Abstract
Heterogeneous nuclear ribonucleoprotein (hnRNP) A1 is involved in pre-mRNA splicing in the nucleus and translational regulation in the cytoplasm. The cytoplasmic redistribution of hnRNP A1 is a regulated process during viral infection and cellular stress. Here we demonstrate that hnRNP A1 not only is an internal ribosome entry site (IRES) trans-acting factor that binds specifically to the 5′ untranslated region (UTR) of enterovirus 71 (EV71) and regulates IRES-dependent translation but also binds to the 5′ UTR of Sindbis virus (SV) and facilitates its translation. The cytoplasmic relocalization of hnRNP A1 in EV71-infected cells leads to the enhancement of EV71 IRES-mediated translation, and its function can be substituted by hnRNP A2, whereas the cytoplasmic relocalization of hnRNP A1 following SV infection enhances the SV translation, but this function cannot be replaced by hnRNP A2. Our study provides the first direct evidence that the cytoplasmic relocalization of hnRNP A1 controls not only the IRES-dependent but also non-IRES-dependent translation initiations of RNA viruses.
hnRNP A1, an RNA binding protein that shuttles between the nucleus and the cytoplasm, belongs to a large group of RNA binding proteins (hnRNPs) that are classified into several families and subfamilies based on conserved structural and functional motifs (13, 25). Several hnRNPs, such as hnRNPs A1, C1/C2, E1/E2, I (polypyrimidine tract binding protein [PTB]), and L, are involved in the translational control of mRNAs containing internal ribosome entry sites (IRESs) through a cap-independent mechanism. These hnRNP proteins constitute IRES trans-acting factors (ITAFs) that modulate the activity of IRES sequences present in the 5′ untranslated region (UTR) of viral or cellular mRNAs (7, 29, 44).
hnRNP A1 is involved in several RNA metabolic processes such as pre-mRNA splicing and trafficking (13, 50). The hnRNP A1 protein is composed of 320 amino acids and contains two RNA binding domains and a glycine-rich domain that is responsible for protein-protein interactions. The signal that mediates shuttling between the nucleus and cytoplasm has been identified as the C-terminal 38 amino acids termed M9 (43, 49). Cytoplasmic hnRNP A1 and nuclear hnRNP A1 have different RNA binding profiles. Cytoplasmic hnRNP A1 has a high affinity for AU-rich elements (18, 20), whereas nuclear hnRNP A1 has a high affinity for a polypyrimidine stretch bordered by AG at the 3′ ends of introns (34, 46). Interestingly, hnRNP A1 has been shown to be involved in the replication of many viruses such as mouse hepatitis virus (MHV), hepatitis C virus (HCV), dengue virus, and human papillomavirus type 16 through an interaction with the transcription-regulatory region of viral mRNA (9, 23, 36, 39, 52). hnRNP A1 has also been reported to interact with the human cytomegalovirus immediate-early gene 2 protein, which plays an important role in the regulation of virus replication (48). Recently, hnRNP A1 was reported to relocalize from the nucleus to the cytoplasm after vesicular stomatitis virus infection and to promote vesicular stomatitis virus-induced apoptosis (38).
Enterovirus 71 (EV71), a member of the family Picornavirus, genus Enterovirus, is the major causative agent of hand-foot-and-mouth disease. EV71-infected children can develop severe neurological complications that lead to rapid clinical deterioration and death (21, 35). Enteroviruses all contain a positive-strand RNA genome of approximately 7,400 nucleotides (nt). After infection of the host cell, the genome is translated in a cap-independent manner into a single polyprotein, which is subsequently processed by virus-encoded proteases into the structural capsid proteins and the nonstructural proteins; the latter are involved mainly in the replication of viral RNA. The coding region of EV71 RNA is flanked by the 5′ UTR and 3′ UTR. The 5′ UTR of EV71 RNA contains two secondary structures, a cloverleaf structure involved in viral RNA replication and an IRES, which directs the initiation of translation in a cap-independent manner (3, 51). IRES-dependent translation requires both canonical translation initiation factors and ITAFs. A number of ITAFs have been identified, which interact with the picornavirus IRES and facilitate the initiation of translation: these include PTB, human lupus autoantigen (La), poly(rC) binding protein 2, hnRNP E2, hnRNP C, hnRNP L, and upstream of N-ras (unr) (2, 5, 10, 22, 24).
Given the small size of the EV71 genome, the virus has to make use of numerous proteins or other molecular machinery in the host cells to complete its infectious cycle. To identify cellular factors involved in EV71 replication, we searched for cellular proteins that interact with the 5′ UTR of EV71 genomic RNA. Streptavidin beads were utilized to capture cellular proteins bound to the biotinylated EV71 5′ UTR, and by means of matrix-assisted laser desorption ionization-time-of-flight analysis, 15 proteins were found. Four of these, PTB, unr, and poly(rC) binding proteins 1 and 2, were previously shown to interact with the picornavirus 5′ UTR. Among the 11 novel proteins, we chose to study the interaction of hnRNP A1 with the EV71 5′ UTR further (32).
In this report, we demonstrate that hnRNP A1 interacts with the EV71 5′ UTR, specifically with stem-loops II and VI of the EV71 5′ UTR. However, the knockdown of hnRNP A1 by short interfering RNA (siRNA) has no effect on viral replication. The knockdown of both hnRNPs A1 and A2 reduced viral RNA synthesis and lowered the virus yield.
hnRNP A1 is known to be involved in the IRES-dependent translation of several viral and cellular mRNAs. To see if hnRNP A1 is also involved in the translation of mRNA that does not contain an IRES at the 5′ UTR, we examined its role in the replication of Sindbis virus (SV), as the 5′ UTR of this virus does not have an IRES.
SV is the prototype virus of the family Togaviridae, genus Alphavirus. Its positive-strand RNA genome, which is approximately 11,700 nt in length, has a 5′ cap and is polyadenylated at its 3′ terminus. Following entry into the host cell, the genome RNA is translated into two polyproteins, P123 and P1234; these are processed by a viral protease, giving rise first to several intermediate cleavage products and then to the four viral nonstructural proteins, nsP1, nsP2, nsP3, and nsP4. A complex of nsP4, the viral RNA-dependent RNA polymerase, and uncleaved P123 uses the genomic RNA as a template to synthesize a negative-strand copy of the genome (17, 27, 45). The negative-strand RNA is then used as a template by a complex of the four processed nsPs, i.e., nsP1, nsP2, nsP3, and nsp4, to make two positive-strand RNAs, the genomic (G) RNA and the subgenomic (SG) RNA (approximately 4,100 nt in length), the sequence of which is identical to the 4,100 nt at the 3′ end of the G RNA. The SG RNA serves as the message for the three structural proteins of the virus, the capsid protein, C, and the two envelope proteins, E1 and E2. Like the G RNA, the SG RNA is capped at its 5′ end and polyadenylated at its 3′ end. Translation of the subgenomic RNA gives rise to the structural proteins, which assemble with the genomic RNA to form progeny viruses.
While the viral components of the SV replicase/transcriptase are known and have been characterized to some extent, the cellular proteins involved in alphavirus RNA synthesis remain poorly understood. The first reported interaction between a host protein and viral RNA was that between the mosquito La autoantigen and the 3′ end of the SV negative-strand RNA (37). Recent work by Burnham et al. (6) demonstrated that the host protein hnRNP K interacts with nsP2 and SG RNA in SV-infected cells. Several reports (1, 12, 14) have identified other cellular proteins associated with the SV replicase/transcriptase. These include cytoskeleton proteins, ribosomal subunits, chaperones, and hnRNPs. However, the roles of these cellular proteins in SV replication remain unclear. hnRNP A1 was not identified in those studies.
To learn whether hnRNP A1 is also involved in SV replication, we used an electrophoretic mobility shift assay (EMSA) to determine whether hnRNP A1 would bind to the 5′ UTR of SV. We found that hnRNP A1 interacted with the SV 5′ UTR and facilitated the translation of viral RNA. In contrast to what we observed with EV71, the knockdown of hnRNP A1 reduced SV RNA synthesis and the yield of progeny virus. Taken together, these results indicate that hnRNP A1 plays an important role in the replication of both EV71 and SV.
MATERIALS AND METHODS
Cells and virus.
HeLa cells were cultured at 37°C in Eagle's minimum essential medium supplemented with 10% fetal calf serum (FCS) (Mediatech). Chicken embryo fibroblast (CEF), human embryonal rhabdomyosarcoma (RD), and Vero (green monkey epithelial) cells were grown in Dulbecco's modified Eagle medium supplemented with 10% FCS. SF268 (human glioblastoma) cells were cultured at 37°C in RPMI 1640 medium supplemented with 10% FCS. EV71 was propagated in RD cells, and SV was propagated in CEF cells. Cells were infected with EV71 at the specified multiplicity of infection (MOI) and then incubated at 37°C for 2 h for adsorption. Unbound virus was removed, and the cells were refed with fresh medium. A similar procedure was carried out to infect cells with SV except that the adsorption step was done for 1 h at 34°C. Media from infected cultures were harvested at the indicated times, and titers of EV71 and SV were measured by plaque formation on Vero and CEF cells, respectively.
Plasmid construction.
Plasmid pT7-EV71-5′ UTR was constructed as follows: the 5′ UTR of EV71 was amplified by PCR from the EV71 full-length infectious cDNA clone (41) using EV71 primers 5′-GCCGGTAATACGACTCACTATAGGGAGATTAAAACAGCCTGTGGGT and 3′-CATGTTTGATTGTGTTGAGGGTCAAAAT and was cloned into the pCRII-TOPO vector by TA cloning (Invitrogen). A bicistronic reporter plasmid, pRF-EV71-5′ UTR, containing the EV71 IRES between Renilla luciferase (RLuc) and firefly luciferase (FLuc) was constructed by ligating a NotI-EV71 5′ UTR-NotI fragment into pRF. Plasmid pRF-EV71-5′ UTR-AS was constructed by inserting the reverse sense of the NotI-EV71 5′ UTR-NotI fragment into pRF (32). Plasmid pMB-Toto-Luc was constructed by cloning the open reading frame for FLuc into the SpeI site in the coding sequence for the carboxyl-terminal portion of nsP3 in pMB-Toto, the infectious cDNA of SV, which is driven by the cytomegalovirus (CMV) promoter (kindly provided by Bill Moyle at Robert Wood Medical School, UMDNJ, NJ). The luciferase gene was cut out from pToto-Luc (4) (kindly provided by Margaret MacDonald, Rockefeller University, NY). Plasmid pMB-Toto-Luc-AS was constructed by inserting the reverse FLuc gene into the same restriction site and used as a control plasmid.
Expression and purification of the recombinant hnRNP A1 protein.
hnRNP A1 cDNA derived from SF268 cellular mRNA was cloned into pET30a, a prokaryotic expression vector containing a histidine tag (Novagen), via restriction sites EcoRI and BamHI. After transforming the constructed plasmid pET30a/hnRNP A1 into competent Escherichia coli BL21(DE3)(pLysS) cells, protein expression was induced by the addition of 40 μM isopropyl-β-d-thiogalactopyranoside at 37°C for 2 h. The recombinant hnRNP A1 protein was then purified using a HisTrap kit (GE). Protein purity was determined by electrophoresis on 12% sodium dodecyl sulfate (SDS)-polyacrylamide gels, and the concentration was determined using the Bio-Rad protein assay.
Preparation of labeled RNA probes and binding assay.
PCR was carried out to amplify the cDNA of the deleted EV71 IRES using pT7-EV71-5′UTR as a template and a positive-sense primer containing the T7 promoter and the first 20 nt of the EV71 IRES and different negative-sense primers to produce the 3′ truncated forms of the EV71 IRES. RNA probes for use in RNA gel mobility shift assays (EMSAs) were generated by runoff transcription using bacteriophage T7 RNA polymerase and then purified by use of an RNeasy minikit (Qiagen) and labeled at their 5′ ends by using T4 polynucleotide kinase and [γ-32P]ATP.
The 3′-terminal 45 nt of SV negative-strand RNA was amplified by PCR from pToto, the infectious clone of SV, using a positive-sense primer containing the T7 promoter and the first 20 nt of SV sequence and a negative-sense primer containing SP6 promoter and nt 33 to 45 of SV. The positive-strand RNA was synthesized by in vitro transcription using T7 polymerase and labeled with [α-32P]GTP. The RNA was capped by adding an m7G(5′)ppp(5′)G cap structural analog (NEB) (final concentration, 5 mM) into the reaction mixture and purified as described above. An EMSA was carried out to determine the interaction between the viral RNA and hnRNP A1 as described previously (31). Briefly, 2 μg of hnRNP A1 was incubated for 30 min at 25°C with one of the 32P-labeled RNA probes (1 × 104 cpm; EV71 or SV). The reaction was carried out in binding buffer (10 mM HEPES [pH 7.5], 150 mM KCl, 0.5 mM EGTA, 2 mM MgCl2, 1 mM dithiothreitol, 1 unit RNasin,10% glycerol), and the final volume of the reaction mixture was 10 μl. The binding of hnRNP A1 to the viral RNA sequence was recognized by a slower migration of the labeled RNA probes.
A nonspecific 31-mer RNA oligonucleotide (5′-UGGCCAAYGCCCUGGCUCACAAAUACCACUG-3′) was end labeled with [γ-32P]ATP and used as a control to test the specificity of the binding between hnRNP A1 and the viral RNAs.
siRNA knockdown.
HeLa cells were seeded in 12-well plates in antibiotic-free medium. A total of 100 nmol of siRNA targeting hnRNP A1 or hnRNP A2 (ON-TARGETplus SMARTpool, catalog number L-008221-00-0005 or L-011690-01-0005; Dharmacon) was transfected along with 3 μl FuGENE HD transfection reagent (Roche) in 0.4 ml minimal essential medium supplemented with 10% FCS according to the manufacturer's directions. Cells were either harvested 3 days after transfection for analysis of protein expression by Western blotting, transfected with CMV-RLuc-EV71 5′UTR-FLuc RNA or MB-Toto-Luc RNA, or infected with virus. The IRES activity was determined 2 days after transfection of CMV-RLuc-EV71 5′UTR-FLuc RNA by measuring the RLuc and FLuc activities in a 20/20 luminometer (Turner Biosystems) using a dual-luciferase reporter assay system (Promega) according to the manufacturer's instructions. The translation activity of the SV 5′ UTR was determined 2 days after transfection of MB-Toto-Luc RNA by measuring the FLuc activity using a luciferase assay kit (Promega). To examine viral RNA replication (EV71 or SV), infected cells were labeled with 10 μCi/well of [3H]uridine from 20 to 24 h postinfection (p.i.). Cells were then lysed, and tricarboxylic acid-precipitated counts were measured as described previously (42). The expression of viral protein was monitored by Western blotting using antibody against EV71 3C or SV E1 according to the procedure described below. Virus yield from the cells was determined by plaque formation on Vero cells for EV71 and on CEF cells for SV.
Western blotting.
The expression of hnRNPs A1 and A2 was examined by Western blotting using antibody against these two proteins (Abcam). Briefly, cells were lysed in sample buffer (10% glycerol, 50 mM Tris HCl [pH 6.8], 2% SDS, 100 mM dithiothreitol, and 0.01% bromophenol blue), and proteins were fractionated by SDS-polyacrylamide gel electrophoresis (PAGE) in 12% polyacrylamide gels and transferred onto polyvinylidene difluoride membranes by wet transfer. Membranes were blocked with phosphate-buffered saline (PBS) containing 5% low-fat dry milk. Anti-hnRNP A1 and/or anti-hnRNP A2 mouse antibody was then added, and the membranes were washed with PBS containing 0.2% Tween 20. Goat anti-mouse horseradish peroxidase-conjugated antibody (Bio-Rad) and the ECL kit (Pierce) were used to detect bound antibodies. Chemiluminescence was detected by exposure to Kodak X-ray film.
Fluorescence microscopic analysis.
RD cells grown on coverslips were infected with EV71 at an MOI of 40 PFU/cell. At 6 h p.i., the culture medium was removed, and the cells were washed three times with PBS. The cells on the coverslip were fixed with 3.7% (wt/vol) formaldehyde (Sigma) at room temperature for 20 min. After being washed three times with PBS, the cells on the coverslip were permeabilized in 0.5% Triton X-100 at room temperature for 5 min and washed another three times with PBS. For staining of hnRNP A1 and EV71 3A, the samples were blocked in solution that contained PBS containing 5% bovine serum albumin for 60 min at room temperature and then incubated with anti-hnRNP A1 antibody (1:200) and anti-EV71 3A antibody (1:200) for 1.5 h at room temperature and washed three times with PBS. The samples were then reacted with fluorescein isothiocyanate-conjugated goat anti-mouse immunoglobulin G (IgG) and rhodamine (tetramethyl rhodamine isothiocyanate)-conjugated goat anti-rabbit IgG (Jackson ImmunoResearch Laboratories) for 1 h at room temperature. After washing with PBS, the samples were treated with Hoechst 33258 stain for 15 min at room temperature and washed again with PBS three times. Finally, the coverslips with adhering cells were placed onto a glass slide and sealed with transparent nail polish. The images were captured using a confocal laser scanning microscope (Zeiss LSM510 Meta). A similar procedure was carried out to examine the localization of hnRNP A1 after SV infection except that HeLa cells were infected with SV for 8 h and rabbit anti-SV E2 antibody was used to detect SV.
RESULTS
hnRNP A1 interacts with the IRES of the EV71 5′ UTR.
Using streptavidin beads to capture cellular proteins bound to the biotinylated EV71 5′ UTR and matrix-assisted laser desorption ionization-time-of-flight analysis, we have identified 15 cellular proteins which interact with the EV71 5′ UTR (32). Among these proteins, we chose to further examine the interaction of the EV71 5′ UTR with hnRNP A1. The interaction of hnRNP A1 with the EV71 5′ UTR was confirmed by EMSA. As shown in Fig. 1A, hnRNP A1 bound the 32P-labeled EV71 5′ UTR, slowing its migration in the nondenaturing gel. hnRNP A1 did not bind to a unrelated 31-mer RNA oligonucleotide, suggesting that the interaction between the EV71 5′ UTR and hnRNP A1 is specific.
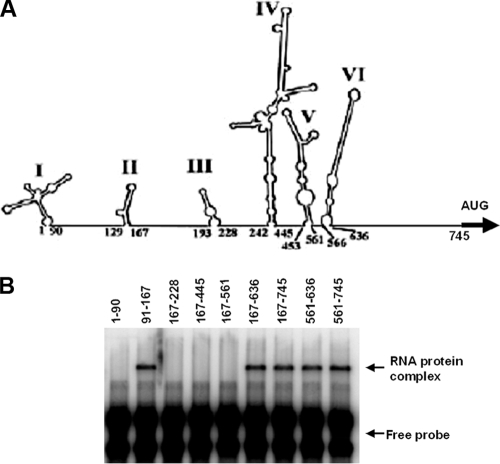
FIG. 1.
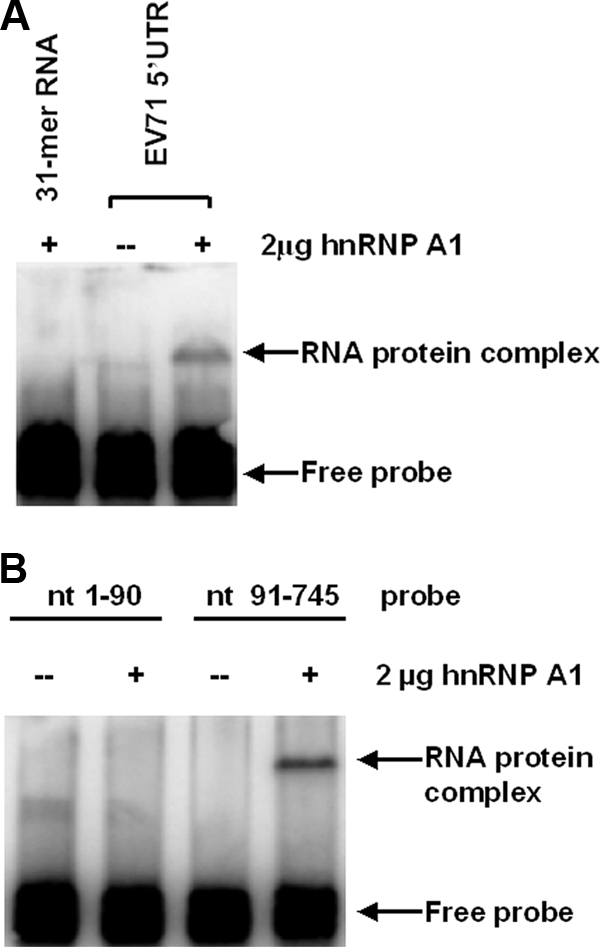
Binding of hnRNP A1 to the EV71 5′ UTR. (A) The binding of hnRNP A1 to the EV71 5′ UTR is specific. EMSA was performed as described in Materials and Methods. The EV71 5′ UTR was labeled with [32P]GTP (1 ×104 cpm per reaction) and incubated at 37°C for 30 min either alone or with 2 μg of hnRNP A1. A nonspecific 31-mer RNA oligonucleotide was end labeled with 32PO4 and reacted with 2 μg of hnRNP A1. (B) hnRNP A1 binds to the IRES of the EV71 5′ UTR. nt 1 to 90 and nt 91 to 745 were labeled with [32P]GTP (1 ×104 cpm per reaction), and EMSA was carried out as described above (A).
The 5′UTR of EV71 contains both a cloverleaf structure (nt 1 to 90) and an IRES (nt 91 to 745), as predicted by MFold (32, 53). We wished to identify which part of the 5′ UTR interacts with hnRNP A1. As shown in Fig. 1B, a 32P-labeled RNA probe containing a sequence from nt 91 to 745 of the EV71 5′ UTR was able to bind to hnRNP A1, while a probe containing a sequence from nt 1 to 90 of the EV71 5′ UTR was not. These results indicate that it is the IRES of the EV71 5′ UTR which interacts with hnRNP A1.
Identification of the hnRNP A1 binding sites on the EV71 IRES.
To identify the RNA sequences in the EV71 IRES which bind to hnRNP A1, serial deletions of the IRES were made to knock out each of the six stem-loops individually; care was taken not to disrupt the secondary structures of the remaining RNA (Fig. 2A). PCR was carried out to amplify the cDNA of the EV71 IRES with various deletions. RNA probes for use in EMSA were then synthesized and labeled at their 5′ ends with [γ-32P]ATP. As shown in Fig. 2B, hnRNP A1 did not interact with the cloverleaf structure (nt 1 to 90) of the 5′ UTR. This finding is consistent with the result presented in Fig. 1B. Within the IRES region, hnRNP A1 did not interact with stem-loop III (nt 167 to 228), stem-loops III and IV (nt 167 to 445), or stem-loops III, IV, and V (nt 167 to 561). However, hnRNP A1 did slow the migration of RNA probes at nt 91 to 167, nt 167 to 636, nt 167 to 745, nt 561 to 636, and nt 561 to 745, indicating that this protein interacts with stem-loops II and VI.
FIG. 2.
Identification of hnRNP A1 binding sites on the EV71 IRES. (A) The secondary structure of the 5′ UTR was predicted by MFold. (B) A serial deletion to eliminate each of the six stem-loops of the IRES was made. RNA probes were labeled with [γ-32P]ATP (1 ×104 cpm per reaction), and EMSA was carried out as described in the legend of Fig. 1A.
The hnRNP A1 function in EV71 replication can be replaced by that of hnRNP A2.
To analyze the role of hnRNP A1 in EV71 replication, we used RNA interference to knock down hnRNP A1 mRNA. siRNA targeting hnRNP A1 was transfected into HeLa cells as described in Materials and Methods. The expression of endogenous hnRNP A1 was examined by Western blotting using anti-hnRNP A1 antibody. Compared with mock-transfected cells, cells transfected with hnRNP A1 siRNA showed a marked reduction in the level of hnRNP A1 (Fig. 3A). In contrast, the level of actin was not affected under the same conditions.
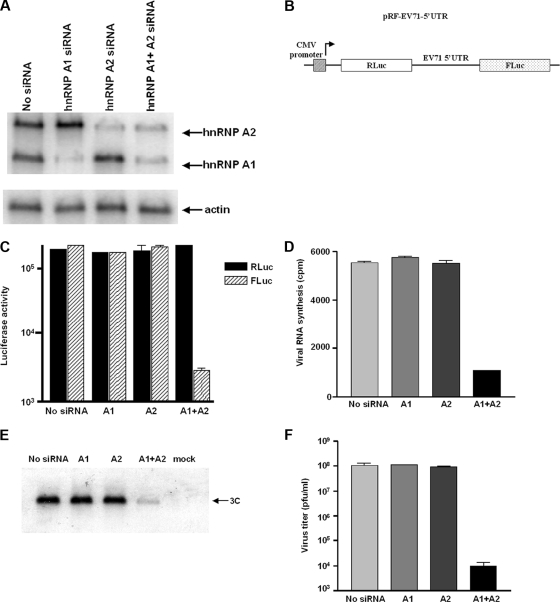
FIG. 3.
Effect of hnRNP A1 and hnRNP A2 on EV71 replication. (A) Western blot to examine the expression of hnRNP A1 and hnRNP A2. hnRNP A1, hnRNP A2, or both were depleted by the siRNA knockdown technique as described in Materials and Methods. HeLa cell lysates were prepared 3 days after transfection of siRNA. Western blotting was carried out using antibodies against hnRNP A1 and hnRNP A2. The expression of actin was examined by Western blotting as a control. (B) Diagram of the bicistronic construct used in the dual-luciferase assay. (C) Effect of knockdown of hnRNP A1 and hnRNP A2 on EV71 IRES activity. HeLa cells were transfected with no siRNA, siRNA targeting hnRNP A1, siRNA targeting hnRNP A2, or siRNA targeting both. Three days after transfection, the CMV-RLuc-EV71 5′ UTR-FLuc RNA transcript was then transfected into the cells. Luciferase activity was measured 2 days after transfection. Mean values and standard errors from triplicate samples are shown in the bar graph. (D) Effect of knockdown of hnRNP A1 and hnRNP A2 on EV71 RNA synthesis. hnRNP A1 and hnRNP A2 were knocked down as described in Materials and Methods. HeLa cells were then infected with EV71 at an MOI of 2 PFU/cell and labeled from 20 to 24 h p.i. with [3H]uridine. Cell lysates were collected and resuspended in a solution of 0.6% NP-40 in PBS. After incubation at 4°C for 30 min, the nuclear fraction was removed by centrifugation, and the acid-insoluble radioactivity was determined in the supernatant (cytoplasmic) fraction. Mean values and standard errors from duplicate samples are shown. (E) Effect of knockdown of hnRNP A1 and hnRNP A2 on EV71 protein synthesis. HeLa cells were transfected with no siRNA or siRNA targeting hnRNP A1, hnRNP A2, or both. Cells were mock infected or infected with EV71 3 days after transfection. Lysates from mock-infected or EV71-infected cells were subjected to 12% SDS-PAGE. Western blotting analysis was carried out using an anti-3C antibody. (F) Replication of EV71 in hnRNP A1- and A2-depleted cells. HeLa cells treated with siRNA targeting hnRNP A1, hnRNP A2, or both were infected with EV71 at an MOI of 2 PFU/cell and incubated at 37°C. Medium was harvested 24 h p.i. and assayed for infectious virus by plaque formation on Vero cells. Mean values and standard errors from duplicate samples are shown.
A bicistronic reporter plasmid (CMV-RLuc-EV71 5′UTR-FLuc, i.e., pRF-EV71-5′UTR) was used as a template for the synthesis of the CMV-RLuc-EV71 5′UTR-FLuc RNA transcript. As hnRNP A1 could possibly be involved in the nuclear export of mRNA, the knockdown of hnRNP A1 could inhibit translation indirectly by blocking the nuclear export of mRNA. To avoid this confounding issue, the CMV-RLuc-EV71 5′UTR-FLuc RNA transcript was transfected into cells to measure the IRES activity of EV71. The bicistronic plasmid contained the EV71 5′ UTR flanked by the RLuc and FLuc open reading frames (Fig. 3B). The translation of RLuc is cap dependent, whereas the translation of the second cistron (FLuc) is IRES dependent. At 48 h after transfection, the RLuc and FLuc expression levels were measured in a 20/20 luminometer (Turner Biosystems) using a dual-luciferase reporter assay (Promega). The relative translation efficiency of the EV71 IRES was determined by a comparison of the level of downstream reporter gene product with the level of upstream reporter gene product. A bicistronic plasmid with the antisense of the EV71 IRES (CMV-RLuc-EV71 5′UTR-AS-FLuc) was used as a control to confirm that the EV71 IRES is actually functional in these assays. The treatment of cells with siRNA targeting hnRNP A1 showed very little effect on the EV71 IRES activity, as indicated by the luciferase activity in the cells (Fig. 3C), even though the level of hnRNP A1 was markedly reduced in the cells treated with hnRNP A1 siRNA (Fig. 3A).
Since there have been reports demonstrating the functional substitutions of hnRNP A1-related proteins for hnRNP A1 in some biological processes (8, 23, 33, 40), we speculated that the lack of any effect of an hnRNP A1 knockdown on EV71 IRES activity might be explained if there was another protein that could substitute for hnRNP A1. To explore this possibility, we tested the effect of the knockdown of hnRNP A2 mRNA on EV71 replication. The knockdown efficiency was examined by Western blotting using antibody to hnRNP A2. As shown in Fig. 3A, the expression of hnRNP A2 was inhibited when cells were transfected with siRNA targeting hnRNP A2. The bicistronic system was used to test the effect of hnRNP A2 on EV71 IRES activity. As was the case for the knockdown of hnRNP A1, the knockdown of hnRNP A2 showed little effect on the IRES activity (Fig. 3C). However, cells transfected with both hnRNP A1 and hnRNP A2 siRNAs showed a dramatic reduction in the EV71 IRES activity compared with the cells transfected without siRNA. A comparison of the RLuc levels of control and hnRNP A1- and A2-depleted cells indicated that the knockdown of both hnRNP A1 and A2 had no effect on cap-dependent translation.
Next, cells were treated with 2 μg/ml actinomycin D and infected with EV71 3 days after transfection of siRNA targeting hnRNP A1, hnRNP A2, or both, and viral RNA synthesis was examined by the incorporation of [3H]uridine into newly synthesized viral RNA. As shown in Fig. 3D, the knockdown of hnRNP A1 alone or hnRNP A2 alone had no effect on viral RNA synthesis. However, when both hnRNP A1 and hnRNP A2 were depleted, a marked reduction in the level of viral RNA synthesis was observed.
To examine the effect of the knockdown of hnRNP A1 and hnRNP A2 on viral protein synthesis, EV71-infected cells were lysed, and Western blotting was performed using an antibody against EV71 3C. As shown in Fig. 3E, the expression of the 3C protein in hnRNP A1- or A2-depleted cells was similar to that in control infected cells. The level of expression of the viral 3C protein was, however, decreased when both hnRNP A1 and hnRNP A2 were knocked down.
To look at the effects of knockdown on virus yield, media were harvested at 24 h p.i. and titrated for infectious virus by plaque formation on Vero cells. Figure 3F shows that the virus yield from hnRNP A1- or A2-depleted cells was similar to that from untreated infected cells. The virus titer was decreased by 4 logs in cells treated with both siRNAs targeting hnRNP A1 and hnRNP A2. This indicates that hnRNP A2 can substitute for the activity of hnRNP A1 in EV71 replication and that both hnRNP A1 and hnRNP A2 likely play important roles in the replication of EV71.
Relocalization of hnRNP A1 during EV71 infection.
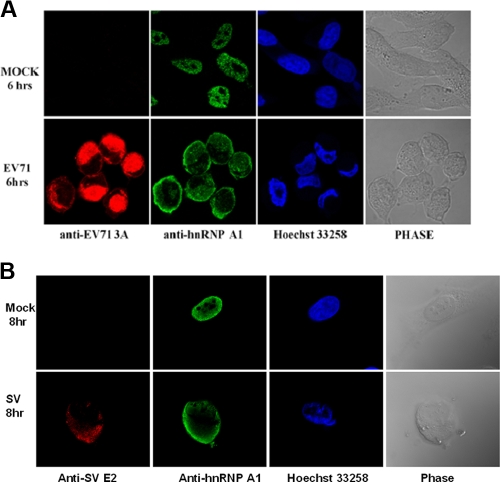
hnRNP A1 is primarily a nuclear protein, although it shuttles back and forth between the nucleus and the cytoplasm (15). Since EV71 replication occurs in the cytoplasm of infected cells, we were interested in knowing whether the subcellular localization of hnRNP A1 was altered by EV71 infection. Figure 4A shows the immunofluorescent staining of hnRNP A1 in EV71-infected RD cells at 6 h p.i. The level of hnRNP A1 in cytoplasm was increased significantly, and a corresponding decrease in levels of nuclear hnRNP A1 was observed in virus-infected cells. As expected, the infected cells expressed the EV71 3A protein in the cytoplasm (Fig. 4A). In comparison, in the uninfected cells, which did not have 3A protein staining, hnRNP A1 was localized predominantly to the nucleus. Hoechst 33258 was used to stain the nuclei. In cells transiently expressing EV71 3C without EV71 replication, no relocalization of hnRNP A1 was detected (data not shown). Thus, the relocalization of hnRNP A1 from the nucleus to the cytoplasm may require EV71 replication or at least one or more of the other proteins. Interestingly, the relocalization of hnRNP A1 was also observed in cells infected with MHV and HCV, both of which require hnRNP A1 for replication (23, 30).
FIG. 4.
Redistribution of hnRNP A1 after EV71 and SV infection. (A) hnRNP A1 relocalized from the nucleus to the cytoplasm after EV71 infection. RD cells were mock infected or infected with EV71 at an MOI of 40 PFU/cell. At 6 h p.i., cells were fixed with formaldehyde and immunostained with antibody against the hnRNP A1 or EV71 3A protein. Fluorescein isothiocyanate-conjugated goat anti-mouse IgG or tetramethyl rhodamine isothiocyanate-conjugated goat anti-rabbit IgG was used as a secondary antibody. Hoechst 33258 dye was used to stain the nuclei. The images were captured by confocal laser scanning microscopy. (B) hnRNP A1 shuttled from the nucleus to the cytoplasm after SV infection. HeLa cells were mock infected or infected with SV at an MOI of 40 PFU/cell for 8 h. Cells were then fixed and immunostained as described above (A) except that antibody to SV E2 was used to detect the expression of the E2 protein.
hnRNP A1 interacts with the 5′ UTR of SV and facilitates its replication.
To learn whether hnRNP A1 also interacts with the 5′ UTR of a virus whose translation is not IRES dependent, we tested the binding of hnRNP A1 to the SV 5′ UTR. An EMSA was carried out as described in Materials and Methods using hnRNP A1 and the labeled 5′ UTR of SV. As shown in Fig. 5A, hnRNP A1 did not bind to a nonspecific 31-mer RNA oligonucleotide but did bind to the capped 5′ UTR of SV, slowing its electrophoretic mobility. These results indicate that the interaction between the SV 5′ UTR and hnRNP A1 is specific.
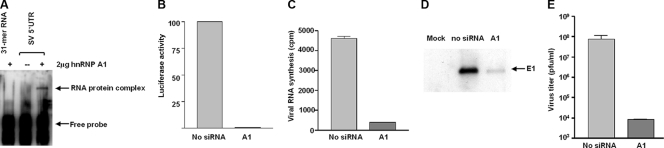
FIG. 5.
Effect of hnRNP A1 on SV replication. (A) The binding of hnRNP A1 to the SV 5′ UTR is specific. The 5′ UTR (nt 1 to 45) was capped and labeled with [32P]GTP. EMSA was carried out as described in the legend of Fig. 1A. (B) Effect of knockdown of hnRNP A1 on SV translation. HeLa cells were transfected with no siRNA or siRNA targeting hnRNP A1. Three days after transfection, cells were then transfected with the MB-Toto-Luc RNA transcript. Firefly luciferase was measured 2 days posttransfection. Mean values and standard errors from triplicate samples are shown in the bar graph. (C) Effect of knockdown of hnRNP A1 on SV RNA synthesis. HeLa cells were transfected with no siRNA or siRNA targeting hnRNP A1. Three days posttransfection, cells were infected with SV at an MOI of 2 PFU/cell and labeled with [3H]uridine from 20 to 24 h p.i. Viral RNA was measured as described in the legend of Fig. 3D. Mean values and standard errors from duplicate samples are shown in the bar graph. (D) Effect of knockdown of hnRNP A1 on SV protein synthesis. HeLa cells were transfected with no siRNA or siRNA targeting hnRNP A1. Three days posttransfection, cells were mock infected or infected with SV at an MOI of 2 PFU/cell for 24 h at 34°C. Lysates from mock-infected or SV-infected cells were subjected to 12% SDS-PAGE. Western blotting analysis was carried out using an anti-E1 antibody. (E) Replication of SV in hnRNP A1-depleted cells. HeLa cells were transfected with no siRNA or siRNA targeting hnRNP A1 and then infected with SV as described above (D). Medium was harvested 24 h p.i. and assayed for infectious virus by plaque formation on CEF cells. Mean values and standard errors from duplicate samples are shown.
To assess the role of hnRNP A1 in SV replication, siRNA targeting hnRNP A1 was used to knock down the mRNA of hnRNP A1 in HeLa cells. The MB-Toto-Luc RNA transcript was synthesized using pMB-Toto-Luc (the SV infectious clone with the insertion of a luciferase gene) as a template. The RNA was transfected into cells 3 days after transfection of siRNA. Treatment with siRNA against hnRNP A1 reduced the level of luciferase activity in the cells by about 95% compared with the activity in the cells transfected without siRNA (Fig. 5B). In addition, hnRNP A1-depleted cells were infected with SV, and viral RNA replication, protein synthesis, and virus yield were monitored as described above. Levels of viral RNA replication, protein synthesis, and virus yield decreased dramatically when hnRNP A1 was knocked down (more than 95% reduction in viral RNA synthesis and more than 3 logs in virus titer) (Fig. 5C and E). These data indicate that the suppression of hnRNP A1 has a detrimental effect on SV replication.
Like EV71, SV replicates in the cytoplasm. We therefore wished to know whether hnRNP A1 redistributes from the nucleus to the cytoplasm after SV infection. We examined the subcellular localization of hnRNP A1 in HeLa cells. Immunocytochemical analyses of SV and hnRNP A1 were performed using a polyclonal antibody against the SV E2 protein and a monoclonal antibody recognizing the hnRNP A1 protein (Fig. 4B). In uninfected cells, hnRNP A1 was found mainly in the nucleus. In SV-infected cells, a redistribution of hnRNP A1 into the cytoplasm was detected in cells expressing the SV E2 protein.
DISCUSSION
In addition to virus-encoded factors, many steps in virus infections involve host factors. Such virus-host interactions are crucial determinants of virus host range, replication, and pathology; knowledge of these interactions offers insights into viral and cellular functions and provides antiviral targets. Thus, the identification of such interactions and the associated host factors is a major frontier in virology (16, 19). Recently, using a human-genome-wide RNA interference screen, Krishnan et al. (26) identified 305 host proteins that affect West Nile virus infection. Functional clustering of these genes revealed a complex dependence of this virus on a wide variety of host molecules and cellular pathways.
To investigate whether hnRNP A1 has a positive role in EV71 replication, the effects of protein knockdown (using siRNAs targeting the mRNA of hnRNP A1) on the synthesis of viral RNA and protein, IRES activity, and viral replication in HeLa cells were assessed. The knockdown of hnRNP A1 alone had no effect on viral RNA and protein synthesis, IRES activity, or viral replication. As it has been known that proteins in the hnRNP A/B family are sometimes functionally interchangeable, we speculated that the function of hnRNP A1 might be substituted by other hnRNP A/B proteins. We therefore examined the effect of knocking down hnRNP A2 on the replication of EV71. As we found with hnRNP A1, the depletion of hnRNP A2 had no effect on EV71 replication. However, when both hnRNPs A1 and A2 were depleted, viral RNA and protein synthesis, IRES activity, and viral replication were markedly inhibited. We therefore conclude that hnRNP A1, an abundant mRNA binding protein present in the nucleus of mammalian cells, is a translation trans-acting factor that interacts with the IRES of EV71 and modulates EV71 replication.
Our finding that the function of hnRNP A1 to facilitate the IRES-dependent translation of EV71 can be substituted by hnRNP A2 suggests that these two members of the hnRNP A/B family of cellular proteins are of particular important for EV71 replication. Since hnRNP A1 and hnRNP A2 are structurally similar and are particularly homologous in their N-terminal RNA binding domains, it is not surprising that these two proteins can participate in similar functions in EV71 replication. In addition, functional substitutions of hnRNP A1-related proteins for hnRNP A1 have been reported for several biological processes. For instance, hnRNP A/B, hnRNP A2/B1, and hnRNP A3, which are highly homologous to hnRNP A1, can substitute for hnRNP A1 in the regulation of the alternative splicing of cellular pre-mRNA (33) and that of human immunodeficiency virus pre-mRNA (8). It was also reported that the role of hnRNP A1 in the replication of MHV and HCV RNA can be substituted by hnRNP A1-related proteins (23, 40).
In the present study, we found that hnRNP A1 specifically interacts with two regions of the EV71 IRES, nt 91 to 167 and nt 561 to 745 (Fig. 2B). It has been known that hnRNP A1 facilitates RNA duplex formation by complementary single-stranded polynucleotides (11); we therefore speculate that it may facilitate intramolecular annealing reactions of the EV71 IRES to form a proper and stable structure for the binding of the 40S ribosome and thus to promote the IRES-dependent translation of EV71. The formation of such a structure may also have a role in recruiting cellular proteins needed for the replication of EV71.
In addition to IRES-dependent translation, some RNA viruses do not have IRES in the 5′ UTR so that their translation is not IRES dependent. We wished to know whether hnRNP A1 can also facilitate the IRES-independent translation of SV. Recent studies from several groups have identified cellular proteins associated with the SV RNA replication complex by isolating nonstructural protein-containing complexes from infected cells (1, 6, 12, 14). However, the roles that most of these proteins play in the replication of SV remain to be elucidated. The effect of knocking down hnRNP A1 on the replication of SV was investigated using procedures similar to those that we used to study the involvement of hnRNP A1 in EV71 replication. We found that after SV infection, hnRNP A1 relocalized from the nucleus to the cytoplasm, where SV replication occurs. hnRNP A1 was able to bind to the capped 5′ UTR of SV, and unlike the case with EV71, the knockdown of hnRNP A1 diminished SV RNA synthesis, translation, and viral replication. We therefore conclude that hnRNP A1 promotes SV replication via binding to the 5′ UTR and facilitates possibly both viral RNA synthesis and translation.
Several hnRNPs, including hnRNPs A1, C1/C2, E1/E2, I (PTB), and L, constitute ITAFs that modulate the activity of IRES sequences generally present in the 5′ UTRs of several viral and cellular mRNAs (7, 28, 29, 44, 47). Our finding that hnRNP A1 enhances the translation of SV RNA is the first report showing that in addition to facilitating IRES-dependent translation, hnRNP A1 is also involved in the non-IRES-dependent translation of a viral RNA.
How does hnRNP A1 contribute to the replication of EV71 and SV? hnRNP A1 is predominantly a nuclear protein. During EV71 and SV infection, it appears that nearly all of hnRNP A1 relocalizes to cytoplasm. Precisely what interactions lead to the recruitment or retention of hnRNP A1 in cytoplasm remain to be characterized. From our current work, it seems that the loss of translation when hnRNP A1 was depleted was sufficient to account for the related loss of viral RNA synthesis and virus yield.
To our knowledge, this is the first report that describes an RNA binding protein (hnRNP A1) actively participating in EV71 and SV replication in molecular detail. These studies not only improve our understanding of the replication of EV71 and SV but also have the potential for use as the basis for developing a drug against these viruses that acts by inhibiting virus replication.
Acknowledgments
This work was supported by U.S. Public Health Service grant AI-70668 from the National Institutes of Health.
Footnotes
Published ahead of print on 1 April 2009.
REFERENCES
- 1.Atasheva, S., R. Gorchakov, R. English, I. Frolov, and E. Frolova. 2007. Development of Sindbis viruses encoding nsP2/GFP chimeric proteins and their application for studying nsP2 functioning. J. Virol. 815046-5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Back, S. H., Y. K. Kim, W. J. Kim, S. Cho, H. R. Oh, J. E. Kim, and S. K. Jang. 2002. Translation of polioviral mRNA is inhibited by cleavage of polypyrimidine tract-binding proteins executed by polioviral 3Cpro. J. Virol. 762529-2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bedard, K. M., and B. L. Semler. 2004. Regulation of picornavirus gene expression. Microbes Infect. 6702-713. [DOI] [PubMed] [Google Scholar]
- 4.Bick, M. J., J.-W. Carroll, G. Gao, S. P. Goff, C. M. Rice, and M. R. MacDonald. 2003. Expression of the zinc-finger antiviral protein inhibits alphavirus replication. J. Virol. 7711555-11562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brunner, J. E., J. H. Nguyen, H. H. Roehl, T. V. Ho, K. M. Swiderek, and B. L. Semler. 2005. Functional interaction of heterogeneous nuclear ribonucleoprotein C with poliovirus RNA synthesis initiation complexes. J. Virol. 793254-3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burnham, A. J., L. Gong, and R. W. Hardy. 2007. Heterogeneous nuclear ribonuclear protein K interacts with Sindbis virus nonstructural proteins and viral subgenomic mRNA. Virology 367212-221. [DOI] [PubMed] [Google Scholar]
- 7.Cammas, A., F. Pileur, S. Bonnal, S. M. Lewis, N. Leveque, M. Holcik, and S. Vagner. 2007. Cytoplasmic relocalization of heterogeneous nuclear ribonucleoprotein A1 controls translation initiation of specific mRNAs. Mol. Biol. Cell 185048-5059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caputi, M., A. Mayeda, A. R. Krainer, and A. M. Zahler. 1999. hnRNP A/B proteins are required for inhibition of HIV-1 pre-mRNA splicing. EMBO J. 184060-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheunim, T., J. Zhang, S. G. Milligan, M. G. McPhillips, and S. V. Graham. 2008. The alternative splicing factor hnRNP A1 is up-regulated during virus-infected epithelial cell differentiation and binds the human papillomavirus type 16 late regulatory element. Virus Res. 131189-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choi, K., J. H. Kim, X. Li, K. Y. Paek, S. H. Ha, S. H. Ryu, E. Wimmer, and S. K. Jang. 2004. Identification of cellular proteins enhancing activities of internal ribosomal entry sites by competition with oligodeoxynucleotides. Nucleic Acids Res. 321308-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cobianchi, F., G. Biamonti, M. Maconi, and S. Riva. 1994. Human hnRNP protein A1: a model polypeptide for a structural and genetic investigation of a broad family of RNA binding proteins. Genetica 94101-114. [DOI] [PubMed] [Google Scholar]
- 12.Cristea, I. M., J. W. Carroll, M. P. Rout, C. M. Rice, B. T. Chait, and M. R. MacDonald. 2006. Tracking and elucidating alphavirus-host protein interactions. J. Biol. Chem. 28130269-30278. [DOI] [PubMed] [Google Scholar]
- 13.Dreyfuss, G., M. J. Matunis, S. Pinol-Roma, and C. G. Burd. 1993. hnRNP proteins and the biogenesis of mRNA. Annu. Rev. Biochem. 62289-321. [DOI] [PubMed] [Google Scholar]
- 14.Frolova, E., R. Gorchakov, N. Garmashova, S. Atasheva, L. A. Vergara, and I. Frolov. 2006. Formation of nsP3-specific protein complexes during Sindbis virus replication. J. Virol. 804122-4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghetti, A., S. Pinol-Roma, W. M. Michael, C. Morandi, and G. Dreyfuss. 1992. hnRNP I, the polypyrimidine tract-binding protein: distinct nuclear localization and association with hnRNAs. Nucleic Acids Res. 203671-3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grdzelishvili, V. Z., H. Garcia-Ruiz, T. Watanabe, and P. Ahlquist. 2005. Mutual interference between genomic RNA replication and subgenomic mRNA transcription in brome mosaic virus. J. Virol. 791438-1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Griffin, D. E. 2007. Alphaviruses, p. 1023-1068. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 18.Hamilton, B. J., E. Nagy, J. S. Malter, B. A. Arrick, and W. F. Rigby. 1993. Association of heterogeneous nuclear ribonucleoprotein A1 and C proteins with reiterated AUUUA sequences. J. Biol. Chem. 2688881-8887. [PubMed] [Google Scholar]
- 19.Hao, L., A. Sakurai, T. Watanabe, E. Sorensen, C. A. Nidom, M. A. Newton, P. Ahlquist, and Y. Kawaoka. 2008. Drosophila RNAi screen identifies host genes important for influenza virus replication. Nature 454890-893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henics, T., A. Sanfridson, B. J. Hamilton, E. Nagy, and W. F. Rigby. 1994. Enhanced stability of interleukin-2 mRNA in MLA 144 cells. Possible role of cytoplasmic AU-rich sequence-binding proteins. J. Biol. Chem. 2695377-5383. [PubMed] [Google Scholar]
- 21.Ho, M., E. R. Chen, K. H. Hsu, S. J. Twu, K. T. Chen, S. F. Tsai, J. R. Wang, S. R. Shih, et al. 1999. An epidemic of enterovirus 71 infection in Taiwan. N. Engl. J. Med. 341929-935. [DOI] [PubMed] [Google Scholar]
- 22.Izumi, R. E., S. Das, B. Barat, S. Raychaudhuri, and A. Dasgupta. 2004. A peptide from autoantigen La blocks poliovirus and hepatitis C virus cap-independent translation and reveals a single tyrosine critical for La RNA binding and translation stimulation. J. Virol. 783763-3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim, C. S., S. K. Seol, O. K. Song, J. H. Park, and S. K. Jang. 2007. An RNA-binding protein, hnRNP A1, and a scaffold protein, septin 6, facilitate hepatitis C virus replication. J. Virol. 813852-3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim, J. H., K. Y. Paek, K. Choi, T.-D. Kim, B. Hahm, K.-T. Kim, and S. K. Jang. 2003. Heterogeneous nuclear ribonucleoprotein C modulates translation of c-myc mRNA in a cell cycle phase-dependent manner. Mol. Cell. Biol. 23708-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krecic, A. M., and M. S. Swanson. 1999. hnRNP complexes: composition, structure, and function. Curr. Opin. Cell Biol. 11363-371. [DOI] [PubMed] [Google Scholar]
- 26.Krishnan, M. N., A. Ng, B. Sukumaran, F. D. Gilfoy, P. D. Uchil, H. Sultana, A. L. Brass, R. Adametz, M. Tsui, F. Qian, R. R. Montgomery, S. Lev, P. W. Mason, R. A. Koski, S. J. Elledge, R. J. Xavier, H. Agaisse, and E. Fikrig. 2008. RNA interference screen for human genes associated with West Nile virus infection. Nature 455242-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuhn, R. J. 2007. Togaviridae, p. 1001-1022. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 28.Lewis, S. M., and M. Holcik. 2008. For IRES trans-acting factors, it is all about location. Oncogene 271033-1035. [DOI] [PubMed] [Google Scholar]
- 29.Lewis, S. M., A. Veyrier, U. N. Hosszu, S. Bonnal, S. Vagner, and M. Holcik. 2007. Subcellular relocalization of a trans-acting factor regulates XIAP IRES-dependent translation. Mol. Biol. Cell 181302-1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li, H. P., X. Zhang, R. Duncan, L. Comai, and M. M. Lai. 1997. Heterogeneous nuclear ribonucleoprotein A1 binds to the transcription-regulatory region of mouse hepatitis virus RNA. Proc. Natl. Acad. Sci. USA 949544-9549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li, M. L., and V. Stollar. 2004. Identification of the amino acid sequence in Sindbis virus nsP4 that binds to the promoter for the synthesis of the subgenomic RNA. Proc. Natl. Acad. Sci. USA 1019429-9434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin, J. Y., M. L. Li, P. N. Huang, K. Y. Chien, J. T. Horng, and S. R. Shih. 2008. Heterogeneous nuclear ribonuclear protein K interacts with the enterovirus 71 5′ untranslated region and participates in virus replication. J. Gen. Virol. 892540-2549. [DOI] [PubMed] [Google Scholar]
- 33.Mayeda, A., S. H. Munroe, J. F. Caceres, and A. R. Krainer. 1994. Function of conserved domains of hnRNP A1 and other hnRNP A/B proteins. EMBO J. 135483-5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mayrand, S. H., and T. Pederson. 1990. Crosslinking of hnRNP proteins to pre-mRNA requires U1 and U2 snRNPs. Nucleic Acids Res. 183307-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McMinn, P. C. 2002. An overview of the evolution of enterovirus 71 and its clinical and public health significance. FEMS Microbiol. Rev. 2691-107. [DOI] [PubMed] [Google Scholar]
- 36.Paranjape, S. M., and E. Harris. 2007. Y box-binding protein-1 binds to the dengue virus 3′-untranslated region and mediates antiviral effects. J. Biol. Chem. 28230497-30508. [DOI] [PubMed] [Google Scholar]
- 37.Pardigon, N., and J. H. Strauss. 1996. Mosquito homolog of the La autoantigen binds to Sindbis virus RNA. J. Virol. 701173-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pettit Kneller, E. L., J. H. Connor, and D. S. Lyles. 2009. hnRNPs relocalize to the cytoplasm following infection with vesicular stomatitis virus. J. Virol. 83770-780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shi, S. T., P. Huang, H. P. Li, and M. M. Lai. 2000. Heterogeneous nuclear ribonucleoprotein A1 regulates RNA synthesis of a cytoplasmic virus. EMBO J. 194701-4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shi, S. T., G. Y. Yu, and M. M. Lai. 2003. Multiple type A/B heterogeneous nuclear ribonucleoproteins (hnRNPs) can replace hnRNP A1 in mouse hepatitis virus RNA synthesis. J. Virol. 7710584-10593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shih, S.-R., M.-C. Tsai, S.-N. Tseng, K.-F. Won, K.-S. Shia, W.-T. Li, J.-H. Chern, G.-W. Chen, C.-C. Lee, Y.-C. Lee, K.-C. Peng, and Y.-S. Chao. 2004. Mutation in enterovirus 71 capsid protein VP1 confers resistance to the inhibitory effects of pyridyl imidazolidinone. Antimicrob. Agents Chemother. 483523-3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shih, S. R., K. F. Weng, V. Stollar, and M. L. Li. 2008. Viral protein synthesis is required for enterovirus 71 to induce apoptosis in human glioblastoma cells. J. Neurovirol. 1453-61. [DOI] [PubMed] [Google Scholar]
- 43.Siomi, H., and G. Dreyfuss. 1995. A nuclear localization domain in the hnRNP A1 protein. J. Cell Biol. 129551-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spriggs, K. A., M. Bushell, S. A. Mitchell, and A. E. Willis. 2005. Internal ribosome entry segment-mediated translation during apoptosis: the role of IRES-trans-acting factors. Cell Death Differ. 12585-591. [DOI] [PubMed] [Google Scholar]
- 45.Strauss, J. H., and E. G. Strauss. 1994. The alphaviruses: gene expression, replication, and evolution. Microbiol. Rev. 58491-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Swanson, M. S., and G. Dreyfuss. 1988. RNA binding specificity of hnRNP proteins: a subset bind to the 3′ end of introns. EMBO J. 73519-3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vagner, S., B. Galy, and S. Pyronnet. 2001. Irresistible IRES. Attracting the translation machinery to internal ribosome entry sites. EMBO Rep. 2893-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang, Y. F., S. C. Chen, F. Y. Wu, and C. W. Wu. 1997. The interaction between human cytomegalovirus immediate-early gene 2 (IE2) protein and heterogeneous ribonucleoprotein A1. Biochem. Biophys. Res. Commun. 232590-594. [DOI] [PubMed] [Google Scholar]
- 49.Weighardt, F., G. Biamonti, and S. Riva. 1995. Nucleo-cytoplasmic distribution of human hnRNP proteins: a search for the targeting domains in hnRNP A1. J. Cell Sci. 108545-555. [DOI] [PubMed] [Google Scholar]
- 50.Weighardt, F., G. Biamonti, and S. Riva. 1996. The roles of heterogeneous nuclear ribonucleoproteins (hnRNP) in RNA metabolism. Bioessays 18747-756. [DOI] [PubMed] [Google Scholar]
- 51.Wimmer, E., and A. Nomoto. 1993. Molecular biology and cell-free synthesis of poliovirus. Biologicals 21349-356. [DOI] [PubMed] [Google Scholar]
- 52.Zhao, X., M. Rush, and S. Schwartz. 2004. Identification of an hnRNP A1-dependent splicing silencer in the human papillomavirus type 16 L1 coding region that prevents premature expression of the late L1 gene. J. Virol. 7810888-10905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zuker, M. 2003. Mfold Web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 313406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]