Abstract
Herpes simplex virus type 1 (HSV-1) acquires its final envelope by budding into cytoplasmic vesicles thought to be derived from trans-Golgi network membranes. This process is facilitated by interactions among the carboxyl termini of viral glycoproteins and tegument proteins. To directly investigate the relative importance of the carboxyl terminus of glycoprotein D (gD) in the presence or absence of gE, a recombinant virus (gDΔct) was constructed to specify a truncated gD lacking the carboxy-terminal 29 amino acids. Furthermore, two additional recombinant viruses were constructed by mutating from ATG to CTG the initiation codons of gE (gEctg) or both gE and gM (gEctg+gMctg), causing lack of expression of gE or both gE and gM, respectively. A fourth mutant virus was constructed to specify the gEctg+gDΔct mutations. The replication properties of these viruses were compared to those of a newly constructed recombinant virus unable to express UL20 due to alteration of the two initiation codons of UL20 (UL20ctgctg). All recombinant viruses were constructed by using the double-Red, site-directed mutagenesis system implemented on the HSV-1(F) genome cloned into a bacterial artificial chromosome. The gEctg, gEctg+gMctg, gDΔct, and gEctg+gDΔct viruses produced viral plaques on African monkey kidney cells (Vero), as well as other cells, that were on average approximately 30 to 50% smaller than those produced by the wild-type virus HSV-1(F). In contrast, the UL20ctgctg virus produced very small plaques containing three to five cells, as reported previously for the ΔUL20 virus lacking the entire UL20 gene. Viral replication kinetics of intracellular and extracellular viruses revealed that all recombinant viruses produced viral titers similar to those produced by the wild-type HSV-1(F) virus intracellularly and extracellularly at late times postinfection, with the exception of the UL20ctgctg and ΔUL20 viruses, which replicated more than two-and-a-half logs less efficiently than HSV-1(F). Electron microscopy confirmed that all viruses, regardless of their different gene mutations, efficiently produced enveloped virions within infected cells, with the exception of the UL20ctgctg and ΔUL20 viruses, which accumulated high levels of unenveloped virions in the cytoplasm. These results show that the carboxyl terminus of gD and the full-length gE, either alone or in a redundant manner, are not essential in cytoplasmic virion envelopment and egress from infected cells. Similarly, gM and gE do not function alone or in a redundant manner in cytoplasmic envelopment and virion egress, confirming previous findings.
Herpes simplex virus type 1 (HSV-1) virion assembly initiates in the nucleus, where procapsid proteins assemble around scaffolding proteins (52, 60), followed by the digestion of these scaffolding proteins, creating empty capsids to be filled with DNA (31, 57, 58). The most-prevalent model for HSV-1 virion egress and viral envelope acquisition calls for primary envelopment of nuclear capsids as they cross the inner nuclear membrane into the perinuclear space and then fusion of enveloped virions with the outer membrane (de-envelopment), delivering nucleocapsids into the cytoplasm (10, 51, 53). In the cytoplasm, viral capsids are coated with tegument proteins and are thought to acquire final viral envelopes by budding into glycoprotein-enriched regions of the trans-Golgi network (TGN) membranes. This final virion morphogenesis step delivers fully enveloped virions into cytoplasmic vesicles, which are ultimately transported out of the cell within vesicles by exocytosis (34).
HSV-1 cytoplasmic virion envelopment is thought to be facilitated by interactions among the cytoplasmic domains of viral glycoproteins and other membrane proteins anchored in TGN-derived membranes with tegument proteins. Such interactions have been described between HSV-1 glycoproteins glycoprotein D (gD [US6]) and VP22 (UL49) (13, 19), gD and VP16 (35), gE (US8) and VP22 (19), and gH (UL22) and VP16 (UL48) (30, 36). Cross-linking experiments have also shown potential interactions of gB, gD, and gH with VP16 (63). In addition, the membrane protein UL11, which intrinsically targets the Golgi apparatus, is believed to direct tegument proteins to the glycoprotein-embedded envelopment site, and it is known to interact with the UL16 tegument protein (4, 42, 43). Mutant viruses with the UL49 (VP22) gene deleted produce smaller-than-wild-type virus plaques, but they can be readily isolated in noncomplementing cells, suggesting redundant roles for VP22 and other tegument proteins in cytoplasmic virion envelopment (17). The tegument protein VP16 serves multiple functions, including transcriptional regulation of viral immediate early, early, and late genes (12, 49, 50) and modulation and downregulation of the virion host shutoff protein vhs (UL41), to which it binds (54). VP16 mutant viruses exhibit drastic defects in infectious virus production and inability to form viral plaques, suggesting a role of VP16 in virion assembly and egress (1, 55, 61). At least part of this defect has been attributed to steps beyond egress from the perinuclear spaces (48).
Deletion of either HSV-1 gD or gH or forced retention of either gD or gH within the endoplasmic reticulum did not appear to cause drastic defects in cytoplasmic virion envelopment and egress, although both glycoproteins are essential for viral infectivity (7, 21, 41, 62). Similarly, gB appeared not to be required for cytoplasmic envelopment and egress, inasmuch as gB-null viruses enveloped and could be rendered infectious after treatment with the fusogen polyethylene glycol (8, 45). However, partial deletion of the carboxyl terminus of gB was recently reported to substantially reduce cytoplasmic envelopment and egress (9). Single or simultaneous deletion of HSV-1 gE and gM genes did not cause any appreciable defects in cytoplasmic virion envelopment and infectious virus production (6). However, simultaneous deletion of HSV-1 gD and gE or gD, gE, and gI was reported to cause drastic accumulation of unenveloped capsids in the cytoplasm, presumably due to the lack of appropriate interactions of the cytoplasmic termini of gD and gE with viral tegument proteins. Because neither of these gene deletions alone caused similar defects, it was concluded that gD and gE function in a redundant manner in cytoplasmic virion egress (18). Other membrane proteins and glycoproteins which traffic to TGN cellular membranes and are known to play important roles in cytoplasmic envelopment include UL11 and the UL20/gK protein complex. Specifically, deletions of the UL11 gene coding for a membrane-bound protein produced mild ultrastructural defects in cytoplasmic virion egress (29), while deletion of either the gK or UL20 gene or specific mutations within these two genes caused drastic inhibition of cytoplasmic virion envelopment (25, 32, 45). Recently, it was reported that simultaneous deletion of HSV-1 UL11 and gM caused drastic inhibition of cytoplasmic virion envelopment (39).
A substantial amount of work on the role of viral tegument proteins and viral glycoproteins and membrane proteins in virion cytoplasmic envelopment has been reported for other viruses belonging to the Alphaherpesvirinae subfamily, notably, the pseudorabies virus (PRV) (46, 47). In most instances, homologous genes appear to retain similar functions; however, there are also important differences among different alphaherpesviruses. For example, the functions of gK and UL20 viral proteins appear to be conserved in both HSV-1 and PRV (25, 28, 32, 37). Furthermore, simultaneous deletion of UL11 and gM for either PRV or HSV produced similar defects in cytoplasmic virion envelopment, although UL11 mutants could not be reciprocally complemented by the heterologous wild-type gene (39). In contrast to these similarities between PRV and HSV-1, simultaneous deletion of the gE, gI, and gM genes drastically inhibited PRV but not HSV-1 cytoplasmic envelopment (5, 6). Recently, it was shown that the HSV-1 UL37 gene, encoding a tegument protein, is essential for replication and plaque formation, although the PRV UL37 homolog is not. However, both UL37 proteins appear to be involved in cytoplasmic virion envelopment (40).
Historically, HSV-1 mutant viruses with mutations in viral glycoproteins and other membrane proteins have been constructed on different genetic backgrounds using diverse methodologies, complicating comparison and interpretation of the resultant defects in virion morphogenesis. To directly compare the role of HSV-1 UL20, gD, gE, and gM in cytoplasmic reenvelopment, we constructed selected single and double mutants in these genes in the same HSV-1(F) genetic background, using similar mutagenesis procedures. The results clearly demonstrated that the carboxyl terminus of gD and full-length gE or gE and gM are not essential either alone or in a redundant manner in cytoplasmic envelopment and virion egress. The gE and gM results confirm previous findings (6). Direct comparison with the UL20ctgctg virus lacking UL20 expression showed that the UL20 protein and, by extension, the UL20/gK protein interactive complex play a primary role in cytoplasmic virion egress.
MATERIALS AND METHODS
Cells and antibodies.
African green monkey kidney (Vero) cells and human endometrium adenocarcinoma cell strain HEC-1-A were obtained from the American Type Culture Collection (Rockville, MD). Cells were maintained in Dulbecco's modified Eagle's medium (Gibco-BRL, Grand Island, NY) supplemented with 10% fetal calf serum and antibiotics. Antibodies used include anti-HSV-1 gE monoclonal antibody (Virusys, Sykesville, MD) and Alexa Fluor 488-conjugated goat anti-mouse immunoglobulin G monoclonal antibody (Invitrogen-Molecular Probes, Carlsbad, CA) for the indirect immunofluorescence assay, as well as anti-HSV gD, anti-HSV gB, and anti-HSV-1 gC monoclonal antibodies (Virusys, Sykesville, MD) for the Western immunoblot assay.
Construction of HSV-1 mutants gDΔct (US6), gEctg (US8), UL20ctgctg, gEctg+gDΔct, and gEctg+gMctg.
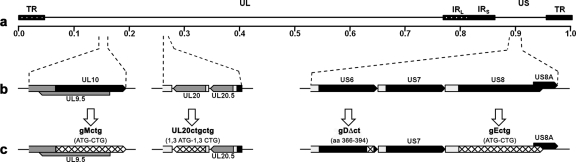
Mutagenesis was accomplished in Escherichia coli by using the markerless two-step Red recombination mutagenesis system with synthetic oligonucleotides (Table 1) (59) implemented on the bacterial artificial chromosome (BAC) plasmid pYEbac102 carrying the HSV-1(F) genome (56) (a kind gift from Y. Kawaguchi, Japan). The gEctg recombinant virus was constructed by changing the initiation codon from ATG to CTG (Fig. 1, Table 1). The gDΔct virus specified an 87-bp deletion coding for the carboxy-terminal 29 gD amino acids while retaining the native stop codon and immediate downstream sequences intact. The UL20ctgctg virus was constructed by altering two potential initiation codon sites (from ATG to CTG) located 6 bp apart at the beginning of the UL20 open reading frame. The gEctg recombinant virus was used as the backbone for construction of the double mutant gEctg+gMctg by mutating the initiation codon for gM (ATG to CTG), as well as to create the gEctg+gDΔct mutant virus by introducing the above-mentioned gD-specific deletion.
TABLE 1.
Red recombination primers used in the study
| Primer | Sequencea | Purpose |
|---|---|---|
| 5′ gE-EnPas | GTTGGGCTCCCATTTTACCCGAAGATCGGCTGCTATCCCCGGGACCTGGATCGCGGGGCGGTGGTGGGGTTTCTTAGGATGACGACGATAAGTAGGG | gEctg (start codon point mutation) |
| 3′ gE-EnPas | AACACAAACACCGAGAAGAAACCCCACCACCGCCCCGCGATCCAGGTCCCGGGGATAGCAGCCGATCTTCGGGTACAACCAATTAACCAATTCTGATTAG | |
| 5′ gD-366Del | GGCAGCCCTGGTCATTTGCGGAATTGTGTACTGGATGCGC_TAGATACCCCCCCTTAATGGAGGATGACGACGATAAGTAGGG | gDΔct (deletion from amino acid 366 but leaving stop codon) |
| 3′ gD-366Del | CAGACCTGACCCCCCCGCACCCATTAAGGGGGGGTATCTA_GCGCATCCAGTACACAATTCCAACCAATTAACCAATTCTGATTAG | |
| 5′ gM-EnPas | GATACGCTCGACGTGTACTGTTCGCACTCGTCGTCCCCACTGGGACGCCCGGCCCCCAGAGGATGACGACGATAAGTAGGG | gMctg (start codon point mutation) |
| 3′ gM-EnPas | GCGCGGAGTCGGGAGATCCTCTGGGGGCCGGGCGTCCCAGTGGGGACGACGAGTGCGAACCAACCAATTAACCAATTCTGATTAG | |
| 5′ UL20-EnPas | GCGACCCTTTGCGGTTTCGGTCTCCCCACCTCCACCGCACACCCCCTGACCCTGCGGGATGACCTTCCTCTGGTGGATCGAAGGATGACGACGATAAGTAGGG | UL20ctgctg (start codon and downstream site point mutations) |
| 3′ UL20-EnPas | CTCGTCGACCAGATCTCGATCCACCAGAGGAAGGTCATCCCGCAGGGTCAGGGGGTGTGCGGTGGAGGTGGGGAGACCGAACAACCAATTAACCAATTCTGATTAG |
The regions homologous to HSV-1 DNA are in standard uppercase, the underlined sequences denote sites of mutation, the underscores represent deletion, and the regions which bind to the marker gene are in bold italics.
FIG. 1.
Genomic map of mutated genes. (a) Represents the prototypic arrangement of the HSV-1 genome with the unique long (UL) and unique short (US) regions flanked by the terminal repeat (TR) and internal repeat (IR) regions. (b) Shows expanded genomic regions of the UL10, UL20, US6, and US8 open reading frames. (c) Shows the effect of ATG mutagenesis on gM, gE, and UL20 gene expression (cross-hatched regions), as well truncation of gD after gDΔct mutagenesis (cross-hatched gD region).
Synthetic oligonucleotides used to mutagenize each targeted gene are shown in Table 1. Specifically, the 5′ end of the forward primer for each mutagenesis contains ∼40 bp of homologous sequence upstream of the site of mutation, followed by the mutant DNA sequence(s). An extra 20 bp downstream of the target site was added. The 3′ end of the forward primer anneals to pEPkans-S, so that it overlaps an I-SceI self-homing endonuclease site. The 5′ end of the reverse primer was designed to contain the reverse complement 40 bp downstream of the target site, which is also homologous to the 5′ end of the forward primer. This DNA sequence was followed by the reverse complement of the targeted mutation site and then an additional 20 bp upstream of the target site. The 3′ end of each primer was designed to anneal to the reverse complement sequences downstream or upstream of the AphA1 gene conferring kanamycin (Kan) resistance, encoded by the pEPkans-S plasmid. Using these primers to amplify the AphA1 gene located on pEPkan-S, a PCR product was produced that contained the desired mutation and the kanamycin resistance cassette.
Maintenance and mutagenesis of the BAC constructs were performed in E. coli strain EL250, which contains a λ prophage encoding recombination enzymes Exo, Beta, and Gam under a heat-inducible promoter (38). Colonies were screened for positive integration of the correct mutant-plus-Kan PCR product by using PCR test primers designed to lie outside of the target mutation sites (not shown). Clones that were confirmed to have the correct integration were used for the resolution step by being grown (from a 1:1,000 dilution) for 2 h in LB medium without kanamycin (with chloramphenicol and ampicillin) at 32°C with shaking at 225 rpm, followed by the addition of arabinose (1% final concentration) and an additional hour of incubation. Cultures were then placed in a 42°C water bath for 30 min, followed by a final 1 h of growth at 32°C. Aliquots of resolved culture were plated on chloramphenicol and ampicillin plates containing 1% arabinose. After 24 h, colonies were screened by replica plating for the loss of kanamycin resistance. DNAs from kanamycin-sensitive colonies were prepared and sent for sequencing to confirm the presence of the desired mutation.
Confirmation of the targeted mutations and recovery of infectious virus.
HSV-1 BAC DNAs (plasmids pgDΔct, pgEctg, pUL20ctgctg, pgEctg/gDΔct, and pgEctg/gMctg) were purified from 50 ml of BAC cultures with a Qiagen large-construct kit (Qiagen, Valencia, CA). Using PCR test primers designed to lie outside of the target mutation site(s), all mutated DNA regions were sequenced to verify the presence of the desired mutations in BACs. In addition, to confirm that no spurious mutations were introduced into the target genes of interest during mutagenesis or during the transfection procedure, cells from cell cultures infected with each mutant virus were collected and DNA was isolated and checked by DNA sequencing to confirm the presence of the desired mutations.
Viruses were recovered from cells transfected with BACs as follows: Vero cells were grown to 95% confluence in 12-well plates. Cells were transfected with BAC DNA mixed with Lipofectamine 2000 in Opti-MEM medium as recommended by the manufacturer (Invitrogen). After 6 h of incubation at 37°C, the medium was removed from the transfected cells, and the cells were washed with phosphate-buffered saline (PBS) before fresh Dulbecco's modified Eagle's medium with 10% fetal calf serum was added. At 48 h posttransfection, virus stocks were collected and designated as passage 0 (P0). Passage P1 viruses were used for all experiments described in the manuscript.
Plaque morphology of mutant viruses.
Visual analysis of plaque morphology of mutant viruses was performed as we have previously described (29, 44, 45). Briefly, near-confluent Vero cell monolayers in six-well plates were infected with the indicated virus at a multiplicity of infection (MOI) of 0.001. After 1 h at room temperature, medium was discarded and cells were washed with PBS before the addition of methylcellulose. After 48 h postinfection (hpi) at 37°C and 5% CO2, cells were washed several times with PBS to remove methylcellulose medium and fixed with ice-cold methanol for 15 min. PBS containing a 1:500 dilution of the polyclonal horseradish peroxidase-conjugated anti-HSV-1 antibody (Dako Cytomation) was added to the cells and placed on a rocker at room temperature for 1 h. Virus plaques were visualized by using a Vector NovaRED peroxidase substrate kit as directed by the manufacturer (Vector, Inc). Plaques were imaged using a Leica DM IRB inverted wide-angle microscope (Leica Microsystems GmbH, Wetzlar, Germany).
Plaque area measurements.
Photographs of viral plaques were taken at ×25 magnification on the Leica model DM IRB inverted wide-angle microscope mentioned above. Fifty randomly selected plaques were imaged in this manner for each of the mutant and wild-type viruses under consideration. Overall, two different sets of 50 viral plaques were measured at 24 and 48 hpi with similar results. The resulting images were analyzed using Zeiss AxioVision SP1 software, release 4.7.1.0 (Carl Zeiss MicroImaging GmbH, Jena, Germany) to determine the area of each plaque in square pixels. The plaque area measurements were natural log transformed and analyzed using the SAS statistical package, version 9.1.3. Distributions were examined for normality using the UNIVARIATE procedure with a Shapiro-Wilk test of normality. The general linear model procedure was used to conduct a one-way analysis of variance on the natural log-transformed data. When overall analyses determined significance (p ≤ 0.05), Tukey's honestly significant differences test was used to examine pairwise differences between the means for each of the five mutants. Results are shown for the viral plaques produced at 48 hpi.
One-step viral growth kinetics.
Analysis of one-step growth kinetics was performed as we have described previously (22, 27). Briefly, viruses were adsorbed on wells of a six-well plate of nearly confluent Vero cell monolayers at 4°C for 1 h at an MOI of 2. Thereafter, plates were incubated at 37°C and 5% CO2 and virus was allowed to penetrate for 1 h at 37°C. Any remaining extracellular virus was inactivated by low-pH treatment (pH 3.0), and cells were incubated at 37°C and 5% CO2. Supernatants and cell pellets were separated at different times postinfection and stored at −80°C. Virus titers were determined by endpoint titration of virus stocks on Vero cells. The experiment was performed a second time, and the titers obtained were averaged, with standard deviation calculated for each time point.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western immunoblotting.
Subconfluent Vero cell monolayers were infected with the indicated virus at an MOI of 5 (Fig. 4). At 24 hpi, cells were collected by low-speed centrifugation, washed with PBS, and lysed at room temperature for 15 min in mammalian protein extraction reagent supplemented with a cocktail of protease inhibitors (Invitrogen-Life Technologies, Carlsbad, CA). Samples were further processed for sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotting as detailed previously (23). Blots were visualized by autoradiography using a Pierce SuperSignal chemiluminescent detection kit (Pierce, Rockford, IL) according to the manufacturer's instructions. All antibody dilutions and buffer washes were performed in Tris-buffered saline supplemented with 0.135 M CaCl2 and 0.11 M MgCl2 (Tris-buffered saline-Ca-Mg) with 0.1% Tween 20.
FIG. 4.
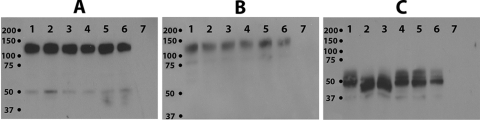
Western immunoblot analysis of glycoproteins specified by mutant viruses. (A) anti-gB monoclonal antibody (MAb) was used to detect gB. (B) gC detection using anti-gC MAb. (C) gD detection using anti-gD MAb. For all panels, lane 1 is wild-type virus cellular extracts; lane 2 is gDΔct; lane 3 is gEctg+gDΔct; lane 4 is gEctg-rescued virus; lane 5 is gEctg; lane 6 is gEctg+gMctg; and lane 7 is mock-infected Vero cell extracts.
For indirect immunofluorescence detection of gE expression, Vero cell monolayers infected with different mutant and wild-type viruses at an MOI of 1 were fixed with 100% methanol for 10 min, washed twice with PBS, and incubated at room temperature for 1 h with the primary anti-gE monoclonal antibody (Virusys, Sykesville, MD) diluted 1:3,000 in PBS-5% goat serum. Cells were then washed three times with PBS and incubated for 1 h at room temperature with Alexa Fluor 488-conjugated goat anti-mouse immunoglobulin G monoclonal antibody (Invitrogen-Molecular Probes, Carlsbad, CA) diluted 1:500 in PBS-10% goat serum. After incubation, excess antibody was removed by washing three times with PBS. Fluorescent micrographs were obtained by using a Leica model DM IRB inverted fluorescence microscope (Leica Microsystems GmbH, Wetzlar, Germany).
Electron microscopy.
Cell monolayers were infected with the indicated virus at an MOI of 5. All cells were prepared for transmission electron microscopy examination at 16 hpi. Infected cells were fixed in a mixture of 2% paraformaldehyde and 1.5% glutaraldehyde in 0.1 M sodium cacodylate buffer, pH 7.3. Following treatment with 1% OsO4 and dehydration in an ethanol series, the samples were embedded in Epon-Araldyte resin and polymerized at 70°C. Thin sections were made on an MTXL ultratome (RMC Products), stained with 5% uranyl acetate and citrate-nitrate-acetate lead, and observed with a Zeiss 10 transmission electron microscope as described previously (24, 25, 32, 45).
RESULTS
Construction and molecular analysis of recombinant viruses carrying one or more mutations.
The availability of the HSV-1(F) genome cloned into a BAC has enabled the rapid and efficient genetic manipulation of the HSV-1 genome in E. coli. To better compare the relative roles of different viral glycoproteins in cytoplasmic virion envelopment and egress, we generated a set of different mutants lacking one or two targeted genes (Fig. 1). Mutagenesis was accomplished in E. coli cells by using the markerless two-step Red recombination mutagenesis system (59) implemented on the pYEbac102 BAC carrying the entire HSV-1(F) genome (56) as described in Materials and Methods. We have previously constructed and characterized a ΔUL20 virus by deleting the UL20 gene without affecting the adjacent UL20.5 gene (25). To eliminate the small possibility that this deletion causes indirect epigenetic effects on adjacent gene expression, we constructed the new UL20ctgctg virus by altering both UL20 ATG codons found 6 bp apart at the 5′ of the UL20 gene. Mutagenesis of either ATG sequence alone produced viable virus, indicating that either or both ATGs may be utilized in virus-infected cells (not shown). Therefore, we mutagenized both ATGs to ensure lack of UL20 gene expression. Similarly, gMctg and gEctg viruses were constructed by mutagenesis of the gM and gE initiation codons, respectively. These mutant viruses were exceptionally stable in cell culture, with no revertants detected by plaque morphology or PCR-assisted DNA sequencing for up to five serial passages in cell culture (not shown). The UL20ctgctg mutant formed wild-type plaques on UL20-complementing cell lines, as was previously shown for the ΔUL20 virus (25), indicating the lack of any secondary mutations within the UL20ctgctg viral genome. A gEctg revertant virus was constructed by site-directed mutagenesis of the BAC containing the gEctg genome (gE rescued), and it replicated and formed plaques in a manner similar to the wild-type virus (not shown).
gD is essential for HSV-1 virus entry and spread (20). However, extracellular domains of gD are necessary and sufficient for gD-mediated virus entry. Different prediction algorithms predicted the cytoplasmic terminus to be approximately 30 amino acids in length (not shown). The gD truncation mutant gDΔct was constructed by deleting an 87-bp DNA fragment encoding the carboxy-terminal 29 amino acids of gD (Fig. 1). The engineered mutations were confirmed via diagnostic PCR and DNA sequencing (see Materials and Methods).
Recovery and plaque morphologies of infectious virus produced by HSV-1 BAC DNA.
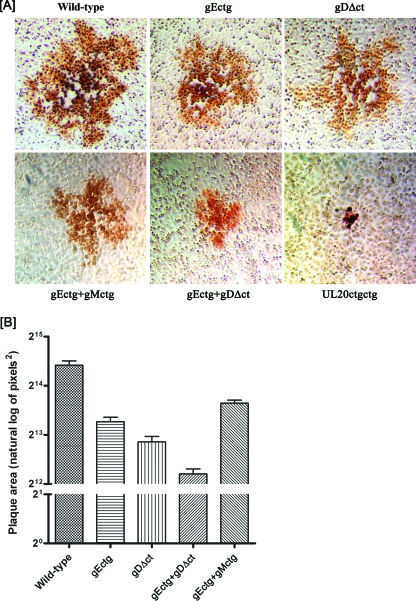
To generate virus stocks from the mutant BAC genomic constructs, individual BAC DNAs were transfected into Vero cells and initial virus stocks recovered and characterized as detailed in Materials and Methods. Passage of these viruses up to five times in Vero cells did not result in any phenotypic revertants or the generation of spurious mutations (not shown). The plaque morphologies of the gDΔct, gEctg, UL20ctgctg, gEctg+gMctg, and gEctg+gDΔct mutant viruses were examined in Vero cells. As expected, the HSV-1(F) wild-type virus produced large plaques (Fig. 2A). The gEctg, gDΔct, and gEctg+gMctg mutant viruses formed plaques that were similar in size to each other and, on average, approximately 50% smaller than the wild-type virus plaques. The gEctg+gDΔct double mutant virus formed plaques which appeared smaller than those formed by either the gEctg or gDΔct mutant virus. Penetration kinetics performed for all mutant viruses revealed that all of the viruses entered with similar entry kinetics into Vero cells (not shown). The UL20ctgctg mutant virus produced the smallest plaques of all mutants, composed of three to five cells per plaque, as has been extensively described previously for a UL20-null mutant virus in which most of the UL20 gene was deleted (Fig. 2A) (25). To better assess the virus plaque sizes produced by individual mutant viruses, 50 randomly chosen viral plaques were selected and statistically analyzed as described in Materials and Methods. This analysis confirmed that deletion of either gE or the carboxyl terminus of gD significantly reduced plaque size in comparison to that of the wild-type HSV-1(F) virus and that the gEctg+gDΔct combined set of mutations reduced plaque size even further than either the gEctg or gDΔct mutation alone (Fig. 2B). Tukey's honestly significant differences test performed after one-way analysis of variance on the natural log-transformed plaque area data showed a statistically significant difference in mean plaque area (P ≤ 0.05) between each of the five mutants (not shown). The plaque morphologies of these viruses were also examined on human endometrial carcinoma (HEC-1-A), human embryonic kidney 293, Syrian hamster, and kidney (BHK-21) cells, and similar results were obtained (not shown).
FIG. 2.
Plaque phenotypes of wild-type, gEctg, gDΔct, gEctg+gMctg, gEctg+gDΔct, and ΔUL20 viruses. (A) Confluent Vero cell monolayers were infected with each virus at an MOI of 0.001, and viral plaques were visualized at 48 hpi by immunohistochemistry as described in Materials and Methods. (B) Fifty different viral plaques were randomly selected, imaged, measured, and statistically analyzed as described in Materials and Methods. Natural log-transformed data for each mutant and the wild-type virus are depicted as bar graphs with standard errors. Similar results were obtained when 50 viral plaques were measured at 24 hpi (not shown).
Replication kinetics of HSV-1 mutants.
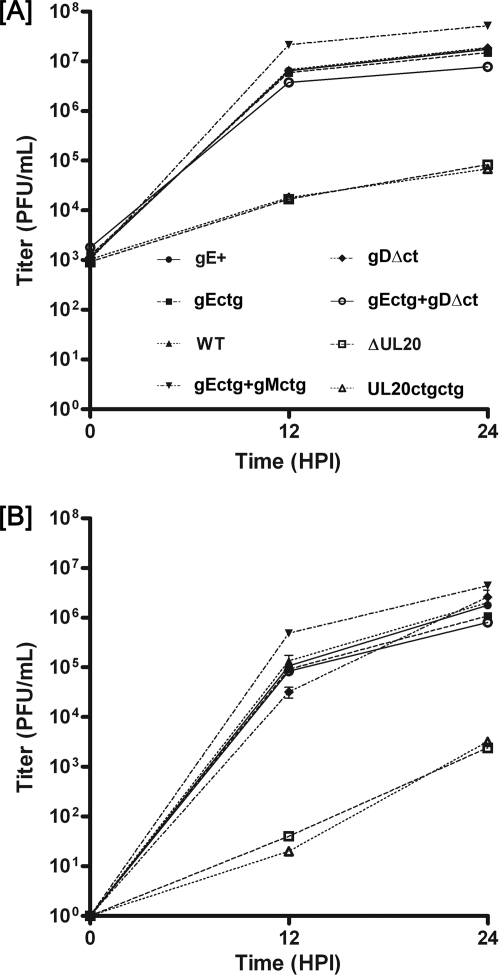
To examine the effect of the various engineered mutations on virus replication, Vero cells were infected at an MOI of 2 with either the wild-type or each mutant virus and virus growth kinetics within infected cells and their supernatants were performed as described in Materials and Methods. All mutant viruses accumulated infectious virions within infected cells at similar rates, approaching titers at 24 hpi similar to those produced by the wild-type virus, with the exception of the gEctg+gMctg double mutant virus, which replicated three- to fivefold more efficiently than the other viruses (Fig. 3A). Both UL20-null mutant viruses, having either the UL20 gene deleted (ΔUL20) or the first two UL20 ATGs altered (UL20ctgctg), replicated more than two-and-a-half logs less efficiently than all other viruses (Fig. 3A). Rates of viral accumulation in extracellular medium were similar to rates of accumulation in infected cells, with the exception that overall viral titers at 24 hpi were up to a log lower than the corresponding titers obtained from infected cells (Fig. 3B). Replication experiments were also performed in HEC-1-A, 293, and BHK-21 cells, with similar results (not shown).
FIG. 3.
Viral replication kinetics. Comparison of virus replication characteristics of wild-type (WT) and mutant viruses. Viral titers at different times after infection of Vero cells at an MOI of 2 were determined for intracellular virus (A) or extracellular virus (B). The experiment was performed a second time, the titers obtained averaged, and the standard deviation calculated for each time point.
Protein expression profiles of viral mutants.
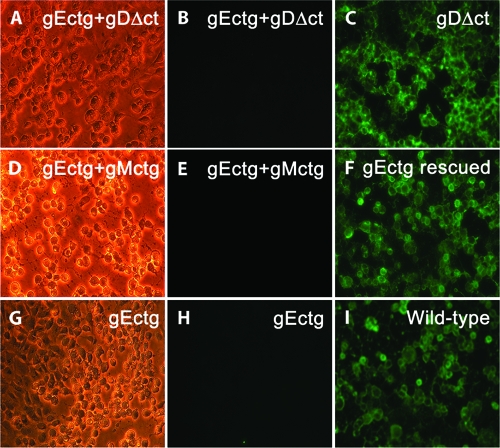
To investigate whether deletion of one or more viral glycoproteins affected the synthesis of other viral glycoproteins, all mutant viruses were tested for the expression of gB, gD, gC, and gE. Western immunoblots revealed that the engineered mutations did not affect the synthesis of gB, gD, and gC (Fig. 4A, B, and C). The mutants specifying the gDΔct mutation expressed gD that migrated with a faster electrophoretic mobility, consistent with the smaller size of the truncated gD (Fig. 4C). The expression of gE was tested by indirect immunofluorescence, since the available antibody did not react strongly enough in immunoblots. Immunofluorescence assay results showed that gE's expression was unaffected by any of the other gene deletions, while no gE expression was detected in cells infected with viruses specifying the gEctg mutations (Fig. 5).
FIG. 5.
Immunofluorescence detection of gE expression. Vero cells were infected at an MOI of 1, and gE expression was detected by indirect immunofluorescence at 36 hpi. The panels in the left column are phase-contrast micrographs of infected cells. The panels in the center column are fluorescent micrographs of mutant viruses lacking gE with anti-gE monoclonal antibody. The panels in the right column are fluorescent micrographs of mutant and wild-type viruses that express gE. All mutant viruses containing the ATG-to-CTG mutations in gE (gEctg) failed to react with anti-gE antibody (panels B, E, and H), while viruses expressing the wild-type gE reacted with the anti-gE antibody (panels C, F, and I).
Ultrastructural characterization of the gDΔct, gEctg, gEctg+gDΔct, and UL20ctgctg mutant viruses.
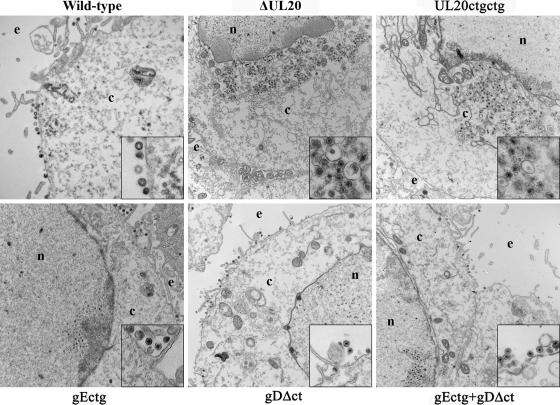
The ultrastructural phenotypes of all viruses relative to that of the wild-type parental virus were investigated by utilizing transmission electron microscopy at 16 hpi, visually examining more than 50 individual cells. As expected, the wild-type virus exhibited no apparent defects in cytoplasmic virion envelopment and egress, as evidenced by the presence of fully enveloped virions intracellularly and extracellularly (Fig. 6). The ultrastructural visualization of Vero cells infected with the different mutant viruses revealed a diverse range of cytoplasmic defects in virion envelopment. The most-pronounced effects were produced by the ΔUL20 and UL20ctgctg mutant viruses, which produced numerous unenveloped capsids in the cytoplasm embedded within morphologically darker stained areas that may be caused by the accumulation of tegument proteins. In contrast, the gEctg, gDΔct, and gEctg+gDΔct ultrastructural morphologies were similar to each other, exhibiting numerous fully enveloped virions in extracellular spaces, while very few viral capsids or enveloped virions were found in the cytoplasm of infected cells (Fig. 6).
FIG. 6.
Ultrastructural morphologies of mutant viruses. Electron micrographs of Vero cells infected at an MOI of 5 with different viruses and processed for electron microscopy at 16 hpi are shown. Enlarged sections of each micrograph are included as insets in each panel. ΔUL20 (constructed previously) is the mutant virus that has the entire UL20 gene deleted (25). UL20ctgctg is the mutant virus containing two ATG-to-CTG codon initiation mutations six bases apart. Nucleus (n), cytoplasm (c), and extracellular space (e) are marked.
DISCUSSION
The main purposes of the work presented herein were to compare the relative importance of gD, gE, gM, gK, and UL20 proteins in cytoplasmic virion envelopment and to specifically assess the potential role of the carboxyl terminus of gD in cytoplasmic envelopment. It was found that a recombinant virus that carried a deletion of the carboxyl terminus of gD (gDΔct) and did not express gE (gEctg) had no appreciable defect in cytoplasmic virion envelopment. In contrast, the UL20ctgctg mutant virus lacking UL20 gene expression exhibited drastic defects in cytoplasmic envelopment, as reported previously for a virus that lacked the entire UL20 gene (26, 44). It was concluded that the carboxyl terminus of gD does not function in a redundant and essential manner with full-length gE in cytoplasmic virion envelopment. The same conclusion was reached for gE and gM, confirming previous findings (6). These data suggest that the gK/UL20 protein complex serves primary functions in cytoplasmic virion envelopment and virion egress.
Over the years, a number of mutant viruses carrying deletions or other mutations within viral genes have been constructed and characterized. These mutant viruses have been engineered utilizing different HSV-1 strains and methodologies, including insertional inactivation, partial gene deletions, and other elaborate methodologies, complicating the direct comparison and interpretation of their phenotypic and replication defects. Recently, the availability of the HSV-1(F) genome cloned into a BAC vector has enabled the rapid construction of recombinant viruses in the same genetic background, thus allowing the construction and direct comparison of recombinant viruses with specific mutations in each targeted gene in the same genomic background, minimizing potential epigenetic effects due to large gene insertions or gene deletions.
Previous work has promoted the hypothesis that gD and gE function in a redundant manner in cytoplasmic virion egress. Specifically, a simultaneous deletion of both gD and gE or gD, gE, and gI genes caused drastic accumulation of unenveloped capsids into the cytoplasm of infected cells (18, 19). Furthermore, studies using gE mutants with progressive deletions of gE's carboxyl terminus showed that the terminal 50 amino acids of gE were dispensable for cytoplasmic morphogenesis in the absence of gD, while conversely, gE amino acids beyond the carboxy-terminal 50 amino acids appeared to be essential for cytoplasmic virion envelopment in the absence of gD (18). We have previously shown that the UL20 protein (UL20p) physically interacts with gK and that this interaction is absolutely essential for UL20p/gK functions in virus-induced cell fusion and cytoplasmic virion envelopment (23, 26, 44). Importantly, while UL20/gK function in both virus-induced cell fusion and cytoplasmic virion envelopment, these functions can be genetically separated, indicating that they involve different domains of gK and UL20 proteins. Therefore, it was of interest to directly compare the egress defects of UL20-null and gD/gE-null viruses.
It has been shown that only the gD extracellular domain, but neither its intramembrane nor its intracellular domain, is essential for virus infectivity and spread (20). In addition, it can be assumed that the carboxyl terminus of gD is the most-probable protein contact domain of gD with tegument proteins during cytoplasmic envelopment, since gD is known to specifically interact with both VP22 and VP16 tegument proteins (19). Therefore, the gD recombinant virus gDΔct was constructed, having the cytoplasmic terminus of gD deleted. The rationale for this mutant virus construction was to preserve the gD receptor domains pertaining to virus-to-cell binding functions while eliminating potential contacts of the gD carboxyl terminus with tegument proteins. As expected, the virus was able to replicate efficiently in comparison to the rate of replication of the wild-type virus (20). Similar results were obtained with the gEctg virus lacking the entire gE gene, in agreement with published observations that gE is not essential for virion egress and spread but can affect the extent of virus spread (14-16, 33).
Ultrastructural examination of cells infected with different mutant viruses revealed that both the gDΔct and gEctg viruses exhibited no appreciable defects in cytoplasmic virion envelopment, as also evidenced by the presence of fully enveloped virions in extracellular spaces and the lack of nonenveloped capsids in the cytoplasm of infected cells. Surprisingly, the double mutant virus gEctg+gDΔct exhibited no appreciable defect in cytoplasmic virion envelopment, while it produced viral plaques that were on average 50% smaller than those of either the gEctg or gDΔct viruses, in agreement with the known effects of gD in viral infectivity and spread, as well as the known role of gE in virus cell-to-cell spread (11). Most likely, the smaller plaques produced by the gEctg+gDΔct virus were due to the combined effect of the individual gDΔct and gEctg mutations. Simultaneous mutations of both gE and gM initiation codons did not affect cytoplasmic virion envelopment and egress, indicating that these two glycoproteins do not function in a redundant manner in cytoplasmic envelopment and egress and confirming previous findings (6). These data combined suggest that simultaneous deletion of the carboxyl terminus of gD (gDΔct) and gE, gI, and gM may not drastically affect cytoplasmic envelopment. Additional work is in progress to address this issue.
Results similar to those obtained in Vero cells with all mutant viruses were also obtained in HEC-1-A, 293, BHK-21, and HEp-2 cells (not shown), indicating that different cell types do not cause appreciable differences in virus replication and spread. We favor the hypothesis that the intramembrane and extracellular domains of gD and gE are indirectly involved in cytoplasmic virion envelopment. This hypothesis is based on recent findings that both gH and gB physically interact with gD (2, 3). These interactions suggest the existence of a coordinated interactive protein complex “fusion machine” that involves complex interactions among gD, gB, and gH and, possibly, other viral proteins and glycoproteins controlling virus entry, virus-induced cell fusion, and virion morphogenesis. In this hypothesis, the absence of gD may cause aberrant gB and gH conformations that directly or indirectly negatively impact cytoplasmic virion envelopment. However, neither gB nor gH is known to be an important determinant in cytoplasmic virion egress, casting some doubt against this hypothesis. Alternatively, given that the gK/UL20p protein complex is exquisitely essential for cytoplasmic virion envelopment, it is possible that lack of gD may affect gK/UL20p functions involved in cytoplasmic virion envelopment by either affecting their transport to TGN membranes or altering their conformations, implying direct or indirect interactions between gD and UL20/gK.
Based on the known and suspected multiple interactions among viral membrane proteins, glycoproteins, and tegument proteins, it is conceivable that the lack of one or more proteins may affect interactions among multiple other proteins involved in virion morphogenesis. Therefore, appropriate caution should be exercised in the assignment of functions to viral proteins on the basis of the deletion of one or more viral structural proteins and glycoproteins. Ideally, domain-specific amino acid changes that affect virion morphogenesis without drastically altering synthesis, intracellular transport, and localization of specific proteins should be preferred in assigning specific protein functions. In this regard, specific amino acid changes in either gK or UL20 that do not appreciably affect their interaction and intracellular localization or the synthesis and intracellular transport of gK and UL20p, as well as other viral glycoproteins, produced gK-null and UL20-null defects in cytoplasmic virion envelopment similar to those generated by deletion of the gK or UL20 genes (24, 44).
In summary, it is clear that the carboxyl terminus of gD does not function in an appreciably redundant manner with either gE or gM in cytoplasmic virion egress in Vero and other cells, although gD, gE, and gM appear to enhance efficient virus spread at a step after cytoplasmic envelopment. Direct comparison of gEctg+gDΔct with viruses lacking UL20 gene expression (ΔUL20 and UL20ctgctg) reveals that the UL20p/gK protein complex appears to play a primary role in cytoplasmic virion envelopment and egress. This observation suggests a model of cytoplasmic envelopment in which gK/UL20 forms protein-protein contacts with tegument proteins, followed by additional secondary interactions among other viral glycoproteins and membrane proteins with tegument proteins. Additional experiments are required to prove this hypothesis and discern the complex interactions between viral membrane protein, glycoproteins, and tegumented capsids in a quest to understand the well-coordinated protein interactions that enable cytoplasmic virion envelopment and virion stability.
Acknowledgments
This work was supported by NIH-NIAID grant AI43000 to K.G.K. D. Chouljenko is a recipient of a Louisiana Board of Regents Economic Development graduate assistantship. We acknowledge financial support by the LSU School of Veterinary Medicine to BIOMMED.
We acknowledge the technical assistance of Olga Borkshenious with electron microscopy and helpful discussions with other BIOMMED staff. We thank Michael Kearney for assistance with the statistical analysis.
Footnotes
Published ahead of print on 8 April 2009.
REFERENCES
- 1.Ace, C. I., T. A. McKee, J. M. Ryan, J. M. Cameron, and C. M. Preston. 1989. Construction and characterization of a herpes simplex virus type 1 mutant unable to transinduce immediate-early gene expression. J. Virol. 632260-2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atanasiu, D., J. C. Whitbeck, T. M. Cairns, B. Reilly, G. H. Cohen, and R. J. Eisenberg. 2007. Bimolecular complementation reveals that glycoproteins gB and gH/gL of herpes simplex virus interact with each other during cell fusion. Proc. Natl. Acad. Sci. USA 10418718-18723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Avitabile, E., C. Forghieri, and G. Campadelli-Fiume. 2007. Complexes between herpes simplex virus glycoproteins gD, gB, and gH detected in cells by complementation of split enhanced green fluorescent protein. J. Virol. 8111532-11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowzard, J. B., R. J. Visalli, C. B. Wilson, J. S. Loomis, E. M. Callahan, R. J. Courtney, and J. W. Wills. 2000. Membrane targeting properties of a herpesvirus tegument protein-retrovirus Gag chimera. J. Virol. 748692-8699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brack, A. R., J. M. Dijkstra, H. Granzow, B. G. Klupp, and T. C. Mettenleiter. 1999. Inhibition of virion maturation by simultaneous deletion of glycoproteins E, I, and M of pseudorabies virus. J. Virol. 735364-5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Browne, H., S. Bell, and T. Minson. 2004. Analysis of the requirement for glycoprotein m in herpes simplex virus type 1 morphogenesis. J. Virol. 781039-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Browne, H., S. Bell, T. Minson, and D. W. Wilson. 1996. An endoplasmic reticulum-retained herpes simplex virus glycoprotein H is absent from secreted virions: evidence for reenvelopment during egress. J. Virol. 704311-4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cai, W. H., B. Gu, and S. Person. 1988. Role of glycoprotein B of herpes simplex virus type 1 in viral entry and cell fusion. J. Virol. 622596-2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calistri, A., P. Sette, C. Salata, E. Cancellotti, C. Forghieri, A. Comin, H. Gottlinger, G. Campadelli-Fiume, G. Palu, and C. Parolin. 2007. Intracellular trafficking and maturation of herpes simplex virus type 1 gB and virus egress require functional biogenesis of multivesicular bodies. J. Virol. 8111468-11478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campadelli-Fiume, G., F. Farabegoli, S. Di Gaeta, and B. Roizman. 1991. Origin of unenveloped capsids in the cytoplasm of cells infected with herpes simplex virus 1. J. Virol. 651589-1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Campadelli-Fiume, G., and T. Gianni. 2006. HSV glycoproteins and their roles in virus entry and egress. In R. M. Sandri-Goldin (ed.), Alpha Herpesviruses, Caister Academic Press, Norfolk, United Kingdom.
- 12.Campbell, M. E., J. W. Palfreyman, and C. M. Preston. 1984. Identification of herpes simplex virus DNA sequences which encode a trans-acting polypeptide responsible for stimulation of immediate early transcription. J. Mol. Biol. 1801-19. [DOI] [PubMed] [Google Scholar]
- 13.Chi, J. H., C. A. Harley, A. Mukhopadhyay, and D. W. Wilson. 2005. The cytoplasmic tail of herpes simplex virus envelope glycoprotein D binds to the tegument protein VP22 and to capsids. J. Gen. Virol. 86253-261. [DOI] [PubMed] [Google Scholar]
- 14.Dingwell, K. S., C. R. Brunetti, R. L. Hendricks, Q. Tang, M. Tang, A. J. Rainbow, and D. C. Johnson. 1994. Herpes simplex virus glycoproteins E and I facilitate cell-to-cell spread in vivo and across junctions of cultured cells. J. Virol. 68834-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dingwell, K. S., L. C. Doering, and D. C. Johnson. 1995. Glycoproteins E and I facilitate neuron-to-neuron spread of herpes simplex virus. J. Virol. 697087-7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dingwell, K. S., and D. C. Johnson. 1998. The herpes simplex virus gE-gI complex facilitates cell-to-cell spread and binds to components of cell junctions. J. Virol. 728933-8942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duffy, C., J. H. Lavail, A. N. Tauscher, E. G. Wills, J. A. Blaho, and J. D. Baines. 2006. Characterization of a UL49-null mutant: VP22 of herpes simplex virus type 1 facilitates viral spread in cultured cells and the mouse cornea. J. Virol. 808664-8675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farnsworth, A., K. Goldsmith, and D. C. Johnson. 2003. Herpes simplex virus glycoproteins gD and gE/gI serve essential but redundant functions during acquisition of the virion envelope in the cytoplasm. J. Virol. 778481-8494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Farnsworth, A., T. W. Wisner, and D. C. Johnson. 2007. Cytoplasmic residues of herpes simplex virus glycoprotein gE required for secondary envelopment and binding of tegument proteins VP22 and UL11 to gE and gD. J. Virol. 81319-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feenstra, V., M. Hodaie, and D. C. Johnson. 1990. Deletions in herpes simplex virus glycoprotein D define nonessential and essential domains. J. Virol. 642096-2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Forrester, A., H. Farrell, G. Wilkinson, J. Kaye, N. Davis-Poynter, and T. Minson. 1992. Construction and properties of a mutant of herpes simplex virus type 1 with glycoprotein H coding sequences deleted. J. Virol. 66341-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Foster, T. P., X. Alvarez, and K. G. Kousoulas. 2003. Plasma membrane topology of syncytial domains of herpes simplex virus type 1 glycoprotein K (gK): the UL20 protein enables cell surface localization of gK but not gK-mediated cell-to-cell fusion. J. Virol. 77499-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Foster, T. P., V. N. Chouljenko, and K. G. Kousoulas. 2008. Functional and physical interactions of the herpes simplex virus type 1 UL20 membrane protein with glycoprotein K. J. Virol. 826310-6323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Foster, T. P., and K. G. Kousoulas. 1999. Genetic analysis of the role of herpes simplex virus type 1 glycoprotein K in infectious virus production and egress. J. Virol. 738457-8468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Foster, T. P., J. M. Melancon, J. D. Baines, and K. G. Kousoulas. 2004. The herpes simplex virus type 1 UL20 protein modulates membrane fusion events during cytoplasmic virion morphogenesis and virus-induced cell fusion. J. Virol. 785347-5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Foster, T. P., J. M. Melancon, T. L. Olivier, and K. G. Kousoulas. 2004. Herpes simplex virus type 1 glycoprotein K and the UL20 protein are interdependent for intracellular trafficking and trans-Golgi network localization. J. Virol. 7813262-13277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Foster, T. P., G. V. Rybachuk, and K. G. Kousoulas. 2001. Glycoprotein K specified by herpes simplex virus type 1 is expressed on virions as a Golgi complex-dependent glycosylated species and functions in virion entry. J. Virol. 7512431-12438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fuchs, W., B. G. Klupp, H. Granzow, and T. C. Mettenleiter. 1997. The UL20 gene product of pseudorabies virus functions in virus egress. J. Virol. 715639-5646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fulmer, P. A., J. M. Melancon, J. D. Baines, and K. G. Kousoulas. 2007. UL20 protein functions precede and are required for the UL11 functions of herpes simplex virus type 1 cytoplasmic virion envelopment. J. Virol. 813097-3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gross, S. T., C. A. Harley, and D. W. Wilson. 2003. The cytoplasmic tail of herpes simplex virus glycoprotein H binds to the tegument protein VP16 in vitro and in vivo. Virology 3171-12. [DOI] [PubMed] [Google Scholar]
- 31.Homa, F. L., and J. C. Brown. 1997. Capsid assembly and DNA packaging in herpes simplex virus. Rev. Med. Virol. 7107-122. [DOI] [PubMed] [Google Scholar]
- 32.Jayachandra, S., A. Baghian, and K. G. Kousoulas. 1997. Herpes simplex virus type 1 glycoprotein K is not essential for infectious virus production in actively replicating cells but is required for efficient envelopment and translocation of infectious virions from the cytoplasm to the extracellular space. J. Virol. 715012-5024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson, D. C., and V. Feenstra. 1987. Identification of a novel herpes simplex virus type 1-induced glycoprotein which complexes with gE and binds immunoglobulin. J. Virol. 612208-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson, D. C., and M. T. Huber. 2002. Directed egress of animal viruses promotes cell-to-cell spread. J. Virol. 761-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson, D. C., M. Wittels, and P. G. Spear. 1984. Binding to cells of virosomes containing herpes simplex virus type 1 glycoproteins and evidence for fusion. J. Virol. 52238-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kamen, D. E., S. T. Gross, M. E. Girvin, and D. W. Wilson. 2005. Structural basis for the physiological temperature dependence of the association of VP16 with the cytoplasmic tail of herpes simplex virus glycoprotein H. J. Virol. 796134-6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klupp, B. G., J. Baumeister, P. Dietz, H. Granzow, and T. C. Mettenleiter. 1998. Pseudorabies virus glycoprotein gK is a virion structural component involved in virus release but is not required for entry. J. Virol. 721949-1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee, E. C., D. Yu, J. Martinez de Velasco, L. Tessarollo, D. A. Swing, D. L. Court, N. A. Jenkins, and N. G. Copeland. 2001. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics 7356-65. [DOI] [PubMed] [Google Scholar]
- 39.Leege, T., W. Fuchs, H. Granzow, M. Kopp, B. G. Klupp, and T. C. Mettenleiter. 2009. Effects of simultaneous deletion of pUL11 and glycoprotein M on virion maturation of herpes simplex virus type 1. J. Virol. 83896-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leege, T., H. Granzow, W. Fuchs, B. G. Klupp, and T. C. Mettenleiter. 18 March 2009. Phenotypic similarities and differences between UL37-deleted pseudorabies virus and herpes simplex virus type 1. J. Gen. Virol. [Epub ahead of print.] doi: 10.1099/vir.0.010322-0. [DOI] [PubMed]
- 41.Ligas, M. W., and D. C. Johnson. 1988. A herpes simplex virus mutant in which glycoprotein D sequences are replaced by beta-galactosidase sequences binds to but is unable to penetrate into cells. J. Virol. 621486-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Loomis, J. S., R. J. Courtney, and J. W. Wills. 2003. Binding partners for the UL11 tegument protein of herpes simplex virus type 1. J. Virol. 7711417-11424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meckes, D. G. J., and J. W. Wills. 2007. Dynamic interactions of the UL16 tegument protein with the capsid of herpes simplex virus. J. Virol. 8113028-13036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Melancon, J. M., T. P. Foster, and K. G. Kousoulas. 2004. Genetic analysis of the herpes simplex virus type 1 UL20 protein domains involved in cytoplasmic virion envelopment and virus-induced cell fusion. J. Virol. 787329-7343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Melancon, J. M., R. E. Luna, T. P. Foster, and K. G. Kousoulas. 2005. Herpes simplex virus type 1 gK is required for gB-mediated virus-induced cell fusion, while neither gB and gK nor gB and UL20p function redundantly in virion de-envelopment. J. Virol. 79299-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mettenleiter, T. C. 2002. Herpesvirus assembly and egress. J. Virol. 761537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mettenleiter, T. C., B. G. Klupp, and H. Granzow. 2006. Herpesvirus assembly: a tale of two membranes. Curr. Opin. Microbiol. 9423-429. [DOI] [PubMed] [Google Scholar]
- 48.Mossman, K. L., R. Sherburne, C. Lavery, J. Duncan, and J. R. Smiley. 2000. Evidence that herpes simplex virus VP16 is required for viral egress downstream of the initial envelopment event. J. Virol. 746287-6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.O'Hare, P., and C. R. Goding. 1988. Herpes simplex virus regulatory elements and the immunoglobulin octamer domain bind a common factor and are both targets for virion transactivation. Cell 52435-445. [DOI] [PubMed] [Google Scholar]
- 50.O'Hare, P., C. R. Goding, and A. Haigh. 1988. Direct combinatorial interaction between a herpes simplex virus regulatory protein and a cellular octamer-binding factor mediates specific induction of virus immediate-early gene expression. EMBO J. 74231-4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reynolds, A. E., E. G. Wills, R. J. Roller, B. J. Ryckman, and J. D. Baines. 2002. Ultrastructural localization of the herpes simplex virus type 1 UL31, UL34, and US3 proteins suggests specific roles in primary envelopment and egress of nucleocapsids. J. Virol. 768939-8952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Singer, G. P., W. W. Newcomb, D. R. Thomsen, F. L. Homa, and J. C. Brown. 2005. Identification of a region in the herpes simplex virus scaffolding protein required for interaction with the portal. J. Virol. 79132-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Skepper, J. N., A. Whiteley, H. Browne, and A. Minson. 2001. Herpes simplex virus nucleocapsids mature to progeny virions by an envelopment → deenvelopment → reenvelopment pathway. J. Virol. 755697-5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smibert, C. A., B. Popova, P. Xiao, J. P. Capone, and J. R. Smiley. 1994. Herpes simplex virus VP16 forms a complex with the virion host shutoff protein vhs. J. Virol. 682339-2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smiley, J. R., and J. Duncan. 1997. Truncation of the C-terminal acidic transcriptional activation domain of herpes simplex virus VP16 produces a phenotype similar to that of the in1814 linker insertion mutation. J. Virol. 716191-6193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tanaka, M., H. Kagawa, Y. Yamanashi, T. Sata, and Y. Kawaguchi. 2003. Construction of an excisable bacterial artificial chromosome containing a full-length infectious clone of herpes simplex virus type 1: viruses reconstituted from the clone exhibit wild-type properties in vitro and in vivo. J. Virol. 771382-1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tatman, J. D., V. G. Preston, P. Nicholson, R. M. Elliott, and F. J. Rixon. 1994. Assembly of herpes simplex virus type 1 capsids using a panel of recombinant baculoviruses. J. Gen. Virol. 751101-1113. [DOI] [PubMed] [Google Scholar]
- 58.Thomsen, D. R., W. W. Newcomb, J. C. Brown, and F. L. Homa. 1995. Assembly of the herpes simplex virus capsid: requirement for the carboxyl-terminal twenty-five amino acids of the proteins encoded by the UL26 and UL26.5 genes. J. Virol. 693690-3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tischer, B. K., J. von Einem, B. Kaufer, and N. Osterrieder. 2006. Two-step Red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. BioTechniques 40191-197. [DOI] [PubMed] [Google Scholar]
- 60.Walters, J. N., G. L. Sexton, J. M. McCaffery, and P. Desai. 2003. Mutation of single hydrophobic residue I27, L35, F39, L58, L65, L67, or L71 in the N terminus of VP5 abolishes interaction with the scaffold protein and prevents closure of herpes simplex virus type 1 capsid shells. J. Virol. 774043-4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weinheimer, S. P., B. A. Boyd, S. K. Durham, J. L. Resnick, and D. R. O'Boyle II. 1992. Deletion of the VP16 open reading frame of herpes simplex virus type 1. J. Virol. 66258-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Whiteley, A., B. Bruun, T. Minson, and H. Browne. 1999. Effects of targeting herpes simplex virus type 1 gD to the endoplasmic reticulum and trans-Golgi network. J. Virol. 739515-9520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhu, Q., and R. J. Courtney. 1994. Chemical cross-linking of virion envelope and tegument proteins of herpes simplex virus type 1. Virology 204590-599. [DOI] [PubMed] [Google Scholar]