Abstract
Many phenotypic differences exist between Homo sapiens and its closest relatives, chimpanzees, and these differences can arise as a result of variations in the regulation of certain genes common to these closely related species. Human-specific endogenous retroviruses (HERVs) and their solitary long terminal repeats (LTRs) are probable candidates for such a role due to the presence of regulatory elements, such as enhancers, promoters, splice sites, and polyadenylation signals. In this study we show for the first time that HERVs can participate in the specific antisense regulation of human gene expression owing to their LTR promoter activity. We found that two HERV LTRs situated in the introns of genes SLC4A8 (for sodium bicarbonate cotransporter) and IFT172 (for intraflagellar transport protein 172) in the antisense orientation serve in vivo as promoters for generating RNAs complementary to the exons of enclosing genes. The antisense transcripts formed from LTR promoter were shown to decrease the mRNA level of the corresponding genes. The human-specific regulation of these genes suggests their involvement in the evolutionary process.
Retroelements (REs) are mobile genetic elements that replicate and transpose through RNA intermediates (11, 13, 22, 39). They constitute more than 42% of human DNA and are the only known class of mobile elements that transpose in mammals in vivo. Four RE families (L1, Alu, SVA, and HERV-K [HML-2]) were transpositionally active after the divergence of human and chimpanzee ancestries, forming a relatively modest fraction of human specific inserts (approximately thousands of copies [33] compared to a total of ∼3 million of human REs [25, 44]). REs are known to be recombination hot spots and provide promoters, polyadenylation signals, and additional splice sites, thus modifying the activity of preexisting human genes (7, 22, 27, 37, 38, 42, 43). At least one-third of all human-specific REs have been mapped within or close to genes (33). Therefore, REs are thought to be one of the causative agents responsible for phenotypic differences between Homo sapiens and its closest relatives, the Pan paniscus and P. troglodytes chimpanzees. Human-specific endogenous retroviruses (HERVs) are probable candidates for such a role due to their powerful regulatory elements located in their long terminal repeats (LTRs) (8, 9, 14, 18, 36).
HERV-K (HML-2) is the only group of endogenous retroviruses known to contain human specific members (10, 31). It occupies ∼5% of the DNA created by insertions of human-specific REs and is one of the best-studied families of human REs. Human-specific elements constitute ∼7% of the group HERV-K (HML-2) (10), which is thought to be the most biologically active human endogenous retroviral family, with 12 members polymorphic in human populations (5, 29, 41). At least 50% of human-specific HML-2 LTRs possess promoter activity and are differentially expressed in normal and cancer tissues (9); some elements are tissue specifically methylated (23, 26).
Previously, it has been shown that LTRs/HERVs in gene introns are preferentially fixed in the antisense position (32, 43), suggesting a strong negative selective pressure on such elements oriented in the hosting gene transcriptional direction. However, some LTRs are also known to possess a bidirectional promoter (15-17). For example, it was recently shown that an antisense-oriented HERV-H LTR serves as an alternative promoter for human gene GSDML, and it was suggested by the authors that its activity is critical for the transcription of GSDML (21). Therefore, antisense orientation of HERVs cannot totally abolish their effect on host gene expression. In the present study, we analyzed another possible mechanism of gene regulation by antisense-oriented HERVs: the alteration of gene expression by cRNA generated from an LTR promoter.
The possibility of LTR involvement in antisense regulation of gene expression has been suggested previously (28). Moreover, recently applied CAGE (for cap analysis of gene expression) technology identified 48,718 human gene antisense transcriptional start sites within transposable elements with 15% located within LTRs (12). However, no experimental evidence for the involvement of any of these antisense transcripts in human gene transcriptional regulation has been obtained to date. Here, we present the first evidence for the human-specific antisense regulation of gene expression occurring due to LTR promoter activity. We found that the human-specific LTRs situated in the introns of the genes SLC4A8 (for sodium bicarbonate cotransporter) and IFT172 (for intraflagellar transport protein 172) in vivo generate transcripts that are complementary to the corresponding mRNAs. These antisense transcripts were shown to reduce the mRNA level of the corresponding genes.
MATERIALS AND METHODS
In silico analysis.
The human-specific HERV-K LTR group (HS) consensus sequence was taken from our previous work (10). LTR flanking regions were investigated with the RepeatMasker program (http://www.repeatmasker.org; A. F. A. Smit and P. Green, unpublished data). Homology searches against GenBank were done by using the BLAST web server at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/BLAST) (2). To determine the genomic locations of LTR flanking regions, the UCSC genome browser and BLAT searches (http://genome.ucsc.edu/cgi-bin/hgGateway) were used. Analysis of alternatively spliced IFT172 and SLC4A8 RNAs was done with the AceView program (http://www.ncbi.nlm.nih.gov/IEB/Research/Acembly/index.html). Potential transcription binding sites were predicted by using SIGNALSCAN (http://www-bimas.cit.nih.gov/molbio/signal/).
Oligonucleotides.
Oligonucleotides were synthesized by using an ASM-102U DNA synthesizer (Biosan, Novosibirsk, Russia). Their structures can be found in Table S2 in the supplemental material.
Tissue sampling.
Samples of human tumor (seminoma) were provided by Blokhin Cancer Research Center, Russian Academy of Medical Sciences, Moscow. Tumor was sampled from orchidectomy specimens with testicular germ cell tumors under nonneoplastic conditions. Representative samples were divided into two parts: one immediately frozen in liquid nitrogen and the other formalin fixed and paraffin embedded for histological analysis. Fetal brain samples were obtained at autopsy after a therapeutic abortion at gestation age of 24 weeks. The sampling was made with a written consent of the patient according to federal law and approved by the ethical committees of the Institute of Bioorganic Chemistry RAS and the Research Center for Obstetrics, Gynaecology and Perinatology of the Russian Academy of Medical Sciences. A chimpanzee brain sample was provided by Tatyana Vinogradova (Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry, Moscow, Russia).
RNA isolation and cDNA synthesis.
Total RNA was isolated from frozen tissues (or collected cells) by using an RNeasy Plus minikit (Qiagen). Full-length cDNA samples were obtained from 1 μg of RNA according to a cap switch effect-based SMART cDNA synthesis protocol (Clontech/BD Biosciences), using an oligo(dT)-containing primer (CDS), PowerScript reverse transcriptase (Clontech/BD Biosciences), and a riboCS oligonucleotide. In the case of orientation-specific PCR, antisense primers for the internal part of the corresponding genes (IFT172for1 or SLC4A8for1) were used for the cDNA synthesis. In each cDNA synthesis reaction, a reverse transcription (RT)-negative control was performed.
RACE (rapid amplification of cDNA ends) was performed as described previously (30). The primers used for 5′RACE are listed in Table S2 in the supplemental material.
Plasmid constructs.
For the generation of stably transfected Tera1 cells, PCR products obtained by using the primer pairs LTR826for-IFT172for3 (“short”IFT-AS transcript), LTR826for-IFT172for8 (“long”IFT-AS LTR), and LTR826for-SLC4A8for5 (SLC-AS) were gel purified, cloned into pGEM-T vector (Promega), digested with NotI (New England Biolabs) and ApaI (Fermentas) restriction endonucleases, and then ligated into NotI/ApaI-digested pcDNA3.1/Hygro(-) vector (Invitrogen). Routine genetic engineering manipulations were carried out according to the published protocols. Primer LTR826for corresponded to the 5′ end of the antisense transcripts determined by RACE, while the primers IFT172for3, IFT172for8, and SLC4A8for5 corresponded to the 3′ ends of the transcripts, determined in a series of RT-PCRs.
To generate IFT172 or SLC4A8 fusions with green fluorescent protein (GFP), RT-PCR products, obtained by using the primer pairs IFT172prot-for/IFT172prot-rev or SLC4A8prot-for/SLC4A8prot-rev, were cloned into pGEM-T vector (Promega), digested with HindIII and SalI restriction endonucleases (Fermentas), and then ligated into HindIII/SalI-digested pTurboGFP-C vector (Evrogen).
Structures of the plasmid constructs used in the study can be found in Fig. S4 in the supplemental material.
Cell culture and establishment of stable transfectants.
The Tera1 (HTB-105 ATCC; human testicular malignant embryonal carcinoma) cell line was cultured in medium (RPMI 1640, Dulbecco modified Eagle medium/F-12; 1:1) with 10% fetal calf serum at 37°C and 5% CO2. At 24 h before transfection, 106 cells were seeded in 25-cm flasks. Transfection was performed with 3 μg of pcDNA3.1/Hygro(-) plasmids, containing “short” IFT-AS, “long” IFT-AS, or SLC-AS sequences, as well as with unmodified pcDNA3.1/Hygro(-) vector. Unifectin-56 was used as the transfection reagent according to the manufacturer's recommendations. Drug selection began 48 h after transfection at 10 μg of hygromycin B (Invitrogen)/ml and continued for 17 to 21 days. Surviving cells were expanded and used for DNA/RNA isolation. In each case, two independent cell lines, stably expressing antisense transcripts, were obtained. Plasmid integration was confirmed by PCR using the primers pcDNA3.1for and pcDNA3.1rev.
Real-time quantitative RT-PCR.
Real-time RT-PCR was performed using MxPro3000 (Stratagene) and an Eva-Green real-time PCR kit (SibEnzyme). Prior to RT-PCR analysis, the priming efficiencies of the primers were examined by genomic PCRs at various temperatures, depending on the primer combination used, with 40 ng of the human genomic DNA. To select LTRs whose transcripts overlap the sequences of the neighboring exons, quantitative RT-PCRs (qRT-PCRs) with the primers Gfor and LTRfor were performed on cDNAs obtained from normal testicular parenchyma and the corresponding cancer (seminoma). Comparison of the IFT-AS and SLC-AS transcriptional levels with the transcriptional levels of the corresponding genes was performed on cDNA samples for 11 human tissues (testicular parenchyma, seminoma, liver, heart, normal lung, lung carcinoma, and embryonic tissues [hippocampus, frontal lobe, occipital cortex, basal ganglion, and thalamus]). cDNAs obtained from Tera1 cells, stably transfected with four different plasmids, were used as templates for the analysis of the effect of IFT-AS and SLC-AS overexpression on the transcription of the corresponding genes. The primers 3′IFT172for and 3′IFT172rev or 3′SLC4A8for and 3′SLC4A8rev were used to determine the level of IFT172 and SLC4A8 RNA; IFT172for1+LTRfor and SLC4A8for1+LTRfor were used to determine the level of antisense transcript expression. To validate 5′RACE data and to measure the level of the potential readthrough transcripts, we performed qRT-PCR with two primers, one of which was complementary to the LTR sequence located upstream of the mapped transcriptional start site (5′LTRfor) and the other of which was complementary to the IFT172 or SLC4A8 exon (IFT172for1 or SLC4A8for1, respectively).
Orientation-specific RT-PCR was performed on cDNAs synthesized with the primers IFT172for1 or SLC4A8for1. The levels of the SLC-AS and IFT-AS transcripts were measured with the primers SLC4A8for2+LTRfor and IFT172for2+LTRfor, respectively. Comparisons of SLC4A8 and IFT172 expression in human and chimpanzee were made by using qRT-PCR on the cDNAs obtained from chimpanzee brain, adult human male brain, and adult human female brain. The primer pairs 3′IFT172for/3′IFT172rev or 3′SLC4A8for/3′SLC4A8rev were used to determine the levels of IFT172 and SLC4A8 RNA; IFT172for11/IFT172-int rev or SLC4A8for1/SLC4A8-int rev were used to determine the levels of intron-containing transcripts.
All measurements were carried out in quadruplicate, and expression levels were normalized to β-actin. No-template (water) and no-reverse transcriptase (RT-) control analyses were performed in all real-time PCRs.
Transient-transfection and GFP fluorescence assay.
Obtained stable transfectants, as well as unmodified Tera1 cells, were transfected with 3 μg of pTurboGFP-C plasmid containing either IFT172 of SLC4A8 or 1 μg of a pCMVβ plasmid (Clontech) as an internal control. Unifectin-56 was used as the transfection reagent according to the manufacturer's recommendations. The cells were incubated with unifectin-DNA complexes for 5 h and then cultured for 24 h in 5 ml of the corresponding growth medium, harvested, and lysed. The GFP activity in the cell lysates was measured with a fluorometer (Turner model 450). The β-galactosidase activity was measured by using an ONPG (o-nitrophenyl-β-d-galactopyranoside) assay (Sigma). The results were normalized with respect to the β-galactosidase activity for each cell line. Transient-transfection experiments were repeated three times.
Western blot analysis.
At 1 day after transfection with GFP-fused constructs, the cells were washed with phosphate-buffered saline (PBS) and lysed with radioimmunoprecipitation assay buffer, containing a protease inhibitor cocktail (Sigma), on ice for 30 min. Lysates were then clarified by centrifugation at 13,000 rpm for 5 min, mixed with protein sample buffer, and subjected to sodium dodecyl sulfate-12% polyacrylamide gel electrophoresis. After electrophoresis, the proteins were transferred to a Hybond-C membrane (Amersham). The membrane was blocked with 5% ECL blocking agent (Amersham) in PBS overnight at 4°C and then incubated with anti-Turbo GFP antibodies (Evrogen; 1:15,000) for 1 h at room temperature. After being washed with PBS, the membrane was incubated with horseradish peroxidase-linked secondary antibodies (Amersham; 1:10,000) for 1 h at room temperature. After extensive washing with PBS, protein bands were visualized by using ECL plus Western blotting detection reagents (Amersham).
Nucleotide sequences for all transcripts used in the present study were submitted to GenBank under accession numbers EU599578 to EU599580.
RESULTS
Identification of novel antisense transcripts, generated from the promoter of human specific LTRs.
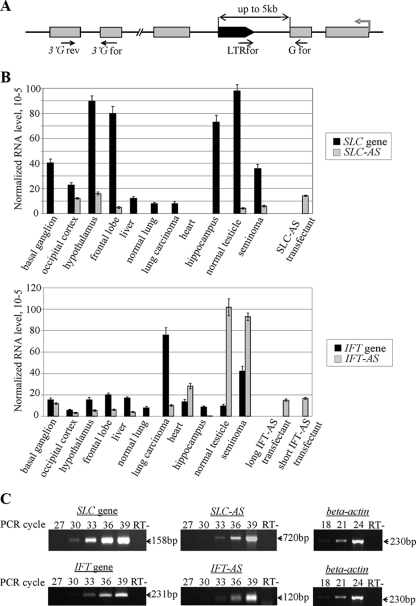
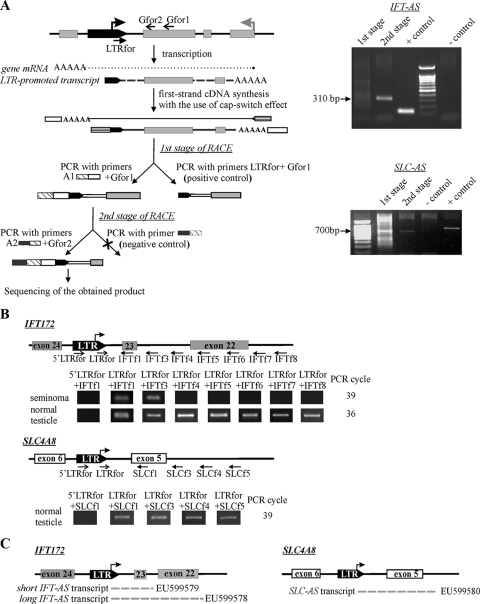
We performed the whole-genome in silico analysis of human specific HERV-K (HML-2) LTRs situated in gene introns in the opposite transcriptional orientation and selected all elements located less than 5 kb away from the nearest exon for further study (Fig. 1A). In total, we identified 13 such cases (the genomic accession numbers, the names of the genes, and the distances from the LTRs to the exons are summarized in Table S1 in the supplemental material). To determine whether these LTRs are transcribed in human tissues, we performed qRT-PCR. To choose only LTRs whose transcripts overlap the sequences of the neighboring exons, we used primers, one of which was designed for the LTR 3′ region (LTRfor, Fig. 1A) and the other of which was designed for the sequence of the gene exon (Gfor, Fig. 1A). The templates were cDNAs obtained from human normal testicular parenchyma sample and the corresponding cancer specimen (seminoma). These tissues contained germ line cells and are known to express HERVs at the highest level (4). Nine of the thirteen selected elements were shown to be transcribed in at least one of the analyzed tissues (data not shown). Using the 5′RACE technique, we found that only two of the nine analyzed LTRs served as promoters and generated transcripts that were complementary at least to the nearest gene exon (Fig. 2A). These are the LTRs located between exons 23 and 24 of the IFT172 gene (LTR-IFT for intraflagellar transport protein [IFT] 172; GenBank accession no. AC074117) and exons 5 and 6 of the gene SLC4A8 (LTR-SLC for sodium bicarbonate cotransporter; GenBank accession no. AC027750). In both cases, the LTR-promoted transcription starts at position 826 of the HS-LTR consensus sequence, which coincides with the previously reported HS-LTR transcriptional start site (24). In a series of qRT-PCRs, we determined two types of transcripts for the LTR-IFT (Fig. 2B and C). One of the amplicons included the sequence complementary to exon 23 (i.e., the “short” transcript) and the other included the sequence complementary to exons 23 and 22 (i.e., the “long” transcript). In the case of the LTR-SLC, we found a unique antisense transcript that included the sequence complementary to the whole of exon 5.
FIG. 1.
(A) Schematic representation of the genomic loci selected for the analysis and positions of the primers used. Gray rectangles represent exons of a gene harboring LTR, the gray arrow indicates the gene transcriptional direction, and the black arrow indicates the human-specific LTR. (B) Comparison of the SLC-AS/IFT-AS RNA level with the mRNA level of the corresponding genes determined by qRT-PCR and normalized to the level of β-actin gene transcripts. For the IFT gene, the observed AS transcript level was a combination of both “short” and “long” transcript concentrations. (C) Representative figure of the RT-PCR results obtained for the frontal lobe sample. The primer pairs 3′SLC4A8for/3′SLC4A8rev, SLC4A8for1/LTRfor, 3′IFT172for/3′IFT172rev, and IFT172for1-LTRfor were used to determine the transcriptional levels of the SLC gene, the SLC-AS gene, the IFT gene, and the IFT-AS gene, respectively. The level of β-actin mRNA was determined using the βact-for and β-act-rev primers. An RT- control was performed in all RT-PCR experiments.
FIG. 2.
(A) Scheme of the 5′RACE technique and its results for the IFT-AS and SLC-AS transcripts. 5′RACE was performed according as described previously (61). The gray arrow indicates the gene transcriptional start site, and the black arrow indicates the transcriptional start site within LTR. White and striped rectangles represent adapter sequences used for cDNA synthesis. cDNA synthesized using the “cap-switch” effect was used as a template for RACE. To achieve the required specificity, the reaction was performed in two stages. In the first PCR, we used a gene-specific primer (Gfor1), complementary to SLC exon 5 or IFT172 exon 23, and the suppression adapter A1, whose 3′-half was complementary to the sequence of the oligonucleotide used for cDNA synthesis. The second PCR was performed with the nested gene-specific primer Gfor2 and the “step-out” suppression adapter A2. Suppression adapters were used to prohibit the amplification of molecules that do not contain annealing site for a gene-specific primer. (B) The extent of the antisense transcripts was determined in a series of qRT-PCRs with pairs of primers, one of which was complementary to the 3′ sequence of the LTR (LTRfor) and the others of which were complementary to different positions within the gene. RT-PCR product size for the primers LTRfor+IFT172for1/IFT172for3-for8 corresponded to theoretically expected lengths of 123, 292, 326, 428, 527, 626, and 712 bp, respectively. The lengths of the RT-PCR products with the primers LTRfor+SLC4A8for1/SLC4A8for3-for5 were 820, 926, 1,050, and 1,155 bp, respectively. To validate 5′RACE data and measure the level of the potential readthrough transcripts initiated somewhere upstream of the LTRs, qRT-PCRs with the primers 5′LTRfor+IFT172for1 or SLC4A8for1 were performed. (C) Types of antisense transcripts found and their corresponding accession numbers.
To validate 5′RACE data, we measured levels of the potential readthrough transcripts initiated somewhere upstream of the LTRs. To this end, we performed qRT-PCRs with primers, one of which was designed for the LTR sequences upstream of the mapped transcriptional start site (5′LTRfor) and the other of which was designed for the gene exon (IFT172for1 or SLC4A8for1), and compared these results to those for LTR-driven transcripts, found using LTR- or downstream sequence-specific primers. For both genes, the levels of potential readthrough transcripts were negligible compared to the LTR-driven RNAs (Fig. 2B).
Orientation of the identified transcripts was confirmed by using strand-specific RT-PCR (see Fig. S1 in the supplemental material). Therefore, it can be concluded that among all human specific LTRs situated in gene introns in the antisense orientation, there are at least two that serve as promoters and generate transcripts complementary to the mRNAs of the corresponding genes. From now on, RNAs that start within the LTR and include sequences complementary to genes IFT172 or SLC4A8 will be correspondingly referred to as transcripts IFT-AS (intraflagellar transport 172, antisense) and SLC-AS (sodium bicarbonate cotransporter, antisense). Nucleotide sequences for all found transcripts were submitted to GenBank under accession numbers EU599578 to EU599580. Detailed structures of the antisense transcripts can be found in Fig. S2 in the supplemental material. In the present study, we investigated both proviral and solitary LTR intronic sequences for their potential effects on the host gene transcription. However, we identified antisense transcripts that overlap with the gene exons and start within the proviral sequence only for the two solitary LTR elements mentioned above.
Comparison of the levels of IFT-AS and SLC-AS transcripts with the mRNA levels of the corresponding genes.
The mRNA levels of the genes IFT172 and SLC4A8 were determined by using qRT-PCR analysis with primers complementary to different gene exons (3′Gfor and 3′Grev, Fig. 1A); the levels of transcripts IFT-AS and SLC-AS were measured with primers, one of which was designed for the 3′ region of the LTR (LTRfor, Fig. 1A) and the other of which was designed for the sequence of the nearest gene exon (Gfor, Fig. 1A). cDNAs obtained from 11 human tissues were used as templates (normal testicular parenchyma, seminoma, liver, heart, normal lung, lung carcinoma, and embryonic tissues [hippocampus, frontal lobe, occipital cortex, basal ganglion, and thalamus]). The SLC-AS transcript was found only in 5 of 11 analyzed tissues (Fig. 1B). In all cases, the level of SLC-AS mRNA was considerably lower than that of the SLC4A8 gene. IFT-AS was transcribed in all analyzed tissues except normal lung. In heart, testicular parenchyma, and seminoma, its mRNA level was higher than that of the IFT172 gene. In normal parenchyma, the IFT-AS level was ∼16-fold higher than IFT172. The “long” IFT-AS transcript, which includes sequence complementary to exons 23 and 22, was found only in testicular parenchyma.
Effect of IFT-AS and SLC-AS overexpression on mRNA level of the corresponding genes.
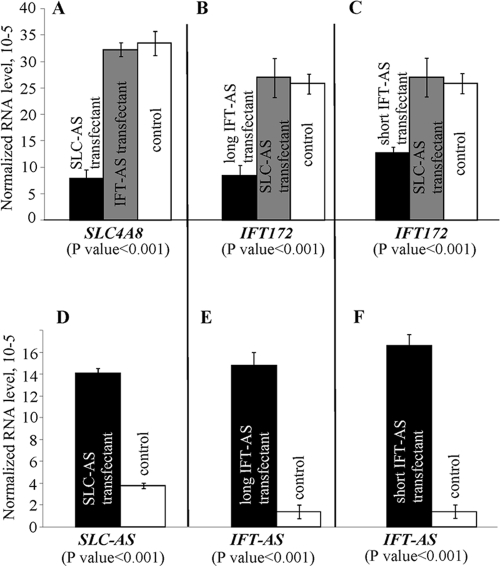
To investigate transcriptional regulation of IFT172 and SLC4A8 genes by the antisense RNAs, we examined this in Tera1 cells since they express IFT172 and SLC4A8 at high levels, whereas IFT-AS and SLC-AS RNAs are under-represented in this cell line. We produced four stably transfected Tera1-derived cell lines: (i) Tera1, stably expressing SLC-AS transcript; (ii) Tera1, expressing long IFT-AS transcript; (iii) Tera1, expressing short IFT-AS transcript; and (iv) Tera1, transfected with plasmid vector without insert (control cell line). Two replicates for each type of transfected cell line were generated.
We analyzed the effect of the antisense transcript overexpression on mRNA level of the corresponding genes by using qRT-PCR. The levels of IFT172 and SLC4A8 RNAs, as well as the transgene expression, in the obtained cell lines were evaluated. The results are summarized in Fig. 3. An ∼4-fold increase in SLC-AS expression led to a 3.9-fold decrease of SLC4A8 mRNA level. Overexpression of the long IFT-AS and short IFT-AS transcripts reduced the levels of IFT172 mRNA by 2.9- and 1.8-fold, respectively. At the same time, overexpression of SLC-AS did not affect the level of IFT172 mRNA and vice versa. The differences observed were found to be statistically significant (P < 0.001). It should be noted that in all cases the levels of the antisense RNAs in the transfected cells were similar to or lower than the levels present in many human tissues (Fig. 1B). This means that gene downregulation by the corresponding natural antisense RNAs, observed in vitro in Tera1 cells, quite probably models the situation occurring in some types of human cells in vivo.
FIG. 3.
Effect of SLC-AS and IFT-AS overexpression on the mRNA levels of the corresponding genes. Relative levels of SLC4A8 (A), IFT172 (B and C), SLC-AS (D), and IFT-AS (E and F) transcripts were determined by using qRT-PCR. All measurements were carried out in quadruplicate, and expression levels were normalized to the β-actin gene transcript. P values were calculated by using a pairwise t test with α = 0.05.
We were unable to measure the concentration of the respective proteins in the transfected cells. The only variant of commercially available polyclonal antibody to IFT172 did not appear to be functional and, at present, there are no available antibodies to SLC4A8 on the market.
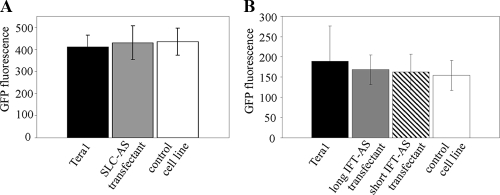
For this reason, we analyzed the possibility of the translational regulation of IFT172/SLC4A8 expression by the IFT-AS and SLC-AS transcripts with the use of two GFP fusion constructs. The first construct (IFT) contained a GFP-fused cDNA sequence for the central tricorn protease domain of the IFT172 protein (amino acids 688 to 1063), including the sequence complementary to the antisense transcripts. The second construct (SLC) contained a GFP fused sequence for the domain IPR013769 (bicarbonate transporter, cytoplasmic; amino acids 42 to 383) of the SLC4A8 protein and also encompassed the target sequence for the antisense transcript (see Fig. S4B in the supplemental material). We did not clone entire protein coding sequences into the GFP fused constructs because of the large sizes of the full-length cDNAs.
These constructs were transiently expressed in cell lines overproducing either IFT-AS or SLC-AS, as well as in the control cell line and in untransfected Tera1 cells. In order to control the GFP fusion protein expression, we performed Western blot hybridization with the antibodies to GFP. In the case of SLC and IFT constructs, we detected major bands corresponding to the expected lengths of the fusion proteins (∼57 and 60 kDa, respectively), whereas a band corresponding to ∼27 kDa was seen for an “empty” control GFP construct. We did not observe any considerable differences in the levels of GFP fluorescence between the tested cell lines (Fig. 4).
FIG. 4.
Synthesis of the fused proteins GFP-SLC (A) and GFP-IFT (B) in cell lines overexpressing antisense transcripts and in control cell lines. Protein levels were determined by measuring the GFP activity in untransfected Tera1 cells, SLC/IFT-AS transfectants, and vector-transfectant. pCMVβ plasmid was used as an internal control. The results were normalized to the β-galactosidase activity in each cell line. Transient-transfection experiments were repeated three times.
Therefore, when the target sequence includes both exons and introns, there is a decrease in gene expression in the cells overproducing AS RNAs (Fig. 3). On the other hand, when the target sequence had only exons but lacked introns, no AS RNA influence on the gene expression was detected (Fig. 4). These data imply that IFT-AS and SLC-AS RNAs probably act on pre-mRNA level (e.g., on the level of unspliced RNA). Detailed analysis of the mechanism(s) of the IFT-AS and SLC-AS RNA activity will be a matter of our further studies.
DISCUSSION
In this study we present the identification of three novel human-specific natural antisense transcripts, generated from the LTR promoters. One of these LTRs is located between exons 23 and 24 of gene IFT172 (for intraflagellar transport 172 protein) and drives the transcription of two different antisense RNAs, whereas the other one is located between exons 5 and 6 of gene SLC4A8 (for sodium bicarbonate cotransporter) and serves as promoter for only one antisense transcript. In the case of IFT172, we detected two types of antisense transcripts started within the LTR: the first one (short IFT-AS) contains sequence complementary to exon 23, whereas the second one (long IFT-AS) contains sequence complementary to both exons 23 and 22. For SLC4A8, we found one antisense RNA (SLC-AS) containing sequence complementary to whole exon 5.
Overexpression of short IFT-AS, long IFT-AS, and SLC-AS transcripts led to 1.8-, 2.9-, and 3.9-fold decreases in the mRNA level, respectively. It should be mentioned that in cell culture experiments the level of antisense RNAs was close to that in human tissues, which suggests that the observed gene transcription downregulation by the antisense RNAs reflects in vivo processes. Promoter regions of the genes IFT172 and SLC4A8 contain multiple binding sites for various nuclear factor proteins (see Fig. S3 in the supplemental material), most of which were reported to participate in a tissue-specific regulation of gene transcription (6, 19, 34, 40). Therefore, the concentration of mRNA for these genes is most likely regulated by the antisense RNAs and by the content of specific protein transcriptional factors, which may differ greatly among the tissues. Antisense regulation, therefore, most probably represents just a part of a more complex mechanism for the control of each gene transcription, as suggested by the rather complex sense-antisense transcript relationships shown in Fig. 1B.
With regard to the functions of the genes, regulated by the activity of human-specific LTRs, the product of the SLC4A8 gene belongs to the family of sodium-coupled bicarbonate transporters and represents an Na+-dependent, Cl−, HCO3− exchanger (NDCBE1) (1). A variant of this gene product has been also described under the name NBC3 (3). Amlal et al. proposed that NBC3 is likely to be involved in cell pH regulation by transporting bicarbonate from blood to the cell, and its enhanced expression in severe acid stress played an important role in cell survival by mediating the influx of HCO3− ions into the cells (3). The product of gene IFT172 is a homologue of the mouse IFT protein. IFT proteins are required for biogenesis of flagella and cilia in multiple organisms, including the green alga Chlamydomonas reinhardtii, Caenorhabditis elegans, insects, and mice (35). Moreover, it has been shown (20) that mouse mutants for IFT proteins exhibit ventral spinal cord patterning defects that appear to result from the reduced hedgehog signaling. The authors of that study concluded that intraflagellar transport machinery has an essential and vertebrate-specific role in the hedgehog signaling, thus participating in growth and developmental processes, e.g., in the development of the neural tube.
Interestingly, according to the public databases (http://genome.ucsc.edu/cgi-bin/hgGateway), IFT172 transcription is upregulated in the brain and testis, whereas maximal SLC4A8 transcription was detected in the brain, in the testis, and in cardiac myocytes. Taking into account gene expression peculiarities of these genes, they may be somehow involved in the development of the human brain. Considering the human-specific regulation of the IFT172 and SLC4A8 shown in the present study, these genes at least theoretically could be suggested as one of the possible causative factors for the phenotypic differences between humans and chimpanzees.
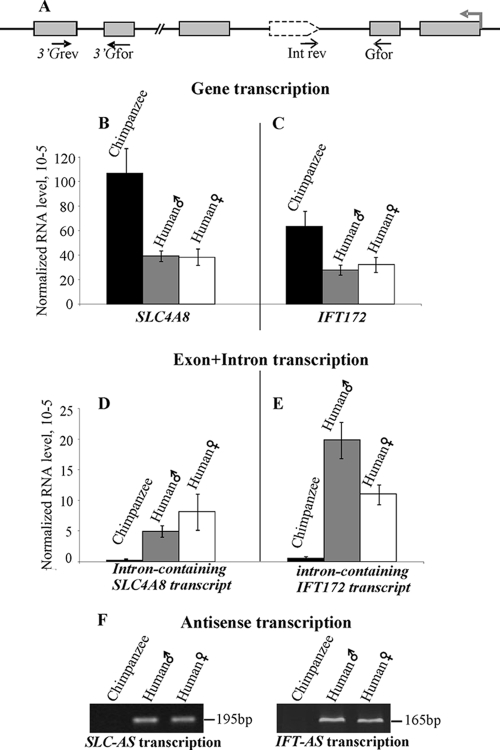
Comparison of SLC4A8 and IFT172 expression in humans and chimpanzees is more complicated due to the reduced number of the appropriate chimpanzee samples available for such an analysis. We compared the transcriptional levels in one tissue sample from the chimpanzee brain and in two human brains. In our experiments, we found that in this very limited sampling genes in the chimpanzee brain were transcribed approximately two- to threefold more often than in human brains (Fig. 5). These data suggest some human-specific modulation of the expression of these genes. The products corresponding to the LTR-driven antisense transcripts were found in both human samples at relatively high levels, and it was also found that these intronic regions corresponding to the human antisense transcripts were transcribed at an ca. 20- to 30-fold lower level in chimpanzees. The presence of such minor transcripts most likely represented unspliced chimpanzee IFT172 and SLC4A8 pre-mRNAs. Indeed, in a series of orientation-specific RT-PCR tests with a cDNA synthesis primer targeted for reverse direction-only transcripts (schematized in Fig. S1 in the supplemental material), we detected no products in chimpanzees versus high transcriptional levels in humans. Therefore, we suggest that there is a human-specific expression of the antisense RNAs targeting exons 22, 23, and 5 of genes IFT172 and SLC4A8, correspondingly.
FIG. 5.
Comparison of the SLC4A8 and IFT172 expression levels in human and chimpanzee brains. (A) Schematic representation of the genomic loci and positions of the primers used. Gray rectangles represent exons of a gene harboring LTR, the gray arrow indicates the gene transcriptional direction, and the dotted arrow indicates the human-specific LTR. (B to E) Relative levels of SLC4A8 (B) and IFT172 (C), as well as intron-containing SLC4A8 (D) and IFT172 (E), transcripts analogous to the SLC-AS and IFT-AS transcripts in humans were measured by using qRT-PCR. All measurements were carried out in quadruplicate, and RNA levels were normalized to the β-actin gene transcript. (C) Results of orientation-specific RT-PCR analyses for human and chimpanzee samples after 39 cycles of amplification (see Fig. S1 in the supplemental material for the scheme of the method).
Overall, it can be concluded that HERV-K (HML-2) LTRs may participate in human-specific regulation of gene expression by generating antisense transcripts. HERVs represent only a small portion (∼5%) of all human-specific REs (33). Therefore, the comprehensive analysis of other REs specific for human populations will further contribute to the understanding of functional differences in human and chimpanzee genome organization.
Supplementary Material
Acknowledgments
We thank Victor Potapov and Nadezhda Skaptsova (Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry, Moscow, Russia) for the synthesis of oligonucleotides and Tatyana Vinogradova for helpful discussions and tissue samples. We are extremely grateful to Linda Scobie (Glasgow Caledonian University, Glasgow, Scotland) for editing and critical reading of the manuscript.
This study was supported by the Molecular and Cellular Biology Program of the Presidium of the Russian Academy of Sciences, by grant MK-4227.2007.4 from the President of the Russian Federation, and by grants 05-04-50770-a and 08-04-00720-a from the Russian Foundation for Basic Research.
Footnotes
Published ahead of print on 1 April 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Aalkjaer, C., S. Frische, J. Leipziger, S. Nielsen, and J. Praetorius. 2004. Sodium coupled bicarbonate transporters in the kidney, an update. Acta Physiol. Scand. 181505-512. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215403-410. [DOI] [PubMed] [Google Scholar]
- 3.Amlal, H., C. E. Burnham, and M. Soleimani. 1999. Characterization of Na+/HCO-3 cotransporter isoform NBC-3. Am. J. Physiol. 276F903-F913. [DOI] [PubMed] [Google Scholar]
- 4.Bannert, N., and R. Kurth. 2004. Retroelements and the human genome: new perspectives on an old relation. Proc. Natl. Acad. Sci. USA 101(Suppl. 2)14572-14579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belshaw, R., A. L. Dawson, J. Woolven-Allen, J. Redding, A. Burt, and M. Tristem. 2005. Genomewide screening reveals high levels of insertional polymorphism in the human endogenous retrovirus family HERV-K(HML2): implications for present-day activity. J. Virol. 7912507-12514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benfante, R., R. A. Antonini, M. Vaccari, A. Flora, F. Chen, F. Clementi, and D. Fornasari. 2005. The expression of the human neuronal α3 Na+,K+-ATPase subunit gene is regulated by the activity of the Sp1 and NF-Y transcription factors. Biochem. J. 38663-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brosius, J. 1999. RNAs from all categories generate retrosequences that may be exapted as novel genes or regulatory elements. Gene 238115-134. [DOI] [PubMed] [Google Scholar]
- 8.Buzdin, A. 2007. Human-specific endogenous retroviruses. Sci. World J. 71848-1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buzdin, A., E. Kovalskaya-Alexandrova, E. Gogvadze, and E. Sverdlov. 2006. At least 50% of human-specific HERV-K (HML-2) long terminal repeats serve in vivo as active promoters for host nonrepetitive DNA transcription. J. Virol. 8010752-10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buzdin, A., S. Ustyugova, K. Khodosevich, I. Mamedov, Y. Lebedev, G. Hunsmann, and E. Sverdlov. 2003. Human-specific subfamilies of HERV-K (HML-2) long terminal repeats: three master genes were active simultaneously during branching of hominoid lineages. Genomics 81149-156. [DOI] [PubMed] [Google Scholar]
- 11.Buzdin, A. A. 2004. Retroelements and formation of chimeric retrogenes. Cell Mol. Life Sci. 612046-2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conley, A. B., W. J. Miller, and I. K. Jordan. 2008. Human cis natural antisense transcripts initiated by transposable elements. Trends Genet. 2453-56. [DOI] [PubMed] [Google Scholar]
- 13.Deininger, P. L., J. V. Moran, M. A. Batzer, and H. H. Kazazian, Jr. 2003. Mobile elements and mammalian genome evolution. Curr. Opin. Genet. Dev. 13651-658. [DOI] [PubMed] [Google Scholar]
- 14.Dewannieux, M., S. Blaise, and T. Heidmann. 2005. Identification of a functional envelope protein from the HERV-K family of human endogenous retroviruses. J. Virol. 7915573-15577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Domansky, A. N., E. P. Kopantzev, E. V. Snezhkov, Y. B. Lebedev, C. Leib-Mosch, and E. D. Sverdlov. 2000. Solitary HERV-K LTRs possess bi-directional promoter activity and contain a negative regulatory element in the U5 region. FEBS Lett. 472191-195. [DOI] [PubMed] [Google Scholar]
- 16.Dunn, C. A., M. T. Romanish, L. E. Gutierrez, L. N. van de Lagemaat, and D. L. Mager. 2006. Transcription of two human genes from a bidirectional endogenous retrovirus promoter. Gene 366335-342. [DOI] [PubMed] [Google Scholar]
- 17.Feuchter, A., and D. Mager. 1990. Functional heterogeneity of a large family of human LTR-like promoters and enhancers. Nucleic Acids Res. 181261-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frank, O., M. Giehl, C. Zheng, R. Hehlmann, C. Leib-Mosch, and W. Seifarth. 2005. Human endogenous retrovirus expression profiles in samples from brains of patients with schizophrenia and bipolar disorders. J. Virol. 7910890-10901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo, I. C., M. C. Shih, H. C. Lan, N. C. Hsu, M. C. Hu, and B. C. Chung. 2007. Transcriptional regulation of human CYP11A1 in gonads and adrenals. J. Biomed Sci. 14509-515. [DOI] [PubMed] [Google Scholar]
- 20.Huangfu, D., A. Liu, A. S. Rakeman, N. S. Murcia, L. Niswander, and K. V. Anderson. 2003. Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature 42683-87. [DOI] [PubMed] [Google Scholar]
- 21.Huh, J. W., D. S. Kim, D. W. Kang, H. S. Ha, K. Ahn, Y. N. Noh, D. S. Min, K. T. Chang, and H. S. Kim. 2008. Transcriptional regulation of GSDML gene by antisense-oriented HERV-H LTR element. Arch. Virol. 1531201-1205. [DOI] [PubMed] [Google Scholar]
- 22.Kazazian, H. H., Jr. 2004. Mobile elements: drivers of genome evolution. Science 3031626-1632. [DOI] [PubMed] [Google Scholar]
- 23.Khodosevich, K., Y. Lebedev, and E. D. Sverdlov. 2004. Large-scale determination of the methylation status of retrotransposons in different tissues using a methylation tags approach. Nucleic Acids Res. 32e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kovalskaya, E., A. Buzdin, E. Gogvadze, T. Vinogradova, and E. Sverdlov. 2006. Functional human endogenous retroviral LTR transcription start sites are located between the R and U5 regions. Virology 346373-378. [DOI] [PubMed] [Google Scholar]
- 25.Lander, E. S., et al. 2001. Initial sequencing and analysis of the human genome. Nature 409860-921. [DOI] [PubMed] [Google Scholar]
- 26.Lavie, L., M. Kitova, E. Maldener, E. Meese, and J. Mayer. 2005. CpG methylation directly regulates transcriptional activity of the human endogenous retrovirus family HERV-K(HML-2). J. Virol. 79876-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lebedev, Y. B., A. L. Amosova, I. Z. Mamedov, G. Y. Fisunov, and E. D. Sverdlov. 2007. Most recent AluY insertions in human gene introns reduce the content of the primary transcripts in a cell type specific manner. Gene 390122-129. [DOI] [PubMed] [Google Scholar]
- 28.Mack, M., K. Bender, and P. M. Schneider. 2004. Detection of retroviral antisense transcripts and promoter activity of the HERV-K(C4) insertion in the MHC class III region. Immunogenetics. 56321-332. [DOI] [PubMed] [Google Scholar]
- 29.Mamedov, I., Y. Lebedev, G. Hunsmann, E. Khusnutdinova, and E. Sverdlov. 2004. A rare event of insertion polymorphism of a HERV-K LTR in the human genome. Genomics 84596-599. [DOI] [PubMed] [Google Scholar]
- 30.Matz, M. V., N. O. Alieva, A. Chenchik, and S. Lukyanov. 2003. Amplification of cDNA ends using PCR suppression effect and step-out PCR. Methods Mol. Biol. 22141-49. [DOI] [PubMed] [Google Scholar]
- 31.Medstrand, P., and D. L. Mager. 1998. Human-specific integrations of the HERV-K endogenous retrovirus family. J. Virol. 729782-9787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Medstrand, P., L. N. van de Lagemaat, and D. L. Mager. 2002. Retroelement distributions in the human genome: variations associated with age and proximity to genes. Genome Res. 121483-1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mills, R. E., E. A. Bennett, R. C. Iskow, C. T. Luttig, C. Tsui, W. S. Pittard, and S. E. Devine. 2006. Recently mobilized transposons in the human and chimpanzee genomes. Am. J. Hum. Genet. 78671-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pham, N. L., A. Franzen, and E. G. Levin. 2004. NF1 regulatory element functions as a repressor of tissue plasminogen activator expression. Arterioscler Thromb. Vasc. Biol. 24982-987. [DOI] [PubMed] [Google Scholar]
- 35.Rosenbaum, J. L., and G. B. Witman. 2002. Intraflagellar transport. Nat. Rev. Mol. Cell. Biol. 3813-825. [DOI] [PubMed] [Google Scholar]
- 36.Ruda, V. M., S. B. Akopov, D. O. Trubetskoy, N. L. Manuylov, A. S. Vetchinova, L. L. Zavalova, L. G. Nikolaev, and E. D. Sverdlov. 2004. Tissue specificity of enhancer and promoter activities of a HERV-K(HML-2) LTR. Virus Res. 10411-16. [DOI] [PubMed] [Google Scholar]
- 37.Sen, S. K., K. Han, J. Wang, J. Lee, H. Wang, P. A. Callinan, M. Dyer, R. Cordaux, P. Liang, and M. A. Batzer. 2006. Human genomic deletions mediated by recombination between Alu elements. Am. J. Hum. Genet. 7941-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smit, A. F. 1999. Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr. Opin. Genet. Dev. 9657-663. [DOI] [PubMed] [Google Scholar]
- 39.Sverdlov, E. D. 2000. Retroviruses and primate evolution. Bioessays 22161-171. [DOI] [PubMed] [Google Scholar]
- 40.Tatewaki, H., H. Tsuda, T. Kanaji, K. Yokoyama, and N. Hamasaki. 2003. Characterization of the human protein S gene promoter: a possible role of transcription factors Sp1 and HNF3 in liver. Thromb. Haemost. 901029-1039. [DOI] [PubMed] [Google Scholar]
- 41.Turner, G., M. Barbulescu, M. Su, M. I. Jensen-Seaman, K. K. Kidd, and J. Lenz. 2001. Insertional polymorphisms of full-length endogenous retroviruses in humans. Curr. Biol. 111531-1535. [DOI] [PubMed] [Google Scholar]
- 42.Ustyugova, S. V., Y. B. Lebedev, and E. D. Sverdlov. 2006. Long L1 insertions in human gene introns specifically reduce the content of corresponding primary transcripts. Genetica 128261-272. [DOI] [PubMed] [Google Scholar]
- 43.van de Lagemaat, L. N., P. Medstrand, and D. L. Mager. 2006. Multiple effects govern endogenous retrovirus survival patterns in human gene introns. Genome Biol. 7R86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Venter, J. C., et al. 2001. The sequence of the human genome. Science 2911304-1351. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.