Abstract
The varicella-zoster virus major transactivator, IE62, contains a potent N-terminal acidic transcriptional activation domain (TAD). Our experiments revealed that the minimal IE62 TAD encompasses amino acids (aa) 19 to 67. We showed that the minimal TAD interacts with the human Mediator complex. Site-specific mutations revealed residues throughout the minimal TAD that are important for both activation and Mediator interaction. The TAD interacts directly with aa 402 to 590 of the MED25 subunit, and site-specific TAD mutations abolished this interaction. Two-dimensional nuclear magnetic resonance spectroscopy revealed that the TAD is intrinsically unstructured. Our studies suggest that transactivation may involve the TAD adopting a defined structure upon binding MED25.
Varicella-zoster virus (VZV) is a ubiquitous pathogen that causes varicella (chickenpox) in primary lytic infection and zoster (shingles) during reactivation from latent infection (5). The viral genome is approximately 125 kb in size and encodes at least 71 unique open reading frames. The VZV IE62 protein, which contains 1,310 amino acids (aa), is the major viral transactivator and is essential for viral growth (5, 21). The focus of this work, the IE62 acidic transcriptional activation domain (TAD), was previously mapped to the extreme N terminus of the protein (4, 19). This acidic domain is not present in herpes simplex virus type 1 ICP4 (23, 29). The IE62 TAD shows some compositional similarity (containing primarily aliphatic and acidic residues) to the 81-aa herpes simplex virus type 1 VP16 TAD; however, the two TADs share minimal sequence homology (3.9% identity and 5.9% similarity). Furthermore, the IE62 TAD is less acidic than the VP16 TAD, with a calculated isoelectric point of ∼4.3, compared to ∼3.4 for the VP16 TAD (http://us.expasy.org/tools/protparam.html). Despite the identification and initial characterization of this important domain some 15 years ago (4, 19), the cellular target or targets of the IE62 TAD remained elusive. Recent work from this laboratory indicates that one target of the IE62 TAD is the human mediator of transcription (31).
Mediator consists of approximately 30 subunits (MED1 to MED30) that are in large part conserved from yeast to human. These subunits form head, middle, tail, and CDK submodules, although the localization of MED23-27 is not yet assigned (2, 17). Mediator is required for most PolII-mediated transcription, as it acts as a bridging molecule between activators and PolII. In the current model of activated transcription, Mediator is recruited to the promoter by the TADs of activators. This allows further recruitment of general transcription factors and PolII, resulting in preinitiation-complex assembly and subsequent activated transcription (17).
Yang et al. (31) found that both full-length IE62 and a 107-aa fragment containing the IE62 TAD can capture Mediator containing ectopically expressed Flag-tagged MED25 and that upon VZV infection, Mediator is recruited to viral replication compartments within infected cell nuclei. Further, both ectopic expression and small interfering RNA-mediated downregulation of MED25 interfered with IE62 TAD-activated transcription. These results suggest a potential direct physical interaction between the IE62 TAD and MED25; however, the presence of a bridging molecule essential for the observed functional interaction could not be ruled out. In this study, we have continued our investigation of the IE62 TAD and its interaction with Mediator.
The minimal IE62 TAD encompasses aa 19 to 67 and interacts with Mediator.
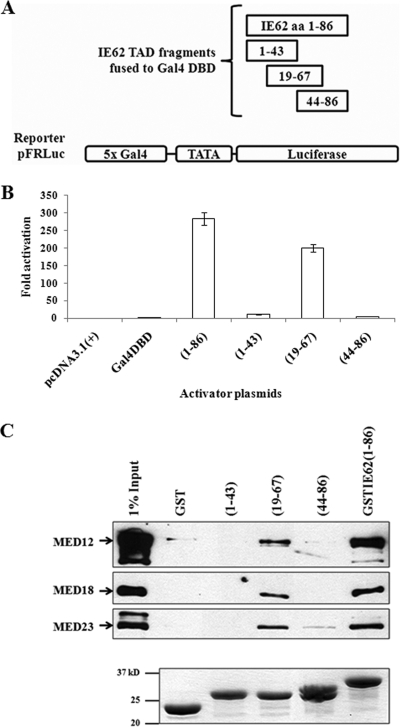
The limits of the IE62 TAD were previously mapped to aa 1 to 90 and 9 to 86 at the N terminus of the protein by Cohen et al. and Perera et al., respectively (4, 19). In order to identify the minimal N-terminal IE62 TAD, plasmids expressing the Gal-4 DNA binding domain (DBD) alone or fused to aa 1 to 43, 19 to 67, and 44 to 86 of IE62 were generated by the insertion of the appropriate coding sequences into the pcDNA3.1(+) plasmid (Invitrogen, Carlsbad, CA) (Fig. 1A). Luciferase reporter assays were then performed via transfection of MeWo cell monolayers with activator plasmids expressing the individual Gal4 DBDIE62 TAD truncation fusions as previously described (31). The reporter plasmid used in these experiments was pFRLuc, which contained a firefly luciferase gene driven by a promoter containing five tandem Gal4 DNA binding sites. The results (Fig. 1B) show that the construct containing the central portion (aa 19 to 67) of the previously mapped TAD exhibited activity at levels approximately 50% of those observed with the full-length TAD (aa 1 to 86). In contrast, neither the N- nor C-terminal halves of the TAD exhibited transactivating activity. Thus, the minimal N-terminal IE62 TAD is contained within this 49-aa central fragment.
FIG. 1.
IE62 aa 19 to 67 constitute the minimal TAD. (A) Schematic of the IE62 truncations fused to the Gal4 DBD. (B) Results from luciferase reporter assays. The pFRLuc reporter plasmid was cotransfected into MeWo cells with activator expression plasmids pcDNA3.1(+)-Gal4DBD-IE62(1-86), -(1-43), -(19-67), or -(44-86). The luciferase activity observed with the empty vector, pcDNA3.1(+), was normalized to 1. The promoter activities resulting from the presence of Gal4 DBD fusion proteins are reported as n-fold activation of the luciferase activity over that observed with pcDNA3.1(+). The experiments were repeated four times with consistent results. (C) IE62 aa 19 to 67, but not aa 1 to 43 or aa 44 to 86, interacts with Mediator. Upper panel: immunoblot analysis following GST pull-down assays showing that the aa 19 to 67 fragment of IE62 TAD interacts with Mediator. The IE62 TAD-Mediator interaction is lost or greatly diminished with the same truncated TADs that showed a loss in transactivation. Bottom panel: Coomassie blue-stained gel showing the levels of GST fusion proteins eluted from the beads.
To test our hypothesis that activation by the IE62 TAD requires interaction with the Mediator complex, protein capture assays were performed as previously described (31), using nuclear extracts and glutathione S-transferase (GST) fusions containing the N-terminal, C-terminal, and central fragments of the IE62 TAD. Eluates from the pull-down assay were immunoblotted, using antibodies against MED12 (CDK submodule), MED18 (head), and MED23 (unassigned). Of the three TAD fragments, only the one containing the central portion captured Mediator, whereas the N- and C-terminal halves of the TAD did not (Fig. 1C). These data indicate that the presence of the intact Mediator complex is required for the transactivating activity of the IE62 TAD.
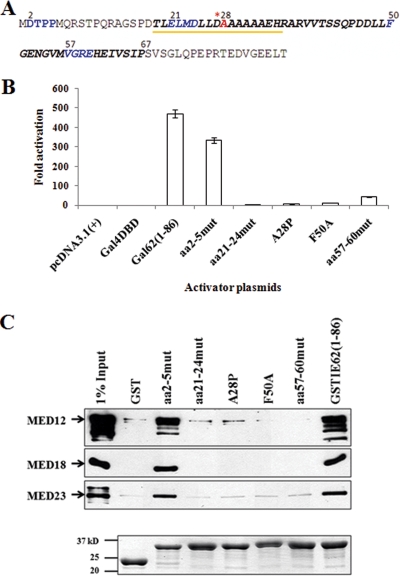
Specific mutations (Fig. 2A) within the Gal4 DBD and GST fusions containing aa 1 to 86 of IE62 were generated using a QuikChange II site-directed mutagenesis kit (Stratagene, La Jolla, CA). These mutations included alanine block mutations encompassing aa 2 to 5, 21 to 24, and 57 to 60. Point mutations included the mutation of a phenylalanine residue at position 50 to alanine (F50A) and the mutation of an alanine at position 28 to proline (A28P). The block mutation of aa 2 to 5 was expected to have a modest effect, as the altered amino acids lie outside of the minimal IE62 TAD mapped above and the first nine residues of IE62 were previously reported to be dispensable for transactivation (19). aa 21 to 24 are located within the first half of the minimal TAD and contain a glutamic acid and an aspartic acid residue. aa 57 to 60 are located within the second half of the minimal TAD and contain an arginine and a glutamic acid residue. Thus, these mutations were designed to disrupt potential electrostatic interactions needed for activation in accord with the current model by which acidic TADs interact with their targets, which involves an initial electrostatic interaction (10, 15, 26). The mutation of alanine at position 28 to proline was designed to disrupt a predicted stable α-helix within the TAD (see below). Finally, the single phenylalanine residue at position 50 was mutated to alanine based on previous findings that phenylalanine residues in the VP16 TAD are critical for transactivating activity (6, 20).
FIG. 2.
Amino acids throughout the aa-19-to-67 sequence are required for the transactivating activity of the IE62 TAD and Mediator interaction. (A) Schematic showing the point and substitution mutations made in the IE62 TAD (aa 1 to 86). Residues altered to alanine are shown in blue. Upon mutation to proline, the alanine at position 28 (shown in red) was designed to disrupt a stable α-helix predicted to be formed by the underlined residues at the amino acid designated with the asterisk. Residues in the minimal TAD (aa 19 to 67) are shown in bold italics. (B) Results from luciferase assays. (C) Upper panel: Immunoblot analysis following GST pull-down assays showing that the IE62 TAD-Mediator interaction is lost or greatly diminished with the mutant TADs that also showed a loss in transactivation. Bottom panel: Coomassie blue-stained gel showing the levels of GST fusion proteins eluted from the beads. mut, mutant.
Luciferase reporter assays were then performed to assess the effects of the mutations. The results (Fig. 2B) show that all of the mutants generated from mutations within the minimal TAD exhibited major (≥90%) losses of transactivating activity compared to the wild type. The aa-57-to-60 block mutant consistently showed the highest residual activity. In contrast, the aa-2-to-5 block mutant retained over 70% of wild-type activity. These experiments were repeated three times with consistent results. Since mutation of the TAD could potentially lead to loss of transactivating activity without a direct correlation to loss of Mediator interaction, the ability of each of the mutant IE62 TADs to interact with Mediator was assessed in GST pull-down assays. Only the wild-type TAD and the aa-2-to-5 mutant, which were capable of transactivation, also showed interaction with Mediator under our experimental conditions (Fig. 2C). These data therefore strongly support the concept that IE62 TAD-mediated transactivation and interaction with Mediator are linked.
The IE62 TAD (aa 1 to 86) is largely unstructured in solution.
Prediction of secondary structure using the PredictProtein servers (http://predictprotein.org) indicated the potential for the presence of an extended α-helix involving aa 19 to 35 of IE62 TAD. The presence of a stable α-helix within the IE62 TAD would be in contrast to our own and previous computer predictions and two-dimensional nuclear magnetic resonance (NMR) studies, which showed a lack of stable secondary structure elements in the activating domains of VP16, the human immunodeficiency virus type 1 Tat protein, and NF-κB in the absence of their binding partners (15, 22, 24, 26). The IE62 TAD exhibits a weaker interaction with Mediator than the VP16 TAD and is a less potent transactivator in the context of transient transfections (29, 31). This finding raised the possibility that the IE62 TAD utilizes a stable preformed α-helix for its target binding, limiting its binding capacity compared to that of the less structured TADs. Therefore, to investigate the structural properties of IE62 TAD, we nominated this protein as an “outreach target” for the Northeast Structural Genomics Consortium ([NESG] http://www.nesg.org; NESG target identification code, OR11).
The pRSET A-IE62TAD plasmid which expresses a double-tagged His-Xpress-IE62 TAD was constructed by insertion of the sequence encoding IE62 aa 1 to 86 into the pRSET A plasmid (Invitrogen, Carlsbad, CA). The protocol to produce both unlabeled and 15N-labeled TAD was based on previously described methods (14) and the protocol used by the protein production site of the NESG at Rutgers University. Briefly, the pRSET A-IE62TAD plasmid was transformed into BL21 Star (DE3) Escherichia coli cells (Invitrogen, Carlsbad, CA), and expression of the protein was induced by the addition of 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) in 15N-containing minimal medium MJ9. Clarified cell lysates were loaded onto a 1-ml HisTrap HP column (GE Healthcare Bio-Sciences Corp., Piscataway, NJ), and bound His-Xpress-IE62 TAD was eluted from the column with 10 ml elution buffer (20 mM sodium phosphate-300 mM imidazole [pH 7.4]). The protein solutions were concentrated to ∼0.6 mM for NMR data acquisition in a buffer containing 25 mM KH2PO4 and 250 mM NaCl (pH 6.0), using Amicon Ultra-4 centrifugal filter units (Millipore, Billerica, MA).
NMR spectra were recorded as described in reference 3 at 25°C on 600-MHz and 750-MHz Varian Inova spectrometers equipped with cryogenic probes (measurement times are indicated in parentheses below). For unlabeled IE62 TAD, a two-dimensional [1H,1H]-nuclear Overhauser effect spectroscopy (NOESY) spectrum (17 h) was acquired on a 600-MHz spectrometer with 512 and 256 complex points in t1 and t2, respectively, a spectral width of 12.7 ppm in both dimensions, and a mixing time of 70 ms. For 15N-labeled TAD, NMR data were acquired on a 750-MHz spectrometer. Two-dimensional [15N,1H]-heteronuclear single-quantum coherence (HSQC) (40 min) was recorded with 512 and 256 complex points in t1 (15N) and t2 (1H), respectively, and spectral widths of 25.6 ppm (15N) and 16 ppm (1H). Three-dimensional 15N-resolved [1H,1H]-NOESY (25 h) and three-dimensional 15N-resolved [1H,1H]-total correlation spectroscopy (25 h) spectra (32) were acquired with 32, 150, and 512 complex points in t1 (15N), t2 (1H), and t3 (1H), respectively, spectral widths of 25.6 ppm (15N) and 13 ppm (1H), and mixing times of 70 ms. Heteronuclear 15N-{1H} NOEs were measured as described in reference 9, with the parameters of two-dimensional [15N,1H]-HSQC and a relaxation delay between scans of 3 s (total, 2 h 20 min). All spectra were processed with the program ProSA (11) and analyzed using the programs XEASY (1) and CARA (16).
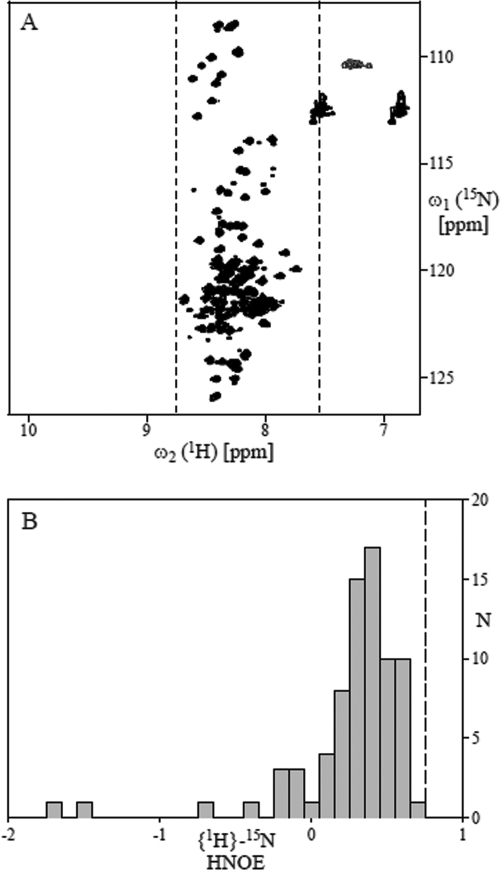
The NMR data revealed that the TAD is largely unstructured in solution and that the predicted α-helix is formed, only transiently, if at all. NMR signal dispersion in two-dimensional [15N,1H]-HSQC is low (Fig. 3A): all polypeptide backbone amide proton shifts are in the comparably narrow range between 7.6 and 8.8 ppm. This is typically observed for proteins lacking a defined tertiary structure (i.e., the amide proton chemical shifts are in the range expected for a “random coil”). Furthermore, all polypeptide backbone heteronuclear 15N-{1H} NOEs are below 0.75 (Fig. 3B), a value that would be expected for residues in a well-defined tertiary fold formed by regular secondary structure elements (3). However, the fact that most of the NOEs are larger than 0 and clustered around 0.4 indicates that some residual structure, possibly an α-helix in the predicted region, is transiently formed. The lack of a defined tertiary structure was further confirmed by (i) inspection of the three-dimensional 15N-resolved [1H,1H]-NOESY and total correlation spectroscopy spectra, which show that sequential NOEs between amide protons, which are strong in α-helical polypeptide segments (3), are virtually absent, and (ii) inspection of the two-dimensional [1H,1H]-NOESY spectrum, which revealed the virtual absence of long-range NOEs between aliphatic protons, which are typically observed between protons in the molecular core of a protein with defined structure (data not shown).
FIG. 3.
NMR data indicating that IE62 TAD (aa 1 to 86) is largely unstructured in solution (pH 7.4). (A) Two-dimensional [15N,1H]-HSQC spectrum recorded at 25°C. The dashed vertical lines at 8.76 and 7.56 ppm indicate the range of amide proton chemical shifts observed for unfolded “random-coil”-like proteins (28). (B) Bar plot showing the distribution of polypeptide backbone heteronuclear {1H}-15N NOEs. The dashed vertical line at 0.75 indicates the value expected for a well-folded protein of about the same size as the TAD; it corresponds to the average of the heteronuclear NOEs (HNOE) measured for backbone amide protons located in regular secondary structure elements of the 76-residue protein of human ubiquitin (Biological Magnetic Resonance Data Bank entry no. 6470).
The IE62 TAD interacts directly with MED25.
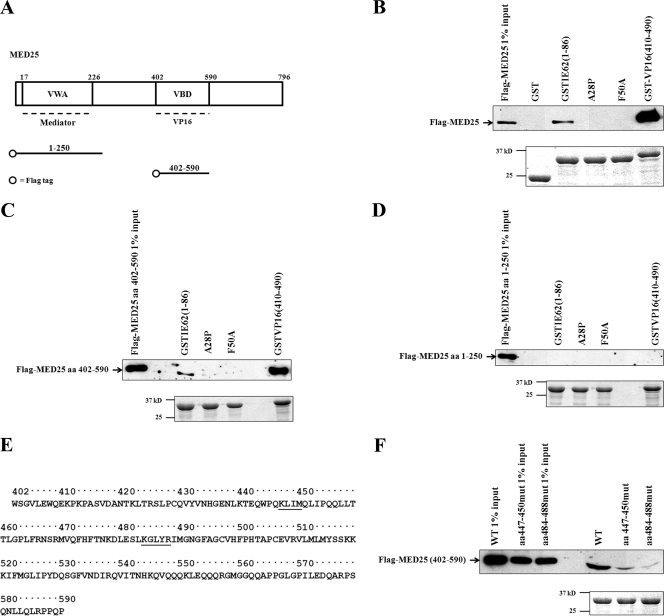
Two functional domains within the primary structure of MED25 have been mapped. The first is similar to the von Willebrand factor A domain (VWA) and has been shown to be important for the incorporation of MED25 into the Mediator complex (18). This domain encompasses aa 17 to 226 of the molecule (Fig. 4A). The second domain, shown to be essential for VP16 TAD binding, has been mapped in two independent studies and designated the activator interaction domain (ACID), or the VP16 binding domain (VBD) (18, 30). The boundaries of the two regions are slightly different, with ACID containing aa 389 to 543 and the VBD containing aa 402 to 590.
FIG. 4.
The functional IE62 TAD interacts with aa 402 to 590 of MED25. (A) MED25 domains and Flag-tagged constructs generated. The VWA domain interacts with Mediator and spans aa 17 to 226. The VP16 TAD interacts with the VBD, consisting of aa 402 to 590 (30). The Flag-tagged MED25 fragments are indicated. (B to D, top panels) Immunoblot analyses following GST pull-down assays showing that full-length MED25 (B) and aa 402 to 590 (C), but not aa 1 to 250 (D), interact with wild-type IE62 TAD. The MED25 aa 402 to 590 interactions were lost with the mutant IE62 TADs that showed loss of transactivation. (B and D, bottom panels) Coomassie blue-stained gels showing the levels of GST fusions eluted from the beads. (E) Amino acid sequence of MED25 aa 402 to 590. Residues altered in block alanine mutations are shown underlined. (F) Upper panel: Immunoblot analysis following GST pull-down assays showing that the MED25 aa-402-to-590 mutants (mut) interact with the IE62 TAD to a significantly lower extent than the wild type (WT). Bottom panel: Coomassie blue-stained gels showing the amounts of GST IE62 TAD fusions eluted from the beads.
In order to determine if the IE62 TAD interacts directly with full-length MED25, GST pull-down assays using whole-cell lysates containing ectopically expressed N-terminal Flag-tagged full-length MED25 and GST fusions containing the wild-type IE62 TAD and the A28P and F50A TAD mutants incorporated into the pGEX-4T-3 vector (GE Healthcare Bio-Sciences Corp., Piscataway, NJ) were performed (Fig. 4B). Immunoblot analysis of the eluates using anti-Flag antibody showed that the wild-type TAD interacts with full-length MED25, whereas the interaction was not detected using the A28P and F50A mutants, indicating a direct interaction between the IE62 TAD and MED25. The questions of the requirement for a bridging molecule and the specific domain of MED25 that interacts with IE62 TAD were addressed using whole-cell lysates expressing Flag-tagged MED25 aa 402 to 590 or aa 1 to 250, which contain the VBD and VWA domains of MED25, respectively. The results (Fig. 4C and D) show that the IE62 TAD had a detectable interaction only with the MED25 VBD. The fusion proteins containing the A28P and F50A TAD mutants did not interact with the MED25 VBD. This result further supports the model of MED25 serving as a direct primary contact site for the IE62 TAD, since the VBD alone, lacking the VWA domain, would not be incorporated into the Mediator complex.
In the next series of experiments, alanine block mutations at aa 447 to 450 and 484 to 488 were introduced into the Flag-tagged MED25 aa-402-to-590 fragment. The mutated residues contain basic amino acids, which could be important for interaction with the acidic IE62 TAD (Fig. 4E). GST pull-down assays showed reductions in the interaction between the IE62 TAD and MED25 VBD fragment resulting from the presence of these mutations, indicating that these two regions in MED25 are involved in the IE62 interaction, with the mutation of aa 484 to 488 having the greater effect (Fig. 4F).
In the work presented here, we have shown that the minimal IE62 TAD consists of aa 19 to 67 of IE62 and that residues throughout this region are important for transactivation. Thus, unlike the VP16 TAD, which contains two subdomains (H1 and H2) that can transactivate independently (13, 15), the minimal IE62 TAD is a single domain. GST pull-down assays showed that the minimal TAD is capable of capturing components representing three different portions of Mediator, suggesting that the entire Mediator complex is recruited by the TAD. Mutations within the minimal TAD that ablated or showed large decreases in transactivation also resulted in loss of binding of all of the Mediator components examined. These data point to a critical role for Mediator in IE62-mediated viral gene expression.
Our NMR data indicate that, contrary to the in silico prediction of a stable α-helix and in accordance with accumulating evidence that acidic TADs generally are largely unstructured (12, 15, 22), the IE62 TAD lacks stable structure in the absence of binding partners. This situation may in part explain the promiscuous nature of IE62 transactivation. It has been suggested that the lack of structure in TADs until they find their binding partner is advantageous in that the intermolecular interactions required for the initiation or propagation of transactivation can be formed with various target proteins, allowing a given TAD to bind to its target with high specificity but comparably low affinity, properties considered to be critical for regulators of gene expression (7, 8, 25, 27).
Finally, our data show that the MED25 VBD domain interacts with the IE62 TAD and that TAD mutations which ablate interaction with this region also result in loss of capture of other Mediator subunits. Thus, MED25 appears to be the major contact point for the IE62 TAD with Mediator. Alanine block mutations introduced at aa 447 to 450 and aa 484 to 488 within the VBD region resulted in a decrease in interaction with the IE62 TAD and suggest that the region of MED25 encompassing aa 484 to 488 is particularly important for the interaction. These data represent the finest mapping to date of the region of MED25 involved in interaction with an acidic TAD. Finally, while MED25 is clearly a direct physical target of the IE62 TAD, this finding does not exclude the possibility of other Mediator contacts with the IE62 molecule. Since no other activation domains have been identified using other fragments of IE62 (4, 19), such regions may be conformational in nature and require the complete native IE62 structure for their existence.
Acknowledgments
This work was supported by grant AI18449 from the National Institute of Allergy and Infectious Diseases and the Protein Structure Initiative of the National Institutes of Health (grant U54-GM074958).
We thank G. T. Montelione, Director of the NESG, for support and Acton and Xiao, Rutgers University, for reagents and advice in preparing the IE62 TAD NMR samples.
Footnotes
Published ahead of print on 8 April 2009.
REFERENCES
- 1.Bartels, C., T. H. Xia, M. Billeter, P. Guntert, and K. Wuthrich. 1995. The program Xeasy for computer-supported NMR spectral analysis of biological macromolecules. J. Biomol. NMR 61-10. [DOI] [PubMed] [Google Scholar]
- 2.Casamassimi, A., and C. Napoli. 2007. Mediator complexes and eukaryotic transcription regulation: an overview. Biochimie 891439-1446. [DOI] [PubMed] [Google Scholar]
- 3.Cavanagh, J., W. J. Fairbrother, A. G. Palmer, M. Rance, and N. J. Skelton. 2006. Protein NMR spectroscopy, 2nd ed. Academic Press, London, United Kingdom.
- 4.Cohen, J. I., D. Heffel, and K. Seidel. 1993. The transcriptional activation domain of varicella-zoster virus open reading frame 62 protein is not conserved with its herpes simplex virus homolog. J. Virol. 674246-4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cohen, J. I., S. E. Straus, and A. M. Arvin. 2007. Varicella-zoster virus replication, pathogenesis and management. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, S. E. Straus (ed.), Fields virology, 5th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, PA.
- 6.Cress, W. D., and S. J. Triezenberg. 1991. Critical structural elements of the VP16 transcriptional activation domain. Science 25187-90. [DOI] [PubMed] [Google Scholar]
- 7.Dunker, A. K., C. J. Brown, J. D. Lawson, L. M. Iakoucheva, and Z. Obradovic. 2002. Intrinsic disorder and protein function. Biochemistry 416573-6582. [DOI] [PubMed] [Google Scholar]
- 8.Dyson, H. J., and P. E. Wright. 2005. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 6197-208. [DOI] [PubMed] [Google Scholar]
- 9.Farrow, N. A., R. Muhandiram, A. U. Singer, S. M. Pascal, C. M. Kay, G. Gish, S. E. Shoelson, T. Pawson, J. D. Forman-Kay, and L. E. Kay. 1994. Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry 335984-6003. [DOI] [PubMed] [Google Scholar]
- 10.Ferreira, M. E., S. Hermann, P. Prochasson, J. L. Workman, K. D. Berndt, and A. P. Wright. 2005. Mechanism of transcription factor recruitment by acidic activators. J. Biol. Chem. 28021779-21784. [DOI] [PubMed] [Google Scholar]
- 11.Guntert, P., V. Dotsch, G. Wider, and K. Wuthrich. 1992. Processing of multidimensional NMR data with the new software Prosa. J. Biomol. NMR 2619-629. [Google Scholar]
- 12.Hermann, S., K. D. Berndt, and A. P. Wright. 2001. How transcriptional activators bind target proteins. J. Biol. Chem. 27640127-40132. [DOI] [PubMed] [Google Scholar]
- 13.Ikeda, K., T. Stuehler, and M. Meisterernst. 2002. The H1 and H2 regions of the activation domain of herpes simplex virion protein 16 stimulate transcription through distinct molecular mechanisms. Genes Cells 749-58. [DOI] [PubMed] [Google Scholar]
- 14.Jansson, M., Y.-C. Li, L. Jendeberg, S. Anderson, G. T. Montelione, and B. Nilsson. 1996. High-level production of uniformly 15N- and 13C-enriched fusion proteins in Escherichia coli. J. Biomol. NMR 7131-141. [DOI] [PubMed] [Google Scholar]
- 15.Jonker, H. R., R. W. Wechselberger, R. Boelens, G. E. Folkers, and R. Kaptein. 2005. Structural properties of the promiscuous VP16 activation domain. Biochemistry 44827-839. [DOI] [PubMed] [Google Scholar]
- 16.Keller, R. L. J. 2004. Optimizing the process of nuclear magnetic resonance spectrum analysis and computer aided resonance assignment. Swiss Federal Institute of Technology Zurich, Zurich, Switzerland.
- 17.Malik, S., and R. G. Roeder. 2005. Dynamic regulation of pol II transcription by the mammalian Mediator complex. Trends Biochem. Sci. 30256-263. [DOI] [PubMed] [Google Scholar]
- 18.Mittler, G., T. Stuhler, L. Santolin, T. Uhlmann, E. Kremmer, F. Lottspeich, L. Berti, and M. Meisterernst. 2003. A novel docking site on Mediator is critical for activation by VP16 in mammalian cells. EMBO J. 226494-6504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perera, L. P., J. D. Mosca, W. T. Ruyechan, G. S. Hayward, S. E. Straus, and J. Hay. 1993. A major transactivator of varicella-zoster virus, the immediate-early protein IE62, contains a potent N-terminal activation domain. J. Virol. 674474-4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Regier, J. L., F. Shen, and S. J. Triezenberg. 1993. Pattern of aromatic and hydrophobic amino acids critical for one of two subdomains of the VP16 transcriptional activator. Proc. Natl. Acad. Sci. USA 90883-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruyechan, W. T. 2004. Mechanism(s) of activation of varicella zoster virus promoters by the VZV IE62 protein. Recent Res. Dev. Virol. 6145-172. [Google Scholar]
- 22.Schmitz, M. L., M. A. dos Santos Silva, H. Altmann, M. Czisch, T. A. Holak, and P. A. Baeuerle. 1994. Structural and functional analysis of the NF-κB p65 C terminus. An acidic and modular transactivation domain with the potential to adopt an α-helical conformation. J. Biol. Chem. 26925613-25620. [PubMed] [Google Scholar]
- 23.Shepard, A. A., A. N. Imbalzano, and N. A. DeLuca. 1989. Separation of primary structural components conferring autoregulation, transactivation, and DNA-binding properties to the herpes simplex virus transcriptional regulatory protein ICP4. J. Virol. 633714-3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shojania, S., and J. D. O'Neil. 2006. HIV-1 Tat is a natively unfolded protein: the solution conformation and dynamics of reduced HIV-1 Tat-(1-72) by NMR spectroscopy. J. Biol. Chem. 2818347-8356. [DOI] [PubMed] [Google Scholar]
- 25.Tompa, P. 2002. Intrinsically unstructured proteins. Trends Biochem. Sci. 27527-533. [DOI] [PubMed] [Google Scholar]
- 26.Uesugi, M., O. Nyanguile, H. Lu, A. J. Levine, and G. L. Verdine. 1997. Induced α helix in the VP16 activation domain upon binding to a human TAF. Science 2771310-1313. [DOI] [PubMed] [Google Scholar]
- 27.Uversky, V. N. 2002. Natively unfolded proteins: a point where biology waits for physics. Protein Sci. 11739-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang, Y., and O. Jardetzky. 2002. Probability-based protein secondary structure identification using combined NMR chemical-shift data. Protein Sci. 11852-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiao, W., L. I. Pizer, and K. W. Wilcox. 1997. Identification of a promoter-specific transactivation domain in the herpes simplex virus regulatory protein ICP4. J. Virol. 711757-1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang, F., R. DeBeaumont, S. Zhou, and A. M. Naar. 2004. The activator-recruited cofactor/Mediator coactivator subunit ARC92 is a functionally important target of the VP16 transcriptional activator. Proc. Natl. Acad. Sci. USA 1012339-2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang, M., J. Hay, and W. T. Ruyechan. 2008. The varicella-zoster virus IE62 protein utilizes the human Mediator complex in promoter activation. J. Virol. 8212154-12163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang, O. W., L. E. Kay, J. P. Olivier, and J. D. Formankay. 1994. Backbone 1H and 15N resonance assignments of the N-terminal Sh3 domain of drk in folded and unfolded states using enhanced-sensitivity pulsed field gradient NMR techniques. J. Biomol. NMR 4845-858. [DOI] [PubMed] [Google Scholar]