Abstract
The parvovirus adeno-associated virus (AAV) contains a small single-stranded DNA genome with inverted terminal repeats that form hairpin structures. In order to propagate, AAV relies on the cellular replication machinery together with functions supplied by coinfecting helper viruses such as adenovirus (Ad). Here, we examined the host cell response to AAV replication in the context of Ad or Ad helper proteins. We show that AAV and Ad coinfection activates a DNA damage response (DDR) that is distinct from that seen during Ad or AAV infection alone. The DDR was also triggered when AAV replicated in the presence of minimal Ad helper proteins. We detected autophosphorylation of the kinases ataxia telangiectasia mutated (ATM) and DNA-dependent protein kinase catalytic subunit (DNA-PKcs) and signaling to downstream targets SMC1, Chk1, Chk2, H2AX, and XRCC4 and multiple sites on RPA32. The Mre11 complex was not required for activation of the DDR to AAV infection. Additionally, we found that DNA-PKcs was the primary mediator of damage signaling in response to AAV replication. Immunofluorescence revealed that some activated damage proteins were found in a pan-nuclear pattern (phosphorylated ATM, SMC1, and H2AX), while others such as DNA-PK components (DNA-PKcs, Ku70, and Ku86) and RPA32 accumulated at AAV replication centers. Although expression of the large viral Rep proteins contributed to some damage signaling, we observed that the full response required replication of the AAV genome. Our results demonstrate that AAV replication in the presence of Ad helper functions elicits a unique damage response controlled by DNA-PK.
Replication of viral genomes produces a large amount of extrachromosomal DNA that may be recognized by the cellular DNA damage machinery. This is often accompanied by activation of DNA damage response (DDR) signaling pathways and recruitment of cellular repair proteins to sites of viral replication. Viruses therefore provide good model systems to study the recognition and response to DNA damage (reviewed in reference 48). The Mre11/Rad50/Nbs1 (MRN) complex functions as a sensor of chromosomal DNA double-strand breaks (DSBs) and is involved in activation of damage signaling (reviewed in reference 41). The MRN complex also localizes to DNA DSBs and is found at viral replication compartments during infection with a number of DNA viruses (6, 40, 47, 70, 75, 77, 87, 93). The phosphatidylinositol 3-kinase-like kinases (PIKKs) ataxia telangiectasia mutated (ATM), ATM and Rad3-related kinase (ATR), and the catalytic subunit of the DNA-dependent protein kinase (DNA-PKcs) are involved in the signal transduction cascades activated by DNA damage (reviewed in references 43, 51, and 71). These kinases respond to distinct types of damage and regulate DSB repair during different phases of the cell cycle (5), either through nonhomologous end-joining (NHEJ) or homologous recombination pathways (reviewed in references 63, 81, and 86). The DNA-PK holoenzyme is composed of DNA-PKcs and two regulatory subunits, the Ku70 and Ku86 heterodimer. DNA-PK functions with XRCC4/DNA ligase IV to repair breaks during NHEJ, and works with Artemis to process DNA hairpin structures during VDJ recombination and during a subset of DNA DSB events (46, 50, 86). While the kinase activity of DNA-PKcs leads to phosphorylation of a large number of substrates in vitro as well as autophosphorylation of specific residues (reviewed in references 16 and 85), it is currently unclear how DNA-PKcs contributes to signaling in cells upon different types of damage.
The adeno-associated virus (AAV) genome consists of a molecule of single-stranded DNA with inverted terminal repeats (ITRs) at both ends that form double-hairpin structures due to their palindromic sequences (reviewed in reference 52). The ITRs are important for replication and packaging of the viral genome and for integration into the host genome. Four viral Rep proteins (Rep78, Rep68, Rep52, and Rep40) are also required for replication and packaging of the AAV genome into virions assembled from the Cap proteins. Although the Rep and Cap genes are replaced in recombinant AAV vectors (rAAV) that retain only the ITRs flanking the gene of interest, these vectors can be replicated by providing Rep in trans (reviewed in reference 7). Productive AAV infection requires helper functions supplied by adenovirus (Ad) or other viruses such as herpes simplex virus (HSV) (reviewed in reference 27), together with components of the host cell DNA replication machinery (54, 55, 58). In the presence of helper viruses or minimal helper proteins from Ad or HSV, AAV replicates in the nucleus at centers where the viral DNA and Rep proteins accumulate (35, 76, 84, 89). Cellular and viral proteins involved in AAV replication, including replication protein A (RPA), Ad DNA-binding protein (DBP), and HSV ICP8, localize with Rep proteins at these viral centers (29, 33, 76).
A number of published reports suggest associations between AAV and the cellular DNA damage machinery. For example, transduction by rAAV vectors is increased by genotoxic agents and DNA damaging treatments (1, 62, 91) although the mechanisms involved remain unclear. Additionally, the ATM kinase negatively regulates rAAV transduction (64, 92), and we have shown that the MRN complex poses a barrier to both rAAV transduction and wild-type AAV replication (11, 67). UV-inactivated AAV particles also appear to activate a DDR involving ATM and ATR kinases that perturbs cell cycle progression (39, 60, 88). It has been suggested that this response is provoked by the AAV ITRs (60) and that UV-treated particles mimic stalled replication forks in infected cells (39). In addition to AAV genome components, the viral Rep proteins have been observed to exhibit cytotoxicity and induce S-phase arrest (3, 65).
The role of cellular repair proteins in AAV genome processing has also been explored by examining the molecular fate of rAAV vectors, which are converted into circular and concatemeric forms that persist episomally (18, 19, 66). Proteins shown to regulate circularization in cell culture include ATM and the MRN complex (14, 64), while in vivo experiments using mouse models have implicated ATM and DNA-PK in this process (14, 20, 72). Additionally, DNA-PKcs and Artemis have recently been shown to cleave the ITR hairpins of rAAV vectors in vivo in a tissue-dependent manner (36). Despite these studies, it is not clear how damage response factors function together and how they impact AAV transduction and replication in human cells.
In this study we examined the cellular response to AAV replication in the context of Ad infection or helper proteins. We show that coinfection with AAV and Ad activates a DDR that is distinct from that seen during infection with Ad alone. The ATM and DNA-PKcs damage kinases are activated and signal to downstream substrates, but the response does not require the MRN complex and is primarily mediated by DNA-PKcs. Although expression of the large Rep proteins induced some DDR events, full signaling appeared to require AAV replication and was accompanied by accumulation of DNA-PK at viral replication compartments. Our results demonstrate that AAV replication induces a unique DNA damage signal transduction response and provides a model system for studying DNA-PK.
MATERIALS AND METHODS
Plasmids and transfections.
The wild-type full-length AAV type 2 genome was supplied by the plasmid pNTC244 (12), and virus was produced by transfection in 293T cells. For production of rAAV vectors, 293T cells were transfected with three plasmids: pXX2, which supplied Rep and Cap proteins (90); pXX6, which contained the Ad helper functions (90); and the vector plasmid pAAV.GFP in which green fluorescent protein (GFP) under the control of the cytomegalovirus (CMV) promoter is cloned between viral ITRs. The Ad helper proteins Ad-DBP and E4orf6 were expressed from the CMV promoter in expression vectors pRK5 or pcDNA3.1 (Clontech). The FLAG-Rep constructs were previously described (59) and were provided by M. Giacca. Rep78 was expressed under the control of the CMV promoter in the pcDNA3.1 plasmid (8). pGL2- and pGL3-based plasmids containing the AAV ITR, p5 promoter, or both elements have been previously described (9). Subconfluent monolayers of cells were transfected by calcium phosphate precipitation according to standard protocols or with Lipofectamine 2000 (Invitrogen) according to the manufacturer's recommendations.
Cell lines and drug treatments.
HeLa, U2OS, and 293 cells were purchased from the American Tissue Culture Collection. A stable cell line derived from HeLa cells that expresses wild-type E1b55K from a retrovirus vector has been described previously (6). A-TLD1 cells and complemented counterparts were previously described (6). NBS cells and complemented counterparts (10) were from P. Concannon. The HCT116-based cell lines (lacking DNA-PKcs or expressing one copy of DNA-PKcs) were provided by E. Hendrickson (61). A-T cells and their complemented counterparts were provided by Y. Shiloh. U2OS cells expressing inducible FLAG-tagged ATR that was kinase dead or wild-type were previously described (57) and were provided by S. Schreiber. All cells, except the MO59J fusion cells, were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10 or 20% fetal bovine serum (FBS) and penicillin/streptomycin, with appropriate selection at 37°C in a humidified atmosphere containing 5% CO2. MO59J fusion cells, Fus9 (lacking DNA-PKcs) and Fus1 (expressing DNA-PKcs), were previously described (34) and were provided by T. Melendy. MO59J fusion cells were maintained in a 1:1 mixture of F10 medium and DMEM supplemented with 10% FBS and penicillin/streptomycin. Where indicated in the figure legends, cells were treated with the ATM inhibitor KU55933, the DNA-PK inhibitor NU7026 (both purchased from Calbiochem and used at a final concentration of 10 μM), or an equal volume of dimethyl sulfoxide (DMSO). Cells were pretreated for 1 h with inhibitors or DMSO before infection and for the duration of infection.
Viruses and infections.
Ad serotype 5 (Ad5) was propagated on 293 cells, purified, and titrated as previously described (6). Wild-type and rAAV vector rAAV.GFP were produced in 293T cells and purified as previously described (30, 90). All AAV titers were determined by quantitative PCR using SYBR Green I double-stranded DNA binding dye and an ABI Prism 7700 Sequence detection system (PE Biosystems). All infections were performed on monolayers of cultured cells in DMEM supplemented with 2% FBS. After 2 h at 37°C, infection medium was replaced with DMEM with 10% or 20% FBS. Infections of Fus cells were performed in a 1:1 mixture of F10 medium and DMEM. Multiplicities of infection (MOIs) are detailed in the figure legends.
Antibodies, immunofluorescence, and immunoblotting.
Commercially available antibodies used in this study were purchased from Abcam (anti-RPA32 phosphorylated at S33 [RPA32-P-533] and RPA32-P-T21), American Research Products, Inc. (Rep clone 303.9), Bethyl (RPA32-P-S4/S8, and Chk1-P-S317), Cell Signaling (Chk2-P-T68 and Chk1-P-S345), Epitomics (ATM), Genetex (Mre11-12D7 and Rad50-13B3), NeoMarkers (DNA-PKcs and Ku70), Novus (Nbs1), Research Diagnostics Inc. (glyceraldehyde-3-phosphate dehydrogenase [GAPDH]), Rockland (ATM phosphorylated at S1981 [ATM-P-S1981] and SMC1 phosphorylated at S957 [SMC1-P-S957]), Santa Cruz (Ku86, ATR, and Chk2), Serotech (XRCC4), Sigma (FLAG-M2), and Upstate (γ-H2AX). The other primary antibodies used in this study were the following: Rep (rabbit polyclonal, a gift from J. Trempe), Rep (IF11, mouse monoclonal; a gift from J. Samulski), Ad-DBP (mouse monoclonal, a gift from A. Levine; and rabbit polyclonal, a gift from P. van der Vliet), E1b55K (mouse monoclonal B-6, a gift from A. Levine), RPA32 (mouse monoclonal, a gift from T. Melendy), and DNA-PK-P-S2056 (DNA-PK phosphorylated at S2056; rabbit polyclonal; a gift from B. Chen). Secondary antibodies for immunofluorescence were coupled to Alexa fluorophores (Invitrogen). Cells grown on coverslips in 24-well plates were processed for immunofluorescence as previously described (6, 75). Images were obtained on a Nikon microscope in conjunction with a charge-coupled-device camera (Cooke Sensicam) in double or triple excitation mode and processed using SlideBook and Adobe Photoshop. Immunoblotting was performed as previously described (6).
RESULTS
AAV and Ad coinfection induces an MRN-independent DDR.
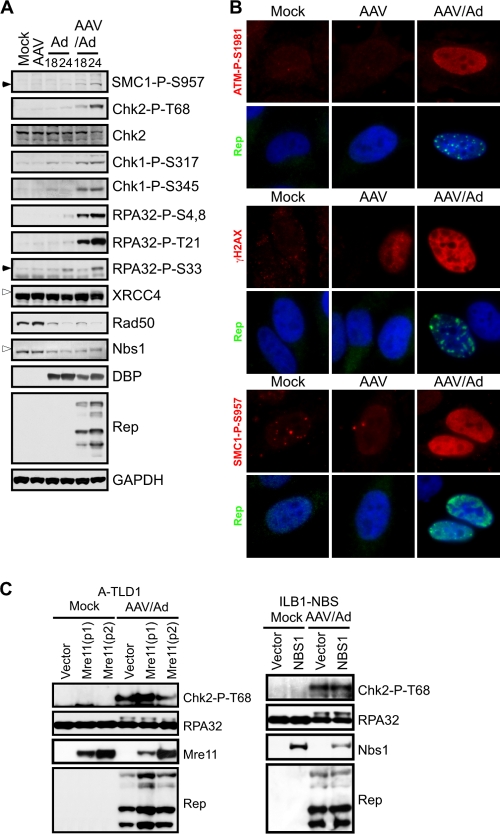
We examined the cellular DDR to replicating AAV in the context of Ad helper virus. U2OS cells were infected with AAV and Ad, either alone or in combination (Fig. 1). Phosphorylation of damage response proteins was detected by immunoblotting (Fig. 1A) and visualized by immunofluorescence (Fig. 1B). Infections were confirmed by immunoblotting with antibodies to the DBP of Ad and Rep proteins of AAV. We have previously demonstrated that wild-type Ad5 infection does not induce significant signaling by the cellular damage machinery due to inactivation of the MRN complex, a sensor of DNA DSBs (6). Infection with Ad or AAV alone did not significantly activate DNA damage signaling. In contrast, we found that AAV and Ad coinfection generated robust signaling to DDR substrates, as revealed by phospho-specific antibodies to SMC1, Chk1, Chk2, H2AX, and RPA32. Consistent with their phosphorylation (26, 42, 49), XRCC4 and Nbs1 exhibited gel mobility shifts. Additionally, we noted the activation of ATM using a phospho-specific antibody recognizing the autophosphorylation site at S1981 (2). Interestingly, immunofluorescence revealed that phosphorylated ATM, SMC1, and H2AX were not localized to AAV replication centers but exhibited diffusely nuclear staining patterns (Fig. 1B). We also found that AAV infection alone induced some H2AX phosphorylation (γ-H2AX) by immunofluorescence although the intensity was much less than that seen during coinfection. Together, these data show that AAV and Ad coinfection elicits a robust DDR not seen significantly with either virus alone.
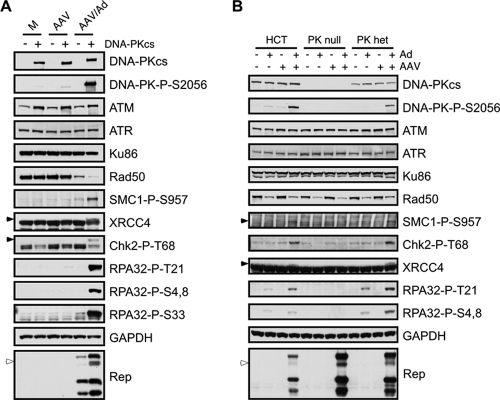
FIG. 1.
AAV and Ad coinfection induces an MRN-independent DDR. (A) DNA damage signaling induced by AAV and Ad coinfection. U2OS cells were infected with Ad (MOI of 25) and AAV (MOI of 2,000) alone or in combination. Cells were harvested at the indicated times and processed for immunoblotting. Open arrowheads indicate slower-migrating phosphorylated proteins and closed arrowheads indicate the phosphorylated protein of interest. GAPDH served as a loading control. (B) HeLa cells were infected with the indicated viruses for approximately 20 h before being fixed and processed for immunofluorescence. Cells were stained with the indicated antibodies to mark activated DDR proteins and Rep centers, and DAPI (4′,6′-diamidino-2-phenylindole) was used to stain the DNA of cell nuclei. (C) DNA damage signaling in response to AAV and Ad coinfection still occurs in cell lines lacking a functional MRN complex. A-TLD1 and NBS cells and their complemented counterparts were uninfected (Mock) or infected with AAV (MOI of 1,000) and Ad5 (MOI of 50) for 30 h. Cells were harvested and processed for immunoblotting with the indicated antibodies. Mre11-P1 and P2 represent two different pools of A-TLD1 cells transduced to express wild-type Mre11.
Since Ad degrades the MRN complex to prevent a DDR during virus infection (6), we examined MRN during coinfection with AAV. We found that AAV coinfection did not affect the ability of Ad to degrade the MRN complex, as revealed by a decrease in total levels of Rad50 and Nbs1 proteins (Fig. 1A). This also suggests that MRN is not required for the AAV-induced damage response. To confirm this observation, we analyzed the damage signaling in response to AAV and Ad coinfection in cells lacking functional MRN. Cells with mutant Mre11 (A-TLD1 cells) and Nbs1 (ILB1-NBS cells) were infected with both viruses and compared to infected cells that were complemented with the respective wild-type cDNAs (Fig. 1C). Immunoblotting demonstrated that signaling to Chk2 (detected with a phospho-specific antibody) and RPA32 (shown by a mobility shift) was observed in mutant cells to a similar extent as in complemented cells. This shows that the DDR to AAV and Ad coinfection occurs independently of a functional MRN complex.
The effects of Rep on the DNA damage response.
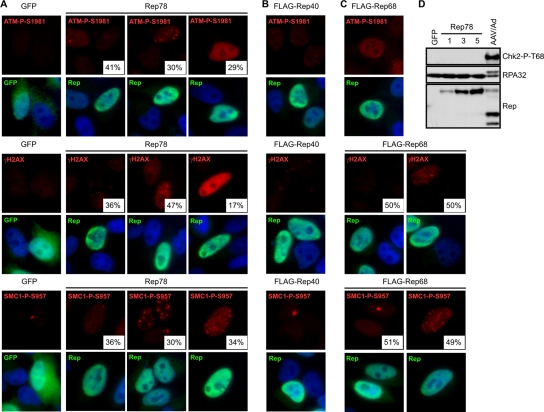
It has been previously shown that the larger AAV Rep proteins (Rep78 and Rep68) can both nick cellular DNA and induce cell cycle arrest (3, 65). We therefore tested whether expression of Rep proteins (Rep78, Rep68, and Rep40) contributes to the damage response we observed. Cells were transfected with plasmids encoding Rep78, FLAG-Rep68, FLAG-Rep40, or GFP as a control, and damage signaling was assessed by immunofluorescence and immunoblotting (Fig. 2). Cells expressing Rep78 displayed some phosphorylation of ATM, H2AX, and SMC1 while GFP-positive cells had none (Fig. 2A). A number of different staining patterns were observed in cells expressing Rep78, and the relative distribution was quantitated. Some cells with robust Rep78 expression had minimal damage signaling while other cells exhibited either distinct nuclear foci or a pan-nuclear staining pattern. Rep40, on the other hand, did not elicit any damage signaling in these experiments (Fig. 2B). Like Rep78, Rep68 induced activation of ATM, H2AX, and SMC1 in some cells; however, these proteins exhibited foci as opposed to the pan-nuclear activation seen during AAV and Ad coinfection (Fig. 2C). When we examined signaling to downstream substrates in the DDR, we found that Chk2 and RPA32 were not significantly phosphorylated upon Rep78 expression (Fig. 2D). Together, these data suggest that the larger Rep proteins can contribute to some DNA damage signaling, but that they do not account for all events observed during coinfection.
FIG. 2.
The effects of Rep proteins on DNA damage signaling. HeLa cells were transfected with plasmids encoding GFP or Rep proteins. Cells were fixed for immunofluorescence approximately 36 h posttransfection and stained with the indicated antibodies to mark activated DDR proteins or Rep and with DAPI (4′,6′-diamidino-2-phenylindole) to stain the nuclei. At least 100 cells were quantitated for the indicated percentages of each phenotype. (A) Rep78 activates DNA damage signaling. (B) FLAG-Rep40 does not activate damage signaling. (C) Damage signaling during FLAG-Rep68 expression. (D) Rep78 does not induce downstream signaling. HeLa cells were transfected with a plasmid encoding GFP or increasing amounts of a plasmid encoding Rep78. Total DNA transfected was the same between samples and was made up with pRK5. Cells were harvested 48 h posttransfection and processed for immunoblotting with the indicated antibodies. Coinfection with AAV and Ad served as a positive control.
AAV replication is sufficient to induce a DDR.
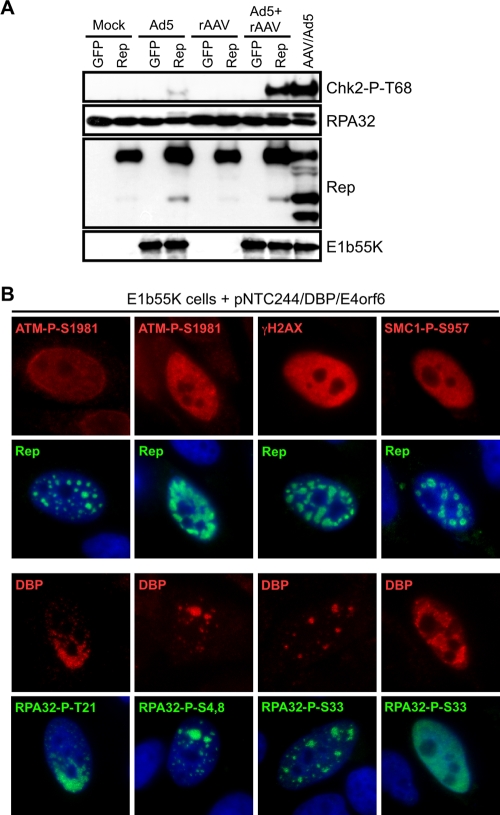
We next examined the contribution of AAV replication to the cellular damage response. Cells were infected with rAAV or Ad5, alone or in combination, and cells were subsequently transfected to express either Rep78 or GFP as a control. DNA damage signaling was analyzed by immunoblotting of cell lysates (Fig. 3A). Phosphoryation of Chk2 and RPA32 was not detected in cells expressing Rep78 or in cells infected with rAAV alone in the presence or absence of Rep expression. Although Ad5 infection alone did not activate damage signaling, Rep78 expression in the presence of Ad5 induced a low level of phosphorylation on Chk2 and RPA32. This may reflect Rep interference with Ad replication (38, 84) or replication of human sequences mediated by Rep protein (79). However, robust DNA damage signaling was observed when rAAV was infected in the presence of Rep78 and Ad5, a condition sufficient to support rAAV replication. Phosphorylation of Chk2 and RPA32 was similar to that observed during wild-type AAV and Ad coinfection, suggesting that replication of the AAV genome may elicit the damage response.
FIG. 3.
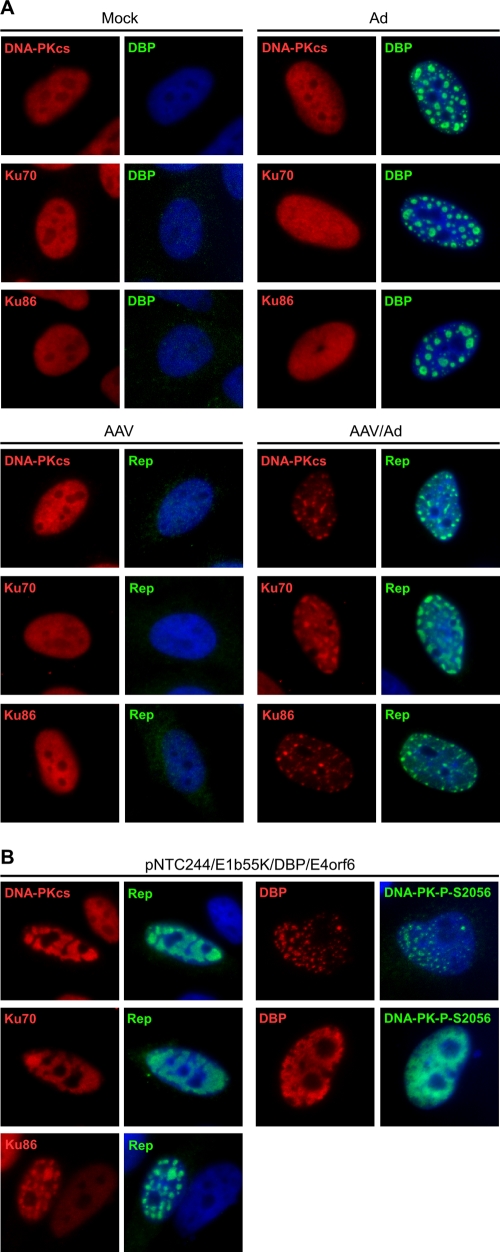
AAV replication induces a DNA damage response. (A) HeLa cells were either uninfected (Mock) or infected with Ad5 (MOI of 50), rAAV-GFP (MOI of 1,000), or a combination of both viruses. At 6 h postinfection, cells were transfected with plasmids encoding GFP or Rep78 and harvested 30 h posttransfection. Lysates were processed for immunoblotting against the indicated proteins. Coinfection with wild-type AAV and Ad5 served as a positive control. (B) HeLa cells transduced to express E1b55K were transfected with pNTC244 and plasmids expressing DBP and E4orf6. At approximately 36 h posttransfection, cells were fixed for immunofluorescence and stained with the indicated antibodies. AAV replication centers are marked by DBP or Rep. DAPI (4′,6′-diamidino-2-phenylindole) was used to mark cell nuclei.
AAV replication can be achieved by transfection of an infectious clone of the AAV genome together with the minimal helper genes from Ad (DBP, E1b55K, and E4orf6 genes) (67, 76). To examine the requirements for damage signaling in more detail, we used immunofluorescence to analyze AAV replication with this minimal helper system (Fig. 3B). A HeLa-derived cell line expressing wild-type E1b55K (6) was transfected with plasmids expressing Ad helpers DBP and E4orf6 and a plasmid encoding the AAV genome, pNTC244 (12). Expression of the helper proteins alone in the E1b55K cell line did not activate a damage response (data not shown). However, cells that contained discrete AAV replication centers displayed phosphorylation of ATM, H2AX, SMC1, and RPA32 (Fig. 3B). Some cells with smaller Rep compartments had weaker staining for phosphorylated ATM. Phosphorylated RPA32 was found colocalized with AAV centers except at late stages, when RPA32-P-S33 staining was spread throughout the nucleoplasm to a greater degree. Together, our data indicate that the damage signaling observed during AAV and Ad coinfection is also activated with minimal helper-mediated AAV replication and demonstrate that replication of the AAV genome is sufficient to activate the cellular DDR.
DNA-PK mediates phosphorylation of damage response proteins during AAV and Ad coinfection.
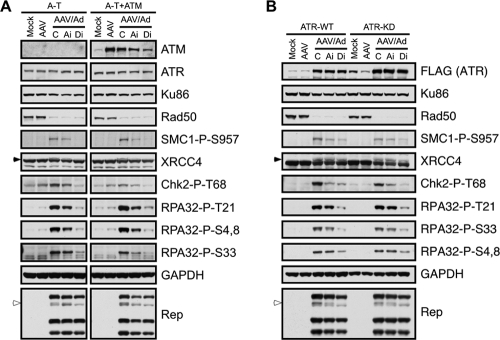
To determine the role of individual PIKKs in virus-induced damage signaling, we examined AAV and Ad coinfections in mutant cell lines and in the presence of chemical inhibitors. Infections were first carried out in ATM-deficient A-T cells and their complemented counterparts (Fig. 4A). In A-T cells, we observed that coinfection induced phosphorylation of SMC1, XRCC4, Chk2, and RPA32, indicating that the signaling was not dependent upon ATM. This conclusion was supported by treatment of matched ATM-expressing cells with a small molecule inhibitor of ATM, which only minimally affected most signaling events (Fig. 4A and data not shown). Phosphorylation of most of these substrates was decreased in the presence of a DNA-PK inhibitor, implicating a role for DNA-PKcs in the signaling response to AAV replication. We next examined the role of ATR in cells expressing doxycycline-inducible wild-type or kinase-dead ATR that acts as a dominant-negative mutant (57). Phosphorylation of SMC1, XRCC4, Chk2, and RPA32 was observed after AAV and Ad coinfection even in the presence of kinase-dead ATR (Fig. 4B), suggesting that ATR is not absolutely required. In further support of this conclusion, we found that caffeine treatment of A-T cells at a concentration that inhibits both ATM and ATR (2.5 mM) had a minimal effect on the DDR during AAV and Ad coinfection (data not shown). Although treatment of kinase-dead ATR cells with the ATM inhibitor slightly diminished some phosphorylation events, they were more drastically affected by the DNA-PK inhibitor. In these experiments we obtained inconsistent results for Chk1 phosphorylation and were therefore unable to conclude which PIKK activated Chk1. In addition to the phosphorylation of DNA damage substrates, we observed slower-migrating forms of the larger Rep proteins. This was most discernible with Rep68, which appeared as a doublet (Fig. 4A and B). In the presence of the DNA-PK inhibitor, this form of Rep68 was lost, with no major effect on the total Rep levels. Together, these data suggest that DNA-PK may be the predominant PIKK responsible for the DDR to AAV replication and that Rep may also be a phosphorylation substrate.
FIG. 4.
ATM and ATR are not the kinases predominantly responsible for signaling in response to AAV and Ad coinfection. Immunoblotting was used to analyze AAV-induced damage signaling. Cells with individual PIKKs inactivated were either uninfected (Mock) or infected with AAV (MOI of 2,000) or AAV and Ad (MOI of 25). Infections were performed in the presence of DMSO as a control (C) or inhibitors to ATM (Ai) and DNA-PKcs (Di). Mock- and AAV-infected cells were also treated with DMSO. Cells were harvested 24 h postinfection and processed for immunoblotting with the indicated antibodies. (A) Infections in A-T cells or a matched line complemented with ATM. Right- and left-hand panels were from the same gels, and lanes not relevant to these results were removed from the figure. (B) Infections in cells expressing inducible ATR that is wild-type (ATR-WT) or kinase-dead (ATR-KD). Cells were induced to express ATR by doxycycline treatment for 48 h before infections, which then proceeded for a further 24 h. The inducible ATR protein is tagged with a FLAG epitope. GAPDH served as a loading control. The open arrowheads indicate a slower-migrating band for Rep68, and the filled arrowheads highlight specific phosphorylated proteins.
To investigate further the dependence on DNA-PK, we infected two different DNA-PKcs-deficient cell lines with both viruses and analyzed AAV-induced damage signaling by immunoblotting. We first compared signaling between two MO59J-derived cell lines that lack or express DNA-PKcs (34) and found that most phosphorylation events during AAV and Ad coinfection were dependent on this kinase (Fig. 5A). Minimal signaling was detected in the absence of DNA-PKcs and during infections with AAV or Ad alone. Additionally, we noted the activation of DNA-PKcs, as measured by a phospho-specific antibody against S2056 (13). Since levels of ATM were slightly lower in the cells that lacked DNA-PKcs (34) (Fig. 5A), we also tested damage signaling in another set of matched DNA-PKcs-deficient cells. We analyzed infections from HCT116-derived cells in which DNA-PKcs was knocked out using homologous recombination to generate cells heterozygous or null for the kinase (61). Immunoblotting of lysates from these cells and the parental HCT116 cells confirmed that AAV and Ad coinfection induced damage signaling mediated by DNA-PKcs (Fig. 5B). Interestingly, one copy of DNA-PKcs was sufficient for this response (Fig. 5B, PK het lanes). We also observed that the doublet of Rep68 was lost in both sets of DNA-PKcs-deficient cells (Fig. 5), further suggesting that Rep is a likely target of this kinase. These data confirm that DNA-PKcs is activated and induces many of the signaling events during the response to AAV and Ad coinfection.
FIG. 5.
DNA-PKcs is required for signaling in response to AAV and Ad coinfection. Immunoblotting was used to analyze damage signaling to AAV and Ad coinfection in cells that lack DNA-PKcs. Cells were either uninfected or infected with AAV (MOI of 1,000 to 2,000), Ad (MOI of 25), or both viruses and harvested 24 h postinfection. Lysates were processed for immunoblotting with the indicated antibodies. The open arrowheads indicate a slower-migrating band for Rep68, and the filled arrowheads highlight specific phosphorylated proteins. (A) Infections in M059J-derived cells lines that lack DNA-PKcs (Fus9) or express the kinase (Fus1). (B) Infections in HCT-derived cells that are heterozygous or null for DNA-PKcs.
We next attempted to determine how DNA-PK activation affects AAV using these two sets of DNA-PKcs-deficient cells. However, we obtained inconsistent results between the mutant cell lines when we measured both rAAV transduction and wild-type AAV replication (data not shown). Additionally, we found that treatment of cells with the small-molecule DNA-PK inhibitor did not affect levels of Rep proteins (Fig. 4) or viral replication (data not shown). While it is unclear from these data how DNA-PK affects AAV transduction and replication, it is evident that DNA-PKcs is activated and induces most of the signaling events during AAV and Ad coinfection.
DNA-PK localizes to AAV replication centers.
Given the involvement of DNA-PK in the damage response to AAV, we next examined the intracellular localization of holoenzyme components (DNA-PKcs, Ku70, and Ku86) during AAV replication. Immunofluorescence of mock-treated cells showed that all three proteins were located diffusely throughout the nucleus, and this pattern was unaffected by infections with Ad or AAV alone (Fig. 6A). In contrast, during AAV and Ad coinfection, the DNA-PKcs, Ku70, and Ku86 proteins all became concentrated at Rep-staining compartments (Fig. 6A). The dramatic redistribution of DNA-PK components to AAV centers was also observed under minimal replication conditions induced by transfection of the AAV genome and Ad helper proteins (DBP and E4orf6) into wild-type E1b55K protein-expressing cells (Fig. 6B). Additionally, we noted that autophosphorylated DNA-PKcs localized with DBP at the AAV replication compartments. These data suggest that AAV replication centers serve as a signal for DNA-PK recognition and activation during the DDR.
FIG. 6.
DNA-PK components localize to AAV replication centers. (A) DNA-PK components localize to viral replication centers during AAV and Ad coinfection. HeLa cells were uninfected (Mock) or infected with Ad5 (MOI of 25), AAV (MOI of 1,000), or both viruses for 24 h before fixing for immunofluorescence. Cells were stained with the indicated antibodies. Viral replication centers are detected with antibodies to DBP and Rep. DAPI (4′,6′-diamidino-2-phenylindole) marks the cell nuclei. (B) HeLa cells expressing E1b55K were transfected with pNTC244, DBP, and E4orf6 expression plasmids for approximately 36 h before cells were fixed for immunofluorescence. Viral Rep centers and DAPI staining are as described in panel A.
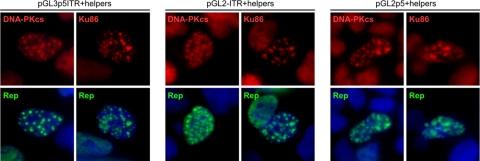
Since AAV replication promoted DNA-PK holoenzyme relocalization, we further examined the requirements for recruitment of these proteins. Previously, it was shown that plasmids containing an AAV ITR or p5 element can establish replication centers in the presence of Rep and HSV proteins (28). To test whether these AAV elements are sufficient to induce DNA-PK localization, we transfected 293 cells with luciferase-based plasmids containing an ITR, p5 promoter, or both components (9) in the presence of Rep and Ad helpers. DNA-PK component accumulation at replication centers was examined by immunofluorescence. When 293 cells were transfected to express Rep, DBP, and E4orf6 alone, there was no effect on DNA-PK distribution (data not shown). However, when replication centers were established using plasmids containing the ITR origin, we found that DNA-PKcs, Ku70, and Ku86 all localized to these compartments (Fig. 7 and data not shown). Although we found the p5 promoter to be a much less efficient origin for Rep-mediated replication, any replication centers that were formed with this plasmid also showed costaining for DNA-PK components. These data demonstrate that replication centers established with AAV cis sequences are sufficient to recruit the DNA-PK holoenzyme.
FIG. 7.
DNA-PK is recruited to Rep-mediated replication compartments. 293 cells were transfected with pGL-based plasmids containing the 5′ ITR (pGL2-ITR), the p5 promoter (pGL3-p5), or both elements (pGL3-p5ITR) in the presence and absence (not shown) of E4orf6, DBP, and Rep78 expression plasmids. Cells were fixed approximately 36 h posttransfection and processed for immunofluorescence with the indicated antibodies. Rep marks replication centers induced by all pGL-based plasmids, and DAPI (4′,6′-diamidino-2-phenylindole) stains the cell nuclei.
DISCUSSION
In this study we examined the cellular response to replicating AAV and found that a robust cellular DDR was induced. Immunoblotting and immunofluorescence demonstrated that a number of DNA damage substrates, including ATM, DNA-PK, Chk1, Chk2, SMC1, and H2AX, were strongly phosphorylated during AAV and Ad coinfection and during AAV replication with minimal Ad helper proteins. Under these conditions, we found that the majority of signaling events were dependent on DNA-PK rather than ATM and ATR. Additionally, the damage response was independent of the MRN complex. This latter observation was surprising, given the requirement for MRN in signaling elicited by mutant Ad infection (6), HSV-1 infection (47), and chromosomal DSBs (6, 41, 80). In the situations where MRN is required, ATM orchestrates damage signaling, whereas we found that in our experiments DNA-PK was the primary kinase mediating the DDR during AAV replication. Interestingly, H2AX is also phosphorylated by DNA-PKcs after DSBs, but only when MRN and ATM are absent (22, 74). MRN and/or ATM may therefore potentially influence DNA-PK signaling to certain substrates. This may be particularly relevant to AAV since ATM is known to have a negative effect on rAAV (64, 92), and MRN negatively impacts AAV transduction and replication (11, 67). Under Ad helper conditions where MRN is present at AAV centers (11, 67), we observed differences in the accumulation of repair factors at AAV centers (R. A. Schwartz and M. D. Weitzman, unpublished data), suggesting that MRN may impact the organization of viral replication compartments. Taken together, these results suggest that the cellular response to AAV replication may be affected by several DNA damage response regulators, including MRN, ATM, and DNA-PK.
In our experiments we also detected the DNA-PK-dependent phosphorylation of RPA32 on multiple sites and localization of these activated forms at AAV centers. RPA, replication factor C (RFC), PCNA, minichromosome maintenance proteins, and DNA polymerase δ have been previously shown to be required for AAV replication in vitro and in cells (54, 55, 58). Interestingly, RPA phosphorylation excludes it from cellular sites of replication (25, 82) and may alter its DNA affinity in order to promote repair (23). We have also previously shown that Rep can bind RPA (76) although we do not know how this interaction may be affected by phosphorylation. In addition to RPA32, it is possible that other cellular replication proteins are modified during the AAV-induced DDR. Cellular minichromosome maintenance proteins are phosphorylated during DNA damage (17, 37, 68) while RFC2/RFC4 and PCNA can become ubiquitinated (44, 78). It will be interesting to determine how AAV-induced signaling impacts these replication factors and their function during productive AAV replication.
We found that activated DNA-PKcs, Ku70, and Ku86 all localized to AAV centers during infection and to compartments formed during Rep-mediated replication of AAV origins (p5 and the ITR), further supporting the role of DNA-PK as a key responder to AAV replication. These data suggest that AAV cis sequences may provide a signal for recognition by DNA-PK, perhaps through the formation of DNA hairpin structures, which are known to activate DNA-PK (50, 85). Protein-protein interactions could also contribute to the recruitment or maintenance of DNA-PK components at viral centers. Indeed, recent proteomic studies identified the Ku70/Ku86 complex and DNA-PKcs as Rep-interacting proteins (56, 59). Additionally, RPA and other proteins that localize to AAV centers may help to stabilize DNA-PK at AAV compartments. This observation is consistent with DNA-PK accumulation at HSV-1 centers via either the viral ICP8 protein or RPA (77, 87). We did not observe DNA-PK localization at Ad replication centers for wild-type or mutant virus (Schwartz and Weitzman, unpublished data), which was surprising given the involvement of DNA-PK in concatemerization of mutant Ad genomes (4, 75). It is possible that association with Ad centers is masked under our immunofluorescence conditions or is too transient to detect in these experiments. Based on our data, recruitment of DNA-PK appears to be specific to AAV replication centers and may be due to a combination of AAV structures and protein interactions.
Multiple viral factors are likely responsible for activation of the DDR during AAV replication. Consistent with a previous report (3), we found that expression of the larger Rep proteins resulted in some damage signaling, perhaps due to their DNA nicking capabilities. However, we also saw that damage signaling was stronger during AAV replication, implying that Rep expression cannot account for all of the DDR events. AAV ITRs have single-strand/double-strand DNA junctions that could resemble stalled replication forks, and it has been suggested that AAV induces a DDR that mimics these structures (39). Although AAV ITRs have been proposed to be sufficient to activate a DDR (60), subsequent work demonstrated that rAAV vectors do not provoke this response (24). Instead, wild-type AAV, UV-inactivated particles, or rAAV genomes with a p5 element were required to induce damage signaling (24). These previous reports have mainly involved very high MOIs (MOIs of 10,000 to 20,000) in the absence of helper proteins (24, 39, 60). In accordance with these studies, we observed that wild-type AAV infection alone resulted in some H2AX phosphorylation even at a much lower MOI. Since damage signaling was far more robust during viral replication, the full DDR is most likely activated in response to a combination of Rep functions, ITRs, and replication intermediates. We detected the damage phosphorylation events with minimal helper proteins in the absence of the E1a gene, and therefore this Ad helper protein is not directly required for the response to replicating AAV. Since E1a activates the p5 promoter to induce Rep expression and thus increase replication (52), the response is likely to be quantitatively increased in the presence of all the Ad helper functions. During natural infection, the MOI of AAV may be lower than that used in our experiments, and only the minimal response will be activated. In the absence of helper functions, the virus will integrate in to the host genome (52), and it is likely that the cellular DNA repair machinery will play an important role in integration.
During the course of our analysis of the AAV-induced DDR, we noticed altered migration of the large Rep proteins. Our results are consistent with other reports showing that all Rep proteins are phosphorylated during AAV and Ad coinfection (15). Although the kinases involved and the sites they modify on Rep are largely unknown, Rep phosphorylation has been correlated with decreased DNA binding and AAV replication (15, 53). Our data suggest that DNA-PKcs may mediate modification of the large Rep proteins. This result draws an interesting parallel to the simian virus 40 (SV40) large tumor antigen, which has similar functions to Rep78/68 during SV40 replication and is phosphorylated by ATM during infection (69). Phosphorylation by DNA-PK also inactivates large tumor antigen during in vitro SV40 DNA replication (83). The preferred target sites for phosphorylation by PIKKs are SQ and TQ motifs. The large Rep proteins share five of these sites while Rep78 contains one additional C-terminal SQ motif. It will be interesting to determine whether these sites are modified by DNA-PKcs during AAV replication and what impact this has on Rep functions.
In addition to AAV, DNA-PK has been previously implicated in the cellular response to infection by a number of other viruses (48). Some viral proteins subvert DNA-PK repair functions, such as the Tax oncoprotein of human T-cell leukemia virus type 1 (21) and the ICP0 protein of HSV-1 which degrades DNA-PKcs (45). The Ad E4orf6 and E4orf3 have also been reported to bind DNA-PK and to inhibit NHEJ (4, 32, 75). E4orf6 may indirectly affect DNA-PK and its autophosphorylation by inhibiting protein phosphatase 2A activity (31, 32). While our studies have established AAV replication as a model system to study DNA-PK, the role of this kinase beyond signaling is not completely clear. One previous report suggested that DNA-PKcs inhibits AAV integration (73), which may benefit replication of the episomal genome. Other in vivo studies have implicated DNA-PKcs in rAAV vector processing in certain murine tissues (14, 20, 36, 64, 72). However, DNA-PKcs did not affect transduction unless gene expression required vector circularization (14). Despite many attempts, we have obtained conflicting AAV transduction and replication data when we compared infections between the two sets of DNA-PKcs-deficient cells employed in the experiment shown in Fig. 5 (data not shown). The impact of DNA-PK signaling on AAV replication and transduction may depend on the cellular context and the viral genome structure. In this report we have uncovered a unique role for DNA-PK in AAV-induced damage signaling, expanding our knowledge of the links between viral replication and cellular DDR pathways.
ADDENDUM IN PROOF
After this paper was accepted, an independent study also observed a cellular DNA damage response activated by AAV and Ad coinfection with DNA-PD as the most prominent kinase mediating these effects (R. F. Collaco, J. M. Bevington, V. Bhrigu, V. Kalman-Maltese, and J. P. Trempe, Virology, in press).
Acknowledgments
We thank A. Berk, B. Chen, D. Chen, P. Concannon, M. Giacca, E. Hendrickson, J. Karlseder, A. Levine, T. Melendy, J. Petrini, R. J. Samulski, S. Schreiber, Y. Shiloh, J. Trempe, and P. van der Vliet for reagents. We are grateful to D. Lee and D. Linfesty for technical support. We thank members of the Weitzman lab for helpful discussions and critical readings of the manuscript. We acknowledge the James B. Pendleton Charitable Trust for providing the microscopy facility.
Work in the Weitzman lab is partially supported by a Pioneer Developmental Chair. This work was supported in part by NIH grants CA97093 and AI43341 (M.D.W.) and by gifts from the Joe W. & Dorothy Dorsett Brown Foundation and the Lebensfeld Foundation to M.D.W. R.A.S. and C.T.C. were supported in part by NIH Training Grants to UCSD and the Salk Institute and by scholarships from the ARCS Foundation.
Footnotes
Published ahead of print on 1 April 2009.
REFERENCES
- 1.Alexander, I. E., D. W. Russell, and A. D. Miller. 1994. DNA-damaging agents greatly increase the transduction of nondividing cells by adeno-associated virus vectors. J. Virol. 688282-8287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bakkenist, C. J., and M. B. Kastan. 2003. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature 421499-506. [DOI] [PubMed] [Google Scholar]
- 3.Berthet, C., K. Raj, P. Saudan, and P. Beard. 2005. How adeno-associated virus Rep78 protein arrests cells completely in S phase. Proc. Natl. Acad. Sci. USA 10213634-13639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyer, J., K. Rohleder, and G. Ketner. 1999. Adenovirus E4 34k and E4 11k inhibit double strand break repair and are physically associated with the cellular DNA-dependent protein kinase. Virology 263307-312. [DOI] [PubMed] [Google Scholar]
- 5.Branzei, D., and M. Foiani. 2008. Regulation of DNA repair throughout the cell cycle. Nat. Rev. Mol. Cell Biol. 9297-308. [DOI] [PubMed] [Google Scholar]
- 6.Carson, C. T., R. A. Schwartz, T. H. Stracker, C. E. Lilley, D. V. Lee, and M. D. Weitzman. 2003. The Mre11 complex is required for ATM activation and the G2/M checkpoint. EMBO J. 226610-6620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carter, B. J. 2004. Adeno-associated virus and the development of adeno-associated virus vectors: a historical perspective. Mol. Ther. 10981-989. [DOI] [PubMed] [Google Scholar]
- 8.Cassell, G. D., and M. D. Weitzman. 2004. Characterization of a nuclear localization signal in the C-terminus of the adeno-associated virus Rep68/78 proteins. Virology 327206-214. [DOI] [PubMed] [Google Scholar]
- 9.Cathomen, T., T. H. Stracker, L. B. Gilbert, and M. D. Weitzman. 2001. A genetic screen identifies a cellular regulator of adeno-associated virus. Proc. Natl. Acad. Sci. USA 9814991-14996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cerosaletti, K. M., A. Desai-Mehta, T. C. Yeo, M. Kraakman-Van Der Zwet, M. Z. Zdzienicka, and P. Concannon. 2000. Retroviral expression of the NBS1 gene in cultured Nijmegen breakage syndrome cells restores normal radiation sensitivity and nuclear focus formation. Mutagenesis 15281-286. [DOI] [PubMed] [Google Scholar]
- 11.Cervelli, T., J. A. Palacios, L. Zentilin, M. Mano, R. A. Schwartz, M. D. Weitzman, and M. Giacca. 2008. Processing of recombinant AAV genomes occurs in specific nuclear structures that overlap with foci of DNA-damage-response proteins. J. Cell Sci. 121349-357. [DOI] [PubMed] [Google Scholar]
- 12.Chejanovsky, N., and B. J. Carter. 1989. Replication of a human parvovirus nonsense mutant in mammalian cells containing an inducible amber suppressor. Virology 171239-247. [DOI] [PubMed] [Google Scholar]
- 13.Chen, B. P., D. W. Chan, J. Kobayashi, S. Burma, A. Asaithamby, K. Morotomi-Yano, E. Botvinick, J. Qin, and D. J. Chen. 2005. Cell cycle dependence of DNA-dependent protein kinase phosphorylation in response to DNA double strand breaks. J. Biol. Chem. 28014709-14715. [DOI] [PubMed] [Google Scholar]
- 14.Choi, V. W., D. M. McCarty, and R. J. Samulski. 2006. Host cell DNA repair pathways in adeno-associated viral genome processing. J. Virol. 8010346-10356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Collaco, R., K. M. Prasad, and J. P. Trempe. 1997. Phosphorylation of the adeno-associated virus replication proteins. Virology 232332-336. [DOI] [PubMed] [Google Scholar]
- 16.Collis, S. J., T. L. DeWeese, P. A. Jeggo, and A. R. Parker. 2005. The life and death of DNA-PK. Oncogene 24949-961. [DOI] [PubMed] [Google Scholar]
- 17.Cortez, D., G. Glick, and S. J. Elledge. 2004. Minichromosome maintenance proteins are direct targets of the ATM and ATR checkpoint kinases. Proc. Natl. Acad. Sci. USA 10110078-10083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duan, D., P. Sharma, L. Dudus, Y. Zhang, S. Sanlioglu, Z. Yan, Y. Yue, Y. Ye, R. Lester, J. Yang, K. J. Fisher, and J. F. Engelhardt. 1999. Formation of adeno-associated virus circular genomes is differentially regulated by adenovirus E4 ORF6 and E2a gene expression. J. Virol. 73161-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duan, D., P. Sharma, J. Yang, Y. Yue, L. Dudus, Y. Zhang, K. J. Fisher, and J. F. Engelhardt. 1998. Circular intermediates of recombinant adeno-associated virus have defined structural characteristics responsible for long-term episomal persistence in muscle tissue. J. Virol. 728568-8577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duan, D., Y. Yue, and J. F. Engelhardt. 2003. Consequences of DNA-dependent protein kinase catalytic subunit deficiency on recombinant adeno-associated virus genome circularization and heterodimerization in muscle tissue. J. Virol. 774751-4759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Durkin, S. S., X. Guo, K. A. Fryrear, V. T. Mihaylova, S. K. Gupta, S. M. Belgnaoui, A. Haoudi, G. M. Kupfer, and O. J. Semmes. 2008. HTLV-1 Tax oncoprotein subverts the cellular DNA damage response via binding to DNA-dependent protein kinase. J. Biol. Chem. 28336311-36320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Falck, J., J. Coates, and S. P. Jackson. 2005. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature 434605-611. [DOI] [PubMed] [Google Scholar]
- 23.Fanning, E., V. Klimovich, and A. R. Nager. 2006. A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res. 344126-4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fragkos, M., M. Breuleux, N. Clement, and P. Beard. 2008. Recombinant adeno-associated viral vectors are deficient in provoking a DNA damage response. J. Virol. 827379-7387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Francon, P., J. M. Lemaitre, C. Dreyer, D. Maiorano, O. Cuvier, and M. Mechali. 2004. A hypophosphorylated form of RPA34 is a specific component of pre-replication centers. J. Cell Sci. 1174909-4920. [DOI] [PubMed] [Google Scholar]
- 26.Gatei, M., D. Young, K. M. Cerosaletti, A. Desai-Mehta, K. Spring, S. Kozlov, M. F. Lavin, R. A. Gatti, P. Concannon, and K. Khanna. 2000. ATM-dependent phosphorylation of nibrin in response to radiation exposure. Nat. Genet. 25115-119. [DOI] [PubMed] [Google Scholar]
- 27.Geoffroy, M. C., and A. Salvetti. 2005. Helper functions required for wild type and recombinant adeno-associated virus growth. Curr. Gene Ther. 5265-271. [DOI] [PubMed] [Google Scholar]
- 28.Glauser, D. L., O. Saydam, N. A. Balsiger, I. Heid, R. M. Linden, M. Ackermann, and C. Fraefel. 2005. Four-dimensional visualization of the simultaneous activity of alternative adeno-associated virus replication origins. J. Virol. 7912218-12230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Glauser, D. L., R. Strasser, A. S. Laimbacher, O. Saydam, N. Clement, R. M. Linden, M. Ackermann, and C. Fraefel. 2007. Live covisualization of competing adeno-associated virus and herpes simplex virus type 1 DNA replication: molecular mechanisms of interaction. J. Virol. 814732-4743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grifman, M., M. Trepel, P. Speece, L. B. Gilbert, W. Arap, R. Pasqualini, and M. D. Weitzman. 2001. Incorporation of tumor-targeting peptides into recombinant adeno-associated virus capsids. Mol. Ther. 3964-975. [DOI] [PubMed] [Google Scholar]
- 31.Hart, L. S., D. Ornelles, and C. Koumenis. 2007. The adenoviral E4orf6 protein induces atypical apoptosis in response to DNA damage. J. Biol. Chem. 2826061-6067. [DOI] [PubMed] [Google Scholar]
- 32.Hart, L. S., S. M. Yannone, C. Naczki, J. S. Orlando, S. B. Waters, S. A. Akman, D. J. Chen, D. Ornelles, and C. Koumenis. 2005. The adenovirus E4orf6 protein inhibits DNA double strand break repair and radiosensitizes human tumor cells in an E1B-55K-independent manner. J. Biol. Chem. 2801474-1481. [DOI] [PubMed] [Google Scholar]
- 33.Heilbronn, R., M. Engstler, S. Weger, A. Krahn, C. Schetter, and M. Boshart. 2003. ssDNA-dependent colocalization of adeno-associated virus Rep. and herpes simplex virus ICP8 in nuclear replication domains. Nucleic Acids Res. 316206-6213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoppe, B. S., R. B. Jensen, and C. U. Kirchgessner. 2000. Complementation of the radiosensitive M059J cell line. Radiat. Res. 153125-130. [DOI] [PubMed] [Google Scholar]
- 35.Hunter, L. A., and R. J. Samulski. 1992. Colocalization of adeno-associated virus Rep and capsid proteins in the nuclei of infected cells. J. Virol. 66317-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Inagaki, K., C. Ma, T. A. Storm, M. A. Kay, and H. Nakai. 2007. The role of DNA-PKcs and Artemis in opening viral DNA hairpin termini in various tissues in mice. J. Virol. 8111304-11321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ishimi, Y., Y. Komamura-Kohno, H. J. Kwon, K. Yamada, and M. Nakanishi. 2003. Identification of MCM4 as a target of the DNA replication block checkpoint system. J. Biol. Chem. 27824644-24650. [DOI] [PubMed] [Google Scholar]
- 38.Jing, X. J., V. Kalman-Maltese, X. Cao, Q. Yang, and J. P. Trempe. 2001. Inhibition of adenovirus cytotoxicity, replication, and E2a gene expression by adeno-associated virus. Virology 291140-151. [DOI] [PubMed] [Google Scholar]
- 39.Jurvansuu, J., K. Raj, A. Stasiak, and P. Beard. 2005. Viral transport of DNA damage that mimics a stalled replication fork. J. Virol. 79569-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kudoh, A., M. Fujita, L. Zhang, N. Shirata, T. Daikoku, Y. Sugaya, H. Isomura, Y. Nishiyama, and T. Tsurumi. 2005. Epstein-Barr virus lytic replication elicits ATM checkpoint signal transduction while providing an S-phase-like cellular environment. J. Biol. Chem. 2808156-8163. [DOI] [PubMed] [Google Scholar]
- 41.Lavin, M. F. 2007. ATM and the Mre11 complex combine to recognize and signal DNA double-strand breaks. Oncogene 267749-7758. [DOI] [PubMed] [Google Scholar]
- 42.Leber, R., T. W. Wise, R. Mizuta, and K. Meek. 1998. The XRCC4 gene product is a target for and interacts with the DNA-dependent protein kinase. J. Biol. Chem. 2731794-1801. [DOI] [PubMed] [Google Scholar]
- 43.Lee, J. H., and T. T. Paull. 2007. Activation and regulation of ATM kinase activity in response to DNA double-strand breaks. Oncogene 267741-7748. [DOI] [PubMed] [Google Scholar]
- 44.Lee, K. Y., and K. Myung. 2008. PCNA modifications for regulation of post-replication repair pathways. Mol. Cells 265-11. [PMC free article] [PubMed] [Google Scholar]
- 45.Lees-Miller, S. P., M. C. Long, M. A. Kilvert, V. Lam, S. A. Rice, and C. A. Spencer. 1996. Attenuation of DNA-dependent protein kinase activity and its catalytic subunit by the herpes simplex virus type 1 transactivator ICP0. J. Virol. 707471-7477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lieber, M. R. 2008. The mechanism of human nonhomologous DNA end joining. J. Biol. Chem. 2831-5. [DOI] [PubMed] [Google Scholar]
- 47.Lilley, C. E., C. T. Carson, A. R. Muotri, F. H. Gage, and M. D. Weitzman. 2005. DNA repair proteins affect the lifecycle of herpes simplex virus 1. Proc. Natl. Acad. Sci. USA 1025844-5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lilley, C. E., R. A. Schwartz, and M. D. Weitzman. 2007. Using or abusing: viruses and the cellular DNA damage response. Trends Microbiol. 15119-126. [DOI] [PubMed] [Google Scholar]
- 49.Lim, D. S., S. T. Kim, B. Xu, R. S. Maser, J. Lin, J. H. Petrini, and M. B. Kastan. 2000. ATM phosphorylates p95/nbs1 in an S-phase checkpoint pathway. Nature 404613-617. [DOI] [PubMed] [Google Scholar]
- 50.Ma, Y., K. Schwarz, and M. R. Lieber. 2005. The Artemis:DNA-PKcs endonuclease cleaves DNA loops, flaps, and gaps. DNA Repair 4845-851. [DOI] [PubMed] [Google Scholar]
- 51.McGowan, C. H., and P. Russell. 2004. The DNA damage response: sensing and signaling. Curr. Opin. Cell Biol. 16629-633. [DOI] [PubMed] [Google Scholar]
- 52.Muzyczka, N., and K. I. Berns. 2001. Parvoviridae: the viruses and their replication, p. 2327-2359. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, vol. 2, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 53.Narasimhan, D., R. Collaco, V. Kalman-Maltese, and J. P. Trempe. 2002. Hyper-phosphorylation of the adeno-associated virus Rep78 protein inhibits terminal repeat binding and helicase activity. Biochim. Biophys. Acta 1576298-305. [DOI] [PubMed] [Google Scholar]
- 54.Nash, K., W. Chen, W. F. McDonald, X. Zhou, and N. Muzyczka. 2007. Purification of host cell enzymes involved in adeno-associated virus DNA replication. J. Virol. 815777-5787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nash, K., W. Chen, and N. Muzyczka. 2008. Complete in vitro reconstitution of adeno-associated virus DNA replication requires the minichromosome maintenance complex proteins. J. Virol. 821458-1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nash, K., W. Chen, M. Salganik, and N. Muzyczka. 2009. Identification of cellular proteins that interact with the adeno-associated virus Rep. protein. J. Virol. 83454-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nghiem, P., P. K. Park, Y. Kim, C. Vaziri, and S. L. Schreiber. 2001. ATR inhibition selectively sensitizes G1 checkpoint-deficient cells to lethal premature chromatin condensation. Proc. Natl. Acad. Sci. USA 989092-9097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ni, T. H., W. F. McDonald, I. Zolotukhin, T. Melendy, S. Waga, B. Stillman, and N. Muzyczka. 1998. Cellular proteins required for adeno-associated virus DNA replication in the absence of adenovirus coinfection. J. Virol. 722777-2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pegoraro, G., A. Marcello, M. P. Myers, and M. Giacca. 2006. Regulation of adeno-associated virus DNA replication by the cellular TAF-I/set complex. J. Virol. 806855-6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Raj, K., P. Ogston, and P. Beard. 2001. Virus-mediated killing of cells that lack p53 activity. Nature 412914-917. [DOI] [PubMed] [Google Scholar]
- 61.Ruis, B. L., K. R. Fattah, and E. A. Hendrickson. 2008. The catalytic subunit of DNA-dependent protein kinase regulates proliferation, telomere length, and genomic stability in human somatic cells. Mol. Cell. Biol. 286182-6195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Russell, D. W., I. E. Alexander, and A. D. Miller. 1995. DNA synthesis and topoisomerase inhibitors increase transduction by adeno-associated virus vectors. Proc. Natl. Acad. Sci. USA 925719-5723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sancar, A., L. A. Lindsey-Boltz, K. Unsal-Kacmaz, and S. Linn. 2004. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 7339-85. [DOI] [PubMed] [Google Scholar]
- 64.Sanlioglu, S., P. Benson, and J. F. Engelhardt. 2000. Loss of ATM function enhances recombinant adeno-associated virus transduction and integration through pathways similar to UV irradiation. Virology 26868-78. [DOI] [PubMed] [Google Scholar]
- 65.Saudan, P., J. Vlach, and P. Beard. 2000. Inhibition of S-phase progression by adeno-associated virus Rep78 protein is mediated by hypophosphorylated pRb. EMBO J. 194351-4361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schnepp, B. C., K. R. Clark, D. L. Klemanski, C. A. Pacak, and P. R. Johnson. 2003. Genetic fate of recombinant adeno-associated virus vector genomes in muscle. J. Virol. 773495-3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schwartz, R. A., J. A. Palacios, G. D. Cassell, S. Adam, M. Giacca, and M. D. Weitzman. 2007. The Mre11/Rad50/Nbs1 complex limits adeno-associated virus transduction and replication. J. Virol. 8112936-12945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shi, Y., G. E. Dodson, P. S. Mukhopadhyay, N. P. Shanware, A. T. Trinh, and R. S. Tibbetts. 2007. Identification of carboxyl-terminal MCM3 phosphorylation sites using polyreactive phosphospecific antibodies. J. Biol. Chem. 2829236-9243. [DOI] [PubMed] [Google Scholar]
- 69.Shi, Y., G. E. Dodson, S. Shaikh, K. Rundell, and R. S. Tibbetts. 2005. Ataxia-telangiectasia-mutated (ATM) is a T-antigen kinase that controls SV40 viral replication in vivo. J. Biol. Chem. 28040195-40200. [DOI] [PubMed] [Google Scholar]
- 70.Shirata, N., A. Kudoh, T. Daikoku, Y. Tatsumi, M. Fujita, T. Kiyono, Y. Sugaya, H. Isomura, K. Ishizaki, and T. Tsurumi. 2005. Activation of ataxia telangiectasia-mutated DNA damage checkpoint signal transduction elicited by herpes simplex virus infection. J. Biol. Chem. 28030336-30341. [DOI] [PubMed] [Google Scholar]
- 71.Shrivastav, M., L. P. De Haro, and J. A. Nickoloff. 2008. Regulation of DNA double-strand break repair pathway choice. Cell Res. 18134-147. [DOI] [PubMed] [Google Scholar]
- 72.Song, S., P. J. Laipis, K. I. Berns, and T. R. Flotte. 2001. Effect of DNA-dependent protein kinase on the molecular fate of the rAAV2 genome in skeletal muscle. Proc. Natl. Acad. Sci. USA 984084-4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Song, S., Y. Lu, Y. K. Choi, Y. Han, Q. Tang, G. Zhao, K. I. Berns, and T. R. Flotte. 2004. DNA-dependent PK inhibits adeno-associated virus DNA integration. Proc. Natl. Acad. Sci. USA 1012112-2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stiff, T., M. O'Driscoll, N. Rief, K. Iwabuchi, M. Lobrich, and P. A. Jeggo. 2004. ATM and DNA-PK function redundantly to phosphorylate H2AX after exposure to ionizing radiation. Cancer Res. 642390-2396. [DOI] [PubMed] [Google Scholar]
- 75.Stracker, T. H., C. T. Carson, and M. D. Weitzman. 2002. Adenovirus oncoproteins inactivate the Mre11-Rad50-NBS1 DNA repair complex. Nature 418348-352. [DOI] [PubMed] [Google Scholar]
- 76.Stracker, T. H., G. D. Cassell, P. Ward, Y. M. Loo, B. van Breukelen, S. D. Carrington-Lawrence, R. K. Hamatake, P. C. van der Vliet, S. K. Weller, T. Melendy, and M. D. Weitzman. 2004. The Rep protein of adeno-associated virus type 2 interacts with single-stranded DNA-binding proteins that enhance viral replication. J. Virol. 78441-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Taylor, T. J., and D. M. Knipe. 2004. Proteomics of herpes simplex virus replication compartments: association of cellular DNA replication, repair, recombination, and chromatin remodeling proteins with ICP8. J. Virol. 785856-5866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tomida, J., Y. Masuda, H. Hiroaki, T. Ishikawa, I. Song, T. Tsurimoto, S. Tateishi, T. Shiomi, Y. Kamei, J. Kim, K. Kamiya, C. Vaziri, H. Ohmori, and T. Todo. 2008. DNA damage-induced ubiquitylation of RFC2 subunit of replication factor C complex. J. Biol. Chem. 2839071-9079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Urcelay, E., P. Ward, S. M. Wiener, B. Safer, and R. M. Kotin. 1995. Asymmetric replication in vitro from a human sequence element is dependent on adeno-associated virus Rep protein. J. Virol. 692038-2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Uziel, T., Y. Lerenthal, L. Moyal, Y. Andegeko, L. Mittelman, and Y. Shiloh. 2003. Requirement of the MRN complex for ATM activation by DNA damage. EMBO J. 225612-5621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.van Gent, D. C., and M. van der Burg. 2007. Non-homologous end-joining, a sticky affair. Oncogene 267731-7740. [DOI] [PubMed] [Google Scholar]
- 82.Vassin, V. M., M. S. Wold, and J. A. Borowiec. 2004. Replication protein A (RPA) phosphorylation prevents RPA association with replication centers. Mol. Cell. Biol. 241930-1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang, Y., X. Y. Zhou, H. Wang, M. S. Huq, and G. Iliakis. 1999. Roles of replication protein A and DNA-dependent protein kinase in the regulation of DNA replication following DNA damage. J. Biol. Chem. 27422060-22064. [DOI] [PubMed] [Google Scholar]
- 84.Weitzman, M. D., K. J. Fisher, and J. M. Wilson. 1996. Recruitment of wild-type and recombinant adeno-associated virus into adenovirus replication centers. J. Virol. 701845-1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Weterings, E., and D. J. Chen. 2007. DNA-dependent protein kinase in nonhomologous end joining: a lock with multiple keys? J. Cell Biol. 179183-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Weterings, E., and D. J. Chen. 2008. The endless tale of non-homologous end-joining. Cell Res. 18114-124. [DOI] [PubMed] [Google Scholar]
- 87.Wilkinson, D. E., and S. K. Weller. 2004. Recruitment of cellular recombination and repair proteins to sites of herpes simplex virus type 1 DNA replication is dependent on the composition of viral proteins within prereplicative sites and correlates with the induction of the DNA damage response. J. Virol. 784783-4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Winocour, E., M. F. Callaham, and E. Huberman. 1988. Perturbation of the cell cycle by adeno-associated virus. Virology 167393-399. [PubMed] [Google Scholar]
- 89.Wistuba, A., A. Kern, S. Weger, D. Grimm, and J. A. Kleinschmidt. 1997. Subcellular compartmentalization of adeno-associated virus type 2 assembly. J. Virol. 711341-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Xiao, X., J. Li, and R. J. Samulski. 1998. Production of high-titer recombinant adeno-associated virus vectors in the absence of helper adenovirus. J. Virol. 722224-2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yalkinoglu, A. O., R. Heilbronn, A. Burkle, J. R. Schlehofer, and H. zur Hausen. 1988. DNA amplification of adeno-associated virus as a response to cellular genotoxic stress. Cancer Res. 483123-3129. [PubMed] [Google Scholar]
- 92.Zentilin, L., A. Marcello, and M. Giacca. 2001. Involvement of cellular double-stranded DNA break binding proteins in processing of the recombinant adeno-associated virus genome. J. Virol. 7512279-12287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhao, X., R. J. Madden-Fuentes, B. X. Lou, J. M. Pipas, J. Gerhardt, C. J. Rigell, and E. Fanning. 2008. Ataxia telangiectasia-mutated damage-signaling kinase- and proteasome-dependent destruction of Mre11-Rad50-Nbs1 subunits in Simian virus 40-infected primate cells. J. Virol. 825316-5328. [DOI] [PMC free article] [PubMed] [Google Scholar]