in many different pathogens, including Brucella melitensis 16M (10), Salmonella enterica serovar Typhi (26), Vibrio cholerae O139 (29), and serovar Typhimurium (6, 22). Although many studies investigated the role of LPS in systemic serovar Typhimurium virulence (22, 30), little is known about the requirement of O-antigen side chains in enteric infection, e.g., induction of colitis. The genomic deletion of the gene coding for the phosphogalactosyltransferase WbaP generated a strain completely devoid of O-antigen side chains. Since ECA, another main surface component of serovar Typhimurium, acts together with LPS as a barrier against the host immune system, it was important that ECA was still present in the generated wbaP mutant strain. Nevertheless, ECA alone does not seem to provide sufficient protection against the innate immune response, since our data show a high susceptibility of the wbaP mutant strain (SKI12) to human complement and the antimicrobial peptide mimic polymyxin B. These findings are supported by data from Salmonella enterica serovar Typhi, showing a reduced serum resistance of a ΔwbaP mutant (SKI12) (14), and by studies with a serovar Typhimurium ΔwaaL mutant strain without O-antigen side chains, resulting in a higher susceptibility to polymyxin B (28). An increased accessibility to the lipid A core, the peptidoglycan layer, and the outer membrane of the serovar Typhimurium ΔwbaP strain (SKI12) for membrane-damaging agents could be an explanation for this result.
Additionally, the ΔwbaP mutant (SKI12) showed a severe defect in motility. There are several other reports connecting LPS mutations with a loss in motility: in serovar Typhimurium, addition of a monoclonal anti-LPS antibody to a diluted bacterial culture led to instant paralysis of the bacteria (8), and in a Pseudomonas aeruginosa ΔwaaL strain, a severe impairment in motility was detected (2). Also, an Escherichia coli mutant lacking O antigen was reported to show a nonmotile phenotype (37).
Studies by Kihlström and Edebo have shown that in vitro invasion into nonphagocytic cells does not necessarily rely on the presence of intact LPS (19). They demonstrated that the adherence of a rough serovar Typhimurium mutant lacking O antigen is not affected (19). The same is true for in vitro invasion studies, in which S. enterica serovar Typhi O-antigen mutants did not show an attenuated phenotype (14). A key mediator of bacterial entry into host cells is the SPI1-encoded TTSS (3, 9, 12). TTSS activity and the length of O-antigen side chains seem to be linked in pathogenic bacteria: in Shigella flexneri, glucosylation of O antigen resulting in a shorter polymeric O-antigen region enhanced TTSS function (45). In P. aeruginosa, increased virulence could be detected in an O-antigen mutant that induced significantly increased lung damage compared to that of the wild type (2). Phospholipase A expression and its secretion into the medium by a TTSS were increased in an O-antigen-negative strain of Yersinia enterocolitica (4). These data are in line with the current model, suggesting that the tip of the TTSS must extend at least to the surface of the O-antigen layer in order to efficiently inject TTSS effectors into the host cells. This suggests that shortening the LPS might enhance TTSS function in serovar Typhimurium. Overall, the ΔwbaP mutant (SKI12) of the serovar Typhimurium strain was slightly more invasive than the isogenic wild-type strain. This was surprising, as motility, which is reduced in this strain, is known to be required for efficient host cell invasion (20). This suggests that the enhanced SPI1 TTSS function may well compensate for the loss of motility of the ΔwbaP mutant (SKI12).
Attenuation in intestinal colonization of the ΔwbaP strain (SKI12) in competitive infection with the wild type was expected, as it has been shown that functional motility and chemotaxis are crucial for serovar Typhimurium in the inflamed intestine (38). Additionally, the reduced resistance of the ΔwbaP mutant (SKI12) to antimicrobial peptides might explain the reduced competitive fitness.
In spite of this attenuation, the ΔwbaP strain (SKI12) was still capable of triggering intestinal inflammation. The SPI1-encoded TTSS is a key virulence factor for triggering colitis (3, 12). It has been shown in cell culture experiments that the serovar Typhimurium strain invades epithelial cells of the cecal mucosa via their SPI1 TTSS. This process results in proinflammatory cytokine induction and in a pronounced inflammatory response (1). In contrast to the in vitro conditions of the cell culture invasion assay, the serovar Typhimurium strain colonizing the guts of streptomycin-treated mice is confronted with a pronounced innate immune defense, comprising a whole arsenal of antimicrobial peptides, lectins, and defensins (31). Strikingly, our data show that an induction of gut inflammation does not necessitate the presence of O-antigen side chains. Although the ΔwbaP mutant (SKI12) is severely attenuated in intestinal colonization in competitive infections and is more susceptible to complement and polymyxin B, it can still elicit colitis.
In summary, we found that the deletion of O-antigen side chains in serovar Typhimurium resulted in a strain competent in triggering gut inflammation. This robust expression of virulence factors by serovar Typhimurium might be the evolutionary prerequisite for the impressive diversification in O-antigen side chain structures without simultaneous loss of pathogenicity.
Acknowledgments
We are grateful to members of the Hardt and the Aebi labs for fruitful discussions, to Manja Barthel and the members of the RCHCI team, and in particular to Susanne Freedrich, Jörg Fehr, and Thomas C. Weber for excellent technical support.
This work was supported by the Swiss National Science Foundation (grant 3100AQ-105541 to M. Aebi and grant 310000-113623/1 to W.-D. Hardt), the European Union (SavinMucoPath grant 032296 to W.-D. Hardt), and ETH, Zurich. K. Endt and K. Ilg are members of Ph.D. programs at the Life Science Zurich Graduate School.
We have no competing financial interests.
Editor: A. J. Bäumler
Footnotes
Published ahead of print on 13 April 2009.
REFERENCES
- 1.Ackermann, M., B. Stecher, N. E. Freed, P. Songhet, W. D. Hardt, and M. Doebeli. 2008. Self-destructive cooperation mediated by phenotypic noise. Nature 454987-990. [DOI] [PubMed] [Google Scholar]
- 2.Augustin, D. K., Y. Song, M. S. Baek, Y. Sawa, G. Singh, B. Taylor, A. Rubio-Mills, J. L. Flanagan, J. P. Wiener-Kronish, and S. V. Lynch. 2007. Presence or absence of lipopolysaccharide O antigens affects type III secretion by Pseudomonas aeruginosa. J. Bacteriol. 1892203-2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barthel, M., S. Hapfelmeier, L. Quintanilla-Martinez, M. Kremer, M. Rohde, M. Hogardt, K. Pfeffer, H. Russmann, and W. D. Hardt. 2003. Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect. Immun. 712839-2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bengoechea, J. A., H. Najdenski, and M. Skurnik. 2004. Lipopolysaccharide O antigen status of Yersinia enterocolitica O:8 is essential for virulence and absence of O antigen affects the expression of other Yersinia virulence factors. Mol. Microbiol. 52451-469. [DOI] [PubMed] [Google Scholar]
- 5.Carpenter, A. E., T. R. Jones, M. R. Lamprecht, C. Clarke, I. H. Kang, O. Friman, D. A. Guertin, J. H. Chang, R. A. Lindquist, J. Moffat, P. Golland, and D. M. Sabatini. 2006. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 7R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Craven, S. E. 1994. Altered colonizing ability for the ceca of broiler chicks by lipopolysaccharide-deficient mutants of Salmonella Typhimurium. Avian Dis. 38401-408. [PubMed] [Google Scholar]
- 7.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 976640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Forbes, S. J., M. Eschmann, and N. J. Mantis. 2008. Inhibition of Salmonella enterica serovar Typhimurium motility and entry into epithelial cells by a protective antilipopolysaccharide monoclonal immunoglobulin A antibody. Infect. Immun. 764137-4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galyov, E. E., M. W. Wood, R. Rosqvist, P. B. Mullan, P. R. Watson, S. Hedges, and T. S. Wallis. 1997. A secreted effector protein of Salmonella dublin is translocated into eukaryotic cells and mediates inflammation and fluid secretion in infected ileal mucosa. Mol. Microbiol. 25903-912. [DOI] [PubMed] [Google Scholar]
- 10.Godfroid, F., B. Taminiau, I. Danese, P. Denoel, A. Tibor, V. Weynants, A. Cloeckaert, J. Godfroid, and J. J. Letesson. 1998. Identification of the perosamine synthetase gene of Brucella melitensis 16M and involvement of lipopolysaccharide O side chain in Brucella survival in mice and in macrophages. Infect. Immun. 665485-5493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166557-580. [DOI] [PubMed] [Google Scholar]
- 12.Hapfelmeier, S., B. Stecher, M. Barthel, M. Kremer, A. J. Muller, M. Heikenwalder, T. Stallmach, M. Hensel, K. Pfeffer, S. Akira, and W. D. Hardt. 2005. The Salmonella pathogenicity island (SPI)-2 and SPI-1 type III secretion systems allow Salmonella serovar typhimurium to trigger colitis via MyD88-dependent and MyD88-independent mechanisms. J. Immunol. 1741675-1685. [DOI] [PubMed] [Google Scholar]
- 13.Herrero, M., V. de Lorenzo, and K. N. Timmis. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 1726557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoare, A., M. Bittner, J. Carter, S. Alvarez, M. Zaldivar, D. Bravo, M. A. Valvano, and I. Contreras. 2006. The outer core lipopolysaccharide of Salmonella enterica serovar Typhi is required for bacterial entry into epithelial cells. Infect. Immun. 741555-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoiseth, S. K., and B. A. Stocker. 1981. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature 291238-239. [DOI] [PubMed] [Google Scholar]
- 16.Hoshino, K., O. Takeuchi, T. Kawai, H. Sanjo, T. Ogawa, Y. Takeda, K. Takeda, and S. Akira. 1999. Cutting edge: Toll-like receptor 4 (TLR4)-deficient mice are hyporesponsive to lipopolysaccharide: evidence for TLR4 as the Lps gene product. J. Immunol. 1623749-3752. [PubMed] [Google Scholar]
- 17.Husmann, M., J. Roth, E. A. Kabat, C. Weisgerber, M. Frosch, and D. Bitter-Suermann. 1990. Immunohistochemical localization of polysialic acid in tissue sections: differential binding to polynucleotides and DNA of a murine IgG and a human IgM monoclonal antibody. J. Histochem. Cytochem. 38209-215. [DOI] [PubMed] [Google Scholar]
- 18.Kaniga, K., J. C. Bossio, and J. E. Galan. 1994. The Salmonella typhimurium invasion genes invF and invG encode homologues of the AraC and PulD family of proteins. Mol. Microbiol. 13555-568. [DOI] [PubMed] [Google Scholar]
- 19.Kihlström, E., and L. Edebo. 1976. Association of viable and inactivated Salmonella typhimurium 395 MS and MR 10 with HeLa cells. Infect. Immun. 14851-857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.La Ragione, R. M., W. A. Cooley, P. Velge, M. A. Jepson, and M. J. Woodward. 2003. Membrane ruffling and invasion of human and avian cell lines is reduced for aflagellate mutants of Salmonella enterica serotype Enteritidis. Int. J. Med. Microbiol. 293261-272. [DOI] [PubMed] [Google Scholar]
- 21.Lawley, T. D., K. Chan, L. J. Thompson, C. C. Kim, G. R. Govoni, and D. M. Monack. 2006. Genome-wide screen for Salmonella genes required for long-term systemic infection of the mouse. PLoS Pathog. 2e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Licht, T. R., K. A. Krogfelt, P. S. Cohen, L. K. Poulsen, J. Urbance, and S. Molin. 1996. Role of lipopolysaccharide in colonization of the mouse intestine by Salmonella typhimurium studied by in situ hybridization. Infect. Immun. 643811-3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 1702575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mirold, S., W. Rabsch, M. Rohde, S. Stender, H. Tschape, H. Russmann, E. Igwe, and W. D. Hardt. 1999. Isolation of a temperate bacteriophage encoding the type III effector protein SopE from an epidemic Salmonella typhimurium strain. Proc. Natl. Acad. Sci. USA 969845-9850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morgan, E., J. D. Campbell, S. C. Rowe, J. Bispham, M. P. Stevens, A. J. Bowen, P. A. Barrow, D. J. Maskell, and T. S. Wallis. 2004. Identification of host-specific colonization factors of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 54994-1010. [DOI] [PubMed] [Google Scholar]
- 26.Mroczenski-Wildey, M. J., J. L. Di Fabio, and F. C. Cabello. 1989. Invasion and lysis of HeLa cell monolayers by Salmonella typhi: the role of lipopolysaccharide. Microb. Pathog. 6143-152. [DOI] [PubMed] [Google Scholar]
- 27.Murray, G. L., S. R. Attridge, and R. Morona. 2003. Regulation of Salmonella typhimurium lipopolysaccharide O antigen chain length is required for virulence; identification of FepE as a second Wzz. Mol. Microbiol. 471395-1406. [DOI] [PubMed] [Google Scholar]
- 28.Nagy, G., V. Danino, U. Dobrindt, M. Pallen, R. Chaudhuri, L. Emody, J. C. Hinton, and J. Hacker. 2006. Down-regulation of key virulence factors makes the Salmonella enterica serovar Typhimurium rfaH mutant a promising live-attenuated vaccine candidate. Infect. Immun. 745914-5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nesper, J., S. Schild, C. M. Lauriano, A. Kraiss, K. E. Klose, and J. Reidl. 2002. Role of Vibrio cholerae O139 surface polysaccharides in intestinal colonization. Infect. Immun. 705990-5996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nevola, J. J., B. A. Stocker, D. C. Laux, and P. S. Cohen. 1985. Colonization of the mouse intestine by an avirulent Salmonella typhimurium strain and its lipopolysaccharide-defective mutants. Infect. Immun. 50152-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ouellette, A. J. 2004. Defensin-mediated innate immunity in the small intestine. Best Pract. Res. Clin. Gastroenterol. 18405-419. [DOI] [PubMed] [Google Scholar]
- 32.Poltorak, A., X. He, I. Smirnova, M. Y. Liu, C. Van Huffel, X. Du, D. Birdwell, E. Alejos, M. Silva, C. Galanos, M. Freudenberg, P. Ricciardi-Castagnoli, B. Layton, and B. Beutler. 1998. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science 2822085-2088. [DOI] [PubMed] [Google Scholar]
- 33.Rabsch, W., H. Tschape, and A. J. Baumler. 2001. Non-typhoidal salmonellosis: emerging problems. Microbes Infect. 3237-247. [DOI] [PubMed] [Google Scholar]
- 34.Raetz, C. R., and C. Whitfield. 2002. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71635-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saldias, M. S., K. Patel, C. L. Marolda, M. Bittner, I. Contreras, and M. A. Valvano. 2008. Distinct functional domains of the Salmonella enterica WbaP transferase that is involved in the initiation reaction for synthesis of the O antigen subunit. Microbiology 154440-453. [DOI] [PubMed] [Google Scholar]
- 36.Schlumberger, M. C., R. Kappeli, M. Wetter, A. J. Muller, B. Misselwitz, S. Dilling, M. Kremer, and W. D. Hardt. 2007. Two newly identified SipA domains (F1, F2) steer effector protein localization and contribute to Salmonella host cell manipulation. Mol. Microbiol. 65741-760. [DOI] [PubMed] [Google Scholar]
- 37.Sheng, H., J. Y. Lim, M. K. Watkins, S. A. Minnich, and C. J. Hovde. 2008. Characterization of an Escherichia coli O157:H7 O-antigen deletion mutant and effect of the deletion on bacterial persistence in the mouse intestine and colonization at the bovine terminal rectal mucosa. Appl. Environ. Microbiol. 745015-5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stecher, B., M. Barthel, M. C. Schlumberger, L. Haberli, W. Rabsch, M. Kremer, and W. D. Hardt. 2008. Motility allows S. Typhimurium to benefit from the mucosal defence. Cell. Microbiol. 101166-1180. [DOI] [PubMed] [Google Scholar]
- 39.Stecher, B., S. Hapfelmeier, C. Muller, M. Kremer, T. Stallmach, and W. D. Hardt. 2004. Flagella and chemotaxis are required for efficient induction of Salmonella enterica serovar Typhimurium colitis in streptomycin-pretreated mice. Infect. Immun. 724138-4150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tough, D. F., S. Sun, and J. Sprent. 1997. T cell stimulation in vivo by lipopolysaccharide (LPS). J. Exp. Med. 1852089-2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsai, C. M., and C. E. Frasch. 1982. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal. Biochem. 119115-119. [DOI] [PubMed] [Google Scholar]
- 42.Valvano, M. A. 2003. Export of O-specific lipopolysaccharide. Front. Biosci. 8s452-s471. [DOI] [PubMed] [Google Scholar]
- 43.Wang, L., D. Liu, and P. R. Reeves. 1996. C-terminal half of Salmonella enterica WbaP (RfbP) is the galactosyl-1-phosphate transferase domain catalyzing the first step of O-antigen synthesis. J. Bacteriol. 1782598-2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang, L., and P. R. Reeves. 1994. Involvement of the galactosyl-1-phosphate transferase encoded by the Salmonella enterica rfbP gene in O-antigen subunit processing. J. Bacteriol. 1764348-4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.West, N. P., P. Sansonetti, J. Mounier, R. M. Exley, C. Parsot, S. Guadagnini, M. C. Prevost, A. Prochnicka-Chalufour, M. Delepierre, M. Tanguy, and C. M. Tang. 2005. Optimization of virulence functions through glucosylation of Shigella LPS. Science 3071313-1317. [DOI] [PubMed] [Google Scholar]