Abstract
The essential proteins NS1 and NS2 of minute virus of mice are encoded by mRNAs R1 and R2, respectively. R2 is derived from R1 by excision of a large intron and thus splicing governs the relative ratios of NS1 and NS2. Excision of the large intron utilizes a nonconsensus 5′ donor site. We identified a U-rich and A-rich intronic sequence immediately downstream of the nonconsensus 5′ donor site that functions as an intronic splicing enhancer (ISE) required for efficient large-intron excision. The ISE binds the cellular RNA-processing proteins TIA-1 and TIAR, which enhance usage of the nonconsensus donor.
The essential nonstructural proteins NS1 and NS2 of minute virus of mice (MVM) are encoded by mRNAs R1 and R2, respectively, utilizing open reading frames (ORFs) in the left half of the genome (6, 8, 21). R1 and R2 are both generated by the P4 promoter and are alternatively spliced at an overlapping small intron in the center of the genome (6, 15). This results in the generation of three forms of NS2; however, NS1 terminates prior to the small intron, so alternative splicing of the small intron from R1 has no known effect on NS1 (6, 7). The larger R1 RNA transcript is further spliced within the nonstructural gene region between nucleotides 514 and 1989 to generate R2. From its initiation at nucleotide (nt) 260, until the large splice donor at nt 514, NS2 is encoded in the same ORF as NS1. After the large splice, translation of NS2 shifts to ORF 2 (8).
The ratio of NS1 to NS2 has been shown to be critical for successful viral infection (7, 24). Since NS1 and NS2 are encoded by RNAs that derive from the same promoter, their relative accumulated levels are controlled by posttranscriptional mechanisms. Alternative splicing of the large intron present in P4-generated RNAs (24) and protein stability (7, 24) have been identified as the major processes governing the relative abundances of NS1 and NS2.
The large-intron 3′ splice site has a poor polypyrimidine tract, and previous studies in our lab have shown that sequences within the large-intron 3′ splice site, exon splicing enhancer elements (ESEs) within the downstream NS2-specific exon, and excision of the downstream small intron participate in the proper excision of the large intron (13, 14, 27-29). In addition, differences in large-intron excision due to differences in the branch point sequences have been shown to play a role in tissue tropism differences between the prototype MVM(p) strain and the lymphotropic variant MVM(i) (4, 9).
The MVM large-intron donor cleavage site is aA/gCaagt rather than the consensus sequence aG/gTaagt (differences are in bold capital letters; the NS1 ORF is underlined), which is fully complementary to the 5′-end of U1 snRNA. Donors utilizing a GC rather than a consensus GT diribonucleotide, although it is the major variant, are still very rare (accounting for less than 1% of the total mammalian introns) and are normally associated with introns that are alternatively spliced (2, 5, 25). In the case of MVM, a T at nucleotide 516 would introduce a termination codon in NS1; thus, the use of the nonconsensus large-intron donor appears necessary for translation of NS1. It seems likely that the nonconsensus donor might also play a role in maintaining the appropriate balance between R1 and R2, and hence between NS1 and NS2. In other cases, additional cis-acting elements and trans-acting factors are typically required to stabilize the spliceosome interaction with a nonconsensus donor (2, 16, 26, 30), and we show here that this is also the case for the MVM large intron.
U- and A-rich intronic sequences immediately downstream of the 5′ splice site of the large intron are required for its efficient excision.
Alternative splicing is often governed by intronic and exonic elements other than those directly comprising the 5′ and 3′ splice sites, which interact with cellular splicing regulators (1, 17). Such elements are often found associated with nonconsensus donor and acceptor sites (11, 16). Inspection of sequences surrounding the MVM large-intron nonconsensus 5′ splice site revealed unusual U- and A-rich regions immediately downstream of this donor.
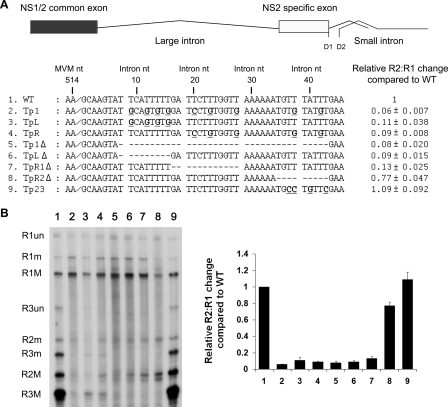
Modifications that reduced the number of U residues in this region between nts 9 and 42 of the large intron (with intron nt 1 being nt 515 of the full-genome sequence) in the Tp1 mutant resulted in more than a 10-fold decrease in splicing of the large intron following transfection of mutants onto murine A9 cells (compare Fig. 1A, lane 2, and B, lane 1). A9 cells were propagated and transfected as previously described (4). Mutations were made in the original infectious clone derived by Merchlinsky et al. (18), using standard PCR techniques as described previously (23), and were sequenced to ensure that they were as expected. RNase protection assays were performed with probes spanning MVM nt 1858 to 2377, and the results were quantified using a Fuji FLA 5000 phosphorimager as previously described (20, 24). These mutations change the coding sequence for NS1 (and thus the capsid P38 promoter, which generates R3 transcripts, is not activated); however, it is important to note that we have previously shown that MVM proteins play no role in the splicing of MVM RNAs (22). When intron nts 8 to 43 were removed to generate the Tp1Δ in-frame deletion mutant, splicing of the large intron was similarly reduced (Fig. 1A, compare lanes 5 and 1, and B), arguing against the possibility that the mutations in Tp1 had introduced an inhibitory sequence. Taken together, these results suggest that U-rich sequences downstream of the nonconsensus donor are required for efficient excision of the large intron.
FIG. 1.
U- and A-rich intronic sequences immediately downstream of the 5′ splice site of the MVM large intron are required for its efficient excision. (A) Diagram of the R2 transcript and the DNA region just downstream of the large-intron donor, showing changes (bold and underlined) or deletions (dashed lines) in the U-rich regions (slashes indicate the 5′ cleavage site between nts 514 and 515 of the published sequence) of the various mutants, as described in the text. MVM and intron numbering is shown. RNase protection analyses quantified using Fujifilm MultiGauge software and performed as described in the text, using an antisense probe spanning nt 1858 to 2377, were used to quantify RNAs generated following transfection of each mutant (as described in the text); the results are shown to the right. Data from at least three experiments, with standard deviations, are presented as the relative R2:R1 changes compared to that of the wild type (WT). (B) Representative RNase protection assay. The identities of the RNA species are given on the left. M, usage of small intron D1 at nt 2280; m, usage of small-intron donor D2 at nt 2317; un, RNA remaining unspliced through the probe region. A graphic analysis of the quantification data shown in panel A is presented in the right panel. (C) Diagram of the DNA region just downstream of the large-intron donor, showing changes (bold and underlined) or deletions (dashed lines) in the A-rich region (slashes indicate the 5′ cleavage site between nts 514 and 515 of the published sequence) of the various mutants, as described in the text. MVM and intron numbering is shown. (D) Left panel: representative RNase protection analysis of RNAs generated from constructs shown in panel C, using a probe spanning nt 1858 to 2377. Right panel: quantification of RNase protection. Data from at least three experiments, with standard deviations, are presented in tabular (bottom) and graphical (top) forms as relative R2:R1 changes compared to that of the wild type. (E) Quantification of RNase protection of constructs, indicated by using probe-spanning nt 1858 to 2377. CD1 refers to consensus donor mutation 1 (see text). The quantifications include data from at least three experiments, with standard deviations, and are presented as relative R2:R1 changes compared to that of the wild type.
Reducing the number of U residues between intron nts 9 and 16 in the TpL mutant or 20 and 42 in the TpR mutant (Fig. 1A) led to decreases in splicing of the large intron similar to that seen for RNAs generated by the larger mutation Tp1 (Fig. 1B, compare lanes 3 and 4 to lanes 2 and 1). Similarly, deletion of intron nts 8 to 16 in the TpLΔ mutant or 17 to 28 in the TpR1Δ mutant (Fig. 1A) led to significant decreases in splicing (Fig. 1B, compare lanes 6 and 7 to lanes 5 and 1). Reducing the number of U residues within intron nts 35 to 43 in the Tp23 mutant or deletion of this region completely in the TpR2Δ mutant (Fig. 1A) had little effect on splicing of the large intron (Fig. 1B, compare lanes 8 and 9 to lane 1). Additional mutations of U-rich residues between intron nts 8 and 28 displayed intermediate phenotypes that did not permit further definition of the essential sequences (data not shown). Based on these results, we suggest that the U-rich region, spanning intron nts 8 to 28, is required for efficient splicing of the large intron and likely for efficient usage of the nonconsensus 5′ donor site.
The small A-rich motif between intron nts 29 to 34, which lies adjacent to the U-rich region, affected large-intron excision, but less dramatically. Deletion of this region in the AGΔ mutant (Fig. 1C) resulted in a reduction of large-intron excision three- to fourfold (Fig. 1D, compare lane 2 to lane 1). Mutations within this region implicated the A residues at nt 29 and 32 as most important; changing these in the AP1 mutant, but not the other nucleotides in this region in the AP2 and AP3 mutants (Fig. 1C), to G residues fully recapitulated the magnitude of splicing reduction seen for the deletion mutant (Fig. 1D, compare lanes 3 to 5 to lane 1).
The presence of a complementary A-rich and U-rich region in this region (Fig. 1) suggested that perhaps RNA secondary structure dependent on complementary base pairing between these elements may have played a role in the function of this region. However, mutations in which the two elements were altered in such a way as to change their sequences yet retain compensatory complementary base pairing still could not support splicing of the large intron (data not shown).
Taken together, the U- and A-rich motifs between intron nts 8 and 34 were shown to be required for large-intron excision and to have the properties of an intronic splicing enhancing element (ISE). Improvement of the nonconsensus intron donor to consensus (in this case, the CD1 mutation, which changed the wild-type aA/gCaagt to aG/gTaagt, terminating NS1), which increased large-intron excision in an otherwise wild-type background, could recover splicing in the Tp mutant to nearly wild-type levels (Fig. 1E), suggesting that the ISE acts to strengthen the nonconsensus 5′ donor site.
The activity of the ISE is distance dependent.
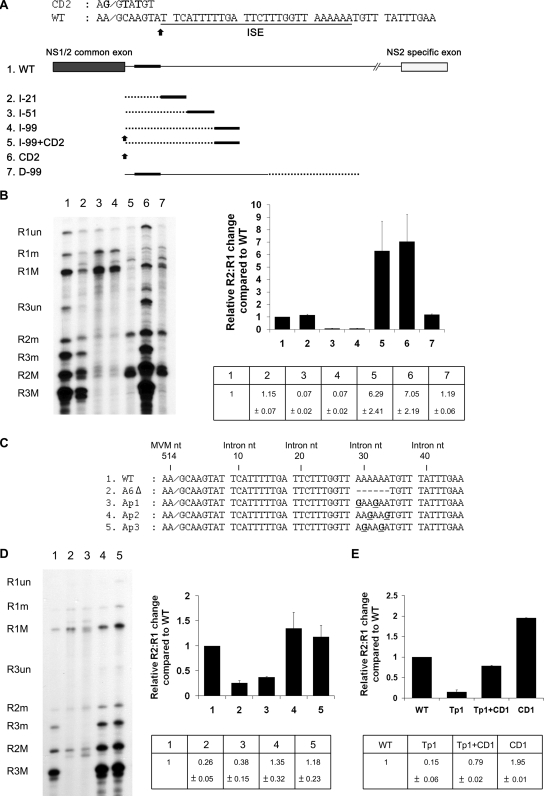
If the intron region from nts 8 to 34 is truly an ISE affecting the nonconsensus donor, it might be expected that its effect is dependent upon its distance from the donor. To test this, we inserted heterologous sequences of increasing lengths taken from a region of the capsid-coding gene of MVM carrying no known cis-acting regulatory elements between intron nts 7 and 8 to expand the distance between the ISE and the donor (MVM DNA nts 3757 to 3777 [21 nts], 3757 to 3807 [51 nts], and 3757 to 3855 [99 nts] were used for insertion mutations I-21, I-51, I-99, and D-99, respectively [Fig. 2A]). As can be seen in Fig. 2B, insertion of 21 nts at this site had no discernible effects on large-intron excision (Fig. 2B, compare lane 2 to lane 1). However, when the ISE was removed further downstream of the donor by 51 and 99 nts, respectively, large-intron excision was reduced significantly (Fig. 2B, compare lanes 3 and 4 to lane 1). The decrease in splicing seen in these experiments was not due merely to an increase in the size of the intron; insertion of the identical 99-nt capsid sequence downstream of the ISE at nt 1250 had no effect on large-intron excision (Fig. 2B, lane 7).
FIG. 2.
The activity of the ISE is distance dependent. (A) Top: Nucleotide sequence of the 5′ splice site region, indicating the position of the ISE and the insertion point for spacer additions. The sequence of the CD2 mutation is also shown (changes from the wild-type [WT] are in bold). Bottom: Diagram of insertion constructs described in the text. Dotted lines indicate heterologous spacer insertion fragments which were taken from a region of the capsid-coding gene of MVM not known to contain RNA-processing signals (see text). The black bars indicate the position of the ISE. Black arrows indicate consensus donor mutation CD2, which is described further in the text. The diagram is not to scale. (B) Left panel: Representative RNase protection analysis of constructs shown in panel A, using probe-spanning nt 1858 to 2377. Right panel: Quantification of RNase protection. The data, taken from at least three experiments, with standard deviations, are presented as the relative R2:R1 changes compared to that of the wild type in both tabular (bottom) and graphical (top) forms.
We generated an additional mutation (CD2) that improved the U1 snRNA-complementarity of the donor site from the wild-type aA/gCaagt to aG/gTaTgt. Rather than introducing a termination mutation in NS1, this alteration changed Gln86 (CAA) to Tyr (TAT). When the nonconsensus wild-type donor in the I-99 insertion mutant was replaced with the CD2 sequence, excision of the large intron was increased, supporting the notion that in its native position the ISE acts to strengthen the nonconsensus 5′ donor site. Interestingly, in an otherwise wild-type background, the CD2 mutation increased large-intron excision to a greater extent than the CD1 mutation (compare Fig. 1E to Fig. 2B, lane 6). Further, in contrast to the results described for the Tp+CD1 mutant described above, in which improvement of the donor to consensus CD1 restored splicing to nearly wild-type levels, improvement of the I-99 donor to the CD2 sequence (I-99+CD2) led to excision of the large intron to levels similar to those seen for the CD2 donor alone, i.e., much greater than for the wild-type (Fig. 2B, lane 5). The basis of the difference between the two results is not clear; however, it likely suggests a complex interaction between the IES and the donor site. Additionally, because NS1 bearing the CD2 mutation (and the change of Gln 86 to Tyr) could efficiently activate P38 (Fig. 2B, lane 6), we placed this mutation back into the wild-type infectious clone. The mutant was nonviable (data not shown). Thus, even though this strong donor did not terminate NS1, it was not tolerated at this site, either because of the Gln-to-Tyr mutation or because of the reduced relative levels of NS1.
TIA-1 and TIAR are important for the efficient use of the MVM nonconsensus large-intron donor.
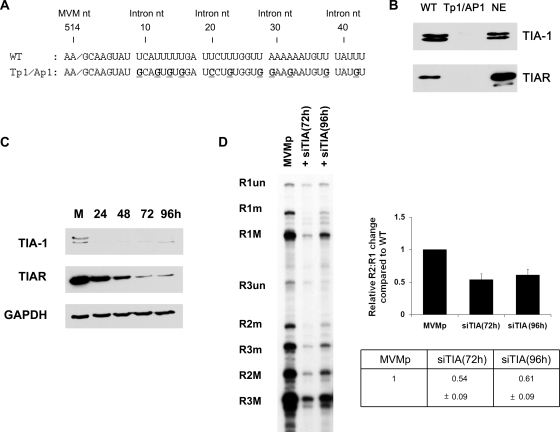
The RNA binding proteins TIA-1 and TIAR are RNA-processing factors that can bind to U-rich regions in RNA and act in a redundant manner to stabilize spliceosomal protein interactions with nonconsensus donor sites (10, 12, 16). TIA-1 and TIAR are abundant in murine and human cells permissive for MVM (data not shown), making these proteins candidates for interaction at the MVM large-intron ISE. As shown in Fig. 3B, both TIA-1 and TIAR efficiently bound to an oligoribonucleotide comprising the wild-type, but not a mutant, MVM large-intron ISE (the sequences of the wild-type and mutant oligoribonucleotide are shown in Fig. 3A). RNA chromatography affinity assays were performed according to published procedures (3), with the following modifications. Biotin-labeled substrate RNAs were synthesized by Integrated DNA Technologies (Corvallis, IA). Substrate RNAs were incubated in a reaction mixture containing 150 μl of HeLa cell nuclear extract (750 to 1,000 μg of protein) in a final volume of 250 μl (20 mM HEPES [pH 7.9], 5% [vol/vol] glycerol, 0.1 M KCl, 0.2 mM EDTA, 0.5 mM dithiothreitol, 4 mM ATP, 4 mM MgCl2, 5 mM creatine phosphate, 2.5% [wt/vol] polyvinyl alcohol) together with 30 μl of streptavidin agarose (Novagen). Beads were then washed three times with 1 ml of washing buffer (20 mM HEPES [pH 7.9], 5% [vol/vol] glycerol, 0.1 M KCl, 0.2 mM EDTA, 0.5 mM dithiothreitol, 4 mM MgCl2). Proteins specifically bound to the substrate RNA were eluted by the addition of 100 μl of protein sample buffer and separated on polyacrylamide sodium dodecyl sulfate gels, electroblotted, and probed with antibodies as indicated in the figure legend. Western immunoblot analysis was performed as previously described (19). The primary antibodies used were as follows: goat anti-human TIA-1, goat anti-human TIAR, rabbit anti-human GAPDH (glyceraldehyde-3-phosphate dehydrogenase) (all from Santa Cruz Biotechnology, Inc.). This result indicated that TIA-1 and TIAR could specifically bind to the U-rich region of the large intron.
FIG. 3.
TIA-1 and TIAR bind to the ISE and govern efficient use of the MVM nonconsensus large-intron donor. (A) Sequences of oligoribonucleotides used for the RNA affinity chromatography assays described in the text. MVM and intron numbering is shown. (B) RNA affinity chromatography assay, performed as described in the text, with oligoribonucleotides shown in panel A. Membranes were probed with anti-TIA-1 or anti-TIAR antibodies. NE, HeLa cell nuclear extract. (C) Western analysis, using anti-TIA-1, anti-TIAR, or anti-GAPDH antibodies, of total protein following siRNA treatment specific to TIA-1 and TIAR. Cells were collected at the indicated times following siRNA transfection and were analyzed as described in the text. M, mock. (D) Left panel: Representative RNase protection analysis of MVM-generated RNA following siRNA transfection, using probe spanning nt 1858 to 2377. Right panel: Quantification of RNase protection; data from at least three experiments, with standard deviations, are presented as relative R2:R1 changes compared to that of the wild type in tabular (bottom) and graphical (top) forms. WT, wild type.
We investigated whether TIA-1 and TIAR could affect the excision of the large intron, using specific small interfering RNAs (siRNAs) to reduce their abundances in host cells. For these assays, A9 cells were transfected with 40-nM siRNA duplexes, using HiPerFect reagent (Qiagen). After 24 h, cells were retransfected with 40 nM siRNA. MVM test plasmids were transfected 24 h after the second siRNA transfection, and RNA was extracted for analysis 24 h later. siRNAs against TIA-1 and TIAR (a set of four siGenome and On-Targetplus reagents, catalog numbers LQ-061591-00-0005 and LQ-047822-01-0005, respectively) were purchased from Dharmacon. As can be seen in Fig. 3C, the endogenous levels of both TIA-1 and TIAR were significantly reduced in murine A9 cells after 72 h of siRNA transfection. At this time point, excision of the large intron from a cotransfected MVM genomic clone was also significantly decreased (Fig. 3D), suggesting that TIA-1 and TIAR were necessary for efficient excision of the large intron. No effect on the splicing of the small intron from P38-generated pre-mRNAs expressed from the same plasmid was observed following siRNA treatment, suggesting that the effect of the TIA-1/TIAR siRNA treatment was not promiscuous.
Taken together, these results suggest that the splicing factors TIA-1 and TIAR, binding specifically to the U-rich region immediately downstream of the donor, directly enhance usage of the nonconsensus site. Overexpression of TIA-1 and TIAR in A9 cells did not enhance large-intron splicing (data not shown), suggesting that the endogenous levels of these two factors in these cells was sufficient to support cleavage of the large-intron donor.
Although depletion of TIA-1 and TIAR via siRNA reduced splicing, the reduction was not as severe as the reduction in response to the cis-acting Tp mutation (see Fig. 1). It is likely that this was due to incomplete depletion of these factors; however, it remains possible that additional cellular factors binding to the ISE play a role in large-intron excision. We have not found differences in the levels of TIA-1 and TIAR in various murine cells of different differentiation statuses (data not shown). Thus, the differential expression of NS2 that influences the replication of MVM variants likely is controlled primarily by the 3′ splice site (4, 9).
We have previously shown that the NS2-specific exon downstream of the large intron contains a bipartite CA-rich enhancer element (13). We have recently identified sequences within the NS1/NS2 shared exon which govern usage of the downstream nonconsensus donor (data not shown), which will be the object of further study. Regulation of the 5′ cleavage event of MVM P4-generated pre-mRNAs is quite complex, involving a nonconsensus donor, both downstream and upstream regulating sequences, and likely also definition of the 5′ exon, involving the 5′-mRNA cap. Together with the complex regulation of large-intron splicing governed at the 3′ splice site, it is clear that excision of the large intron from R1 is an exquisitely controlled process that highlights the importance of the relative ratios of NS1 and NS2 for successful infection.
Acknowledgments
We thank Lisa Burger for outstanding technical assistance; Jianming Qiu, Chris Lorson, and Mark Hannink for helpful discussions; and Mariano Garcia-Blanco and Chris Lorson for valuable reagents.
This study was supported by Public Health Service grants RO1 AI21302 and RO1 AI46458 from the NIAID to D.P.
Footnotes
Published ahead of print on 1 April 2009.
REFERENCES
- 1.Black, D. L. 2003. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 72291-336. [DOI] [PubMed] [Google Scholar]
- 2.Burset, M., I. A. Seledtsov, and V. V. Solovyev. 2000. Analysis of canonical and non-canonical splice sites in mammalian genomes. Nucleic Acids Res. 284364-4375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caputi, M., A. Mayeda, A. R. Krainer, and A. M. Zahler. 1999. hnRNP A/B proteins are required for inhibition of HIV-1 pre-mRNA splicing. EMBO J. 184060-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choi, E. Y., A. E. Newman, L. Burger, and D. Pintel. 2005. Replication of minute virus of mice DNA is critically dependent on accumulated levels of NS2. J. Virol. 7912375-12381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Churbanov, A., S. Winters-Hilt, E. V. Koonin, and I. B. Rogozin. 2008. Accumulation of GC donor splice signals in mammals. Biol. Direct 330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clemens, K. E., D. R. Cerutis, L. R. Burger, C. Q. Yang, and D. J. Pintel. 1990. Cloning of minute virus of mice cDNAs and preliminary analysis of individual viral proteins expressed in murine cells. J. Virol. 643967-3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cotmore, S. F., and P. Tattersall. 1990. Alternate splicing in a parvoviral nonstructural gene links a common amino-terminal sequence to downstream domains which confer radically different localization and turnover characteristics. Virology 177477-487. [DOI] [PubMed] [Google Scholar]
- 8.Cotmore, S. F., and P. Tattersall. 1986. Organization of nonstructural genes of the autonomous parvovirus minute virus of mice. J. Virol. 58724-732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.D'Abramo, A. M., Jr., A. A. Ali, F. Wang, S. F. Cotmore, and P. Tattersall. 2005. Host range mutants of minute virus of mice with a single VP2 amino acid change require additional silent mutations that regulate NS2 accumulation. Virology 340143-154. [DOI] [PubMed] [Google Scholar]
- 10.Del Gatto-Konczak, F., C. F. Bourgeois, C. Le Guiner, L. Kister, M. C. Gesnel, J. Stevenin, and R. Breathnach. 2000. The RNA-binding protein TIA-1 is a novel mammalian splicing regulator acting through intron sequences adjacent to a 5′ splice site. Mol. Cell. Biol. 206287-6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fairbrother, W. G., R. F. Yeh, P. A. Sharp, and C. B. Burge. 2002. Predictive identification of exonic splicing enhancers in human genes. Science 2971007-1013. [DOI] [PubMed] [Google Scholar]
- 12.Förch, P., O. Puig, N. Kedersha, C. Martinez, S. Granneman, B. Seraphin, P. Anderson, and J. Valcarcel. 2000. The apoptosis-promoting factor TIA-1 is a regulator of alternative pre-mRNA splicing. Mol. Cell 61089-1098. [DOI] [PubMed] [Google Scholar]
- 13.Gersappe, A., and D. J. Pintel. 1999. CA- and purine-rich elements form a novel bipartite exon enhancer which governs inclusion of the minute virus of mice NS2-specific exon in both singly and doubly spliced mRNAs. Mol. Cell. Biol. 19364-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haut, D. D., and D. J. Pintel. 1999. Inclusion of the NS2-specific exon in minute virus of mice mRNA is facilitated by an intronic splicing enhancer that affects definition of the downstream small intron. Virology 25884-94. [DOI] [PubMed] [Google Scholar]
- 15.Jongeneel, C. V., R. Sahli, G. K. McMaster, and B. Hirt. 1986. A precise map of splice junctions in the mRNAs of minute virus of mice, an autonomous parvovirus. J. Virol. 59564-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Le Guiner, C., F. Lejeune, D. Galiana, L. Kister, R. Breathnach, J. Stevenin, and F. Del Gatto-Konczak. 2001. TIA-1 and TIAR activate splicing of alternative exons with weak 5′ splice sites followed by a U-rich stretch on their own pre-mRNAs. J. Biol. Chem. 27640638-40646. [DOI] [PubMed] [Google Scholar]
- 17.Matlin, A. J., F. Clark, and C. W. Smith. 2005. Understanding alternative splicing: towards a cellular code. Nat. Rev. Mol. Cell Biol. 6386-398. [DOI] [PubMed] [Google Scholar]
- 18.Merchlinsky, M. J., P. J. Tattersall, J. J. Leary, S. F. Cotmore, E. M. Gardiner, and D. C. Ward. 1983. Construction of an infectious molecular clone of the autonomous parvovirus minute virus of mice. J. Virol. 47227-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller, C. L., and D. J. Pintel. 2002. Interaction between parvovirus NS2 protein and nuclear export factor Crm1 is important for viral egress from the nucleus of murine cells. J. Virol. 763257-3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Naeger, L. K., R. V. Schoborg, Q. Zhao, G. E. Tullis, and D. J. Pintel. 1992. Nonsense mutations inhibit splicing of MVM RNA in cis when they interrupt the reading frame of either exon of the final spliced product. Genes Dev. 61107-1119. [DOI] [PubMed] [Google Scholar]
- 21.Pintel, D., D. Dadachanji, C. R. Astell, and D. C. Ward. 1983. The genome of minute virus of mice, an autonomous parvovirus, encodes two overlapping transcription units. Nucleic Acids Res. 111019-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pintel, D. J., A. Gersappe, D. Haut, and J. Pearson. 1995. Determinants that govern alternative splicing of parvovirus pre-mRNAs. Semin. Virol. 6283-290. [Google Scholar]
- 23.Qiu, J., and D. J. Pintel. 2002. The adeno-associated virus type 2 Rep protein regulates RNA processing via interaction with the transcription template. Mol. Cell. Biol. 223639-3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schoborg, R. V., and D. J. Pintel. 1991. Accumulation of MVM gene products is differentially regulated by transcription initiation, RNA processing and protein stability. Virology 18122-34. [DOI] [PubMed] [Google Scholar]
- 25.Thanaraj, T. A., and F. Clark. 2001. Human GC-AG alternative intron isoforms with weak donor sites show enhanced consensus at acceptor exon positions. Nucleic Acids Res. 292581-2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu, Y., Y. Zhang, and J. Zhang. 2005. Distribution of exonic splicing enhancer elements in human genes. Genomics 86329-336. [DOI] [PubMed] [Google Scholar]
- 27.Zhao, Q., A. Gersappe, and D. J. Pintel. 1995. Efficient excision of the upstream large intron from P4-generated pre-mRNA of the parvovirus minute virus of mice requires at least one donor and the 3′ splice site of the small downstream intron. J. Virol. 696170-6179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao, Q., S. Mathur, L. R. Burger, and D. J. Pintel. 1995. Sequences within the parvovirus minute virus of mice NS2-specific exon are required for inclusion of this exon into spliced steady-state RNA. J. Virol. 695864-5868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao, Q., R. V. Schoborg, and D. J. Pintel. 1994. Alternative splicing of pre-mRNAs encoding the nonstructural proteins of minute virus of mice is facilitated by sequences within the downstream intron. J. Virol. 682849-2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zuccato, E., E. Buratti, C. Stuani, F. E. Baralle, and F. Pagani. 2004. An intronic polypyrimidine-rich element downstream of the donor site modulates cystic fibrosis transmembrane conductance regulator exon 9 alternative splicing. J. Biol. Chem. 27916980-16988. [DOI] [PubMed] [Google Scholar]