Abstract
Thalassemia is a heritable human anemia caused by a variety of mutations that affect expression of the α- or the β-chain of hemoglobin. The expressivity of the phenotype is likely to be influenced by unlinked modifying genes. Indeed, by using a mouse model of α-thalassemia, we find that its phenotype is strongly influenced by the genetic background in which the α-thalassemia mutation resides [129sv/ev/129sv/ev (severe) or 129sv/ev/C57BL/6 (mild)]. Linkage mapping indicates that the modifying gene is very tightly linked to the β-globin locus (Lod score = 13.3). Furthermore, the severity of the phenotype correlates with the size of β-chain-containing inclusion bodies that accumulate in red blood cells and likely accelerate their destruction. The β-major globin chains encoded by the two strains differ by three amino acids, one of which is a glycine-to-cysteine substitution at position 13. The Cys-13 should be available for interchain disulfide bridging and consequent aggregation between excess β-chains. This normal polymorphic variation between murine β-globin chains could account for the modifying action of the unlinked β-globin locus. Here, the variation in severity of the phenotype would not depend on a change in the ratio between α- and β-chains but on the chemical nature of the normal β-chain, which is in excess. This work also indicates that modifying genes can be normal variants that—absent an apparent physiologic rationale—may be difficult to identify on the basis of structure alone.
Keywords: hemoglobin
The severity of thalassemia, one of the most prevalent of the heritable human anemias, is brought about by the insufficient production of functional hemoglobin and the degree of quantitative imbalance between α- or β-globin chains (1–8). Needless to say, there are a large number of genetic alterations that can cause such a disturbance in globin chain production, and many of these have been characterized at the molecular level (see review in ref. 1). Thus, for example, thalassemias arise through mutations that induce alterations in gene transcription, pre-mRNA splicing, mRNA and protein stability, polyadenylation, and even translation (1). The disease offers a virtual textbook of potential molecular genetic lesions.
In addition to the great variety of mutations that affect the expressivity of the phenotype, the phenotype is likely to be further influenced by unlinked modifying genes that could account for the variable severity of the disease seen in certain families wherein the primary genetic lesion is likely to be identical by descent (9–11). To date, save for pathologic hemaglobinopathies, no such modifying genes have been identified.
An opportunity to address the issue of modifying genes arose when we created a mouse model of α-thalassemia and noted that the severity of the disease was greatly influenced by the genetic background in which the mutation was expressed [129sv/ev/129sv/ev (severe) or 129sv/ev/C57BL/6 (mild)] (12). In the work described below, we have identified the modifying gene that ameliorates the α-thalassemia phenotype as the normal β-globin allele present in the C57BL/6 strain of mouse. The severe form of thalassemia can be related to a Cys-13 residue present in the β-globin gene of the 129sv/ev strain and the milder form to a glycine in this position in the C57BL/6 strain. The aggregates of excess β-chains that likely form via cysteine sulfhydryl bridges would account for the very large inclusion bodies seen in the red blood cells in α-thalassemic mice bearing two copies of the 129sv/ev β-globin allele. This mechanism indicates that the severity of thalassemia can depend not only on the imbalance between α- and β-chains but on the nature of the allelic form of the chain in excess.
MATERIALS AND METHODS
Animals.
The α1 knockout mice on 129sv/ev background used for the genetic crosses were generated in our laboratory (12). The inbred strains C57BL-6 and BALB/c were obtained from Taconic Farms and were housed in our animal facility until they were old enough to breed.
Genotyping.
Southern blot analysis. Tail DNAs were prepared from tail biopsies as described (12). DNAs were then digested with EcoRI restriction endoclease run on 1% agarose gel, transferred to nitrocellulose membrane, and hybridized with β-globin cDNA derived from a mouse spleen cDNA library. Analysis of the α-globin genotype was as described (12).
PCR.
D7Mit-40 primers were used in a PCR reaction to distinguish between β-globins of C57BL/6, 129sv/ev, and BALB/c alleles. The amplification reaction contained 1 μl of diluted DNA (1:10 vol/vol in H20), 17 μl of PCR supermix (GIBCO/BRL), 1.5 μl of 5′ oligonucleotides and 1.5 μl of 3′ oligonucleotides in a 21-μl total reaction volume. The thermal cycle conditions were: 94°C for 1 min followed by 35 cycles of 94°C for 30 sec, 52°C for 30 sec, and extension at 72°C for 1 min. The PCR reactions were analyzed by gel electrophoresis in a 4% agarose (NuSieve GTG) gel, prepared, and run with 1× TBE (0.1 M Tris/0.08 M borate/0.001 M EDTA, pH 8.6).
Peripheral Blood Analysis.
Blood was collected from tails in potassium EDTA-treated microtubules. Hematologic indices were determined by using standard hospital methods in the clinical laboratory at Children’s Hospital, Boston.
Cloning Adult β-Globins by PCR.
Tail DNA from 129sv/ev mice was used as a template for a PCR reaction. Primers were designed according to a published BALB/c β-globin major (13). An EcoRI site was added to the primers (forward) 5′-ACGAATTCATGGTGCACCTGACTGAT-3′ and (reverse) 5′-ACGAATTCGTGGTACTTGTGAGCCAAGCA-3′.
The primer reaction contained 1 μl of diluted 129sv/ev genomic tail DNA (1:10 vol/vol in H2O), 0.2 mM dNTP, 1 mM MgCl2, 0.7 μl of forward primer and 0.7 μl of reverse primer (0.5 mg/ml), 5 units of Pwo DNA polymerase (Boehringer Mannheim) in a total reaction volume of 50 μl at pH 8.3. The thermal cycle conditions were: 94°C for 1 min, followed by 35 cycles of 94°C for 30 sec, 56°C for 30 sec, and extension at 72°C for 1 min. The PCR product was restricted with EcoRI and purified by using a Qiagen (Chatsworth, CA) column. It was then incubated for 10 min at 70°C with dNTP and Taq polymerase to add deoxyadenosine(A) to the 3′ ends. It was then ligated to a linearized pCR2.1-Topo vector containing 3′ deoxythymidine(T) overhang (Invitrogen kit). Competent cells for transformation were provided by the kit. Several clones were analyzed with HindIII digestion; only β-major had the site. Two clones of β-major and two of β-minor were sequenced in our core facility by using standard methods.
Electron Microscopy and Immunostaining.
Marrow fragments and blood cells from four mice of each genetic background were fixed in 2.5% glutaraldehyde in 0.1 M phosphate buffer (pH 7.3) at room temperature for 2 hours, washed in PBS (PBS), postfixed in 1% osmium tetroxide in PBS, and stained with aqueous uranyl acetate (2% wt/vol). The samples were taken through propylene oxide and embedded in Araldite (Agar Sci., Essex, United Kingdom). Sections were cut to silver interference colors and floated onto nickel grids. After counterstaining with aqueous uranyl acetate and lead citrate, the sections were viewed in a Philips EM400 electron microscope (Eindhoven, The Netherlands). In each animal, at least 80 consecutive bone marrow reticulocytes and 100 consecutive peripheral blood erythrocytes were examined for the presence and appearance of inclusions. Rounded inclusions with a minimum transverse diameter of 0.7 nm were classified as being large inclusions.
Ultrastructural immunocytochemical studies were performed on sections of bone marrow and blood cells. Rabbit polyclonal antibody against human hemoglobin was purchased from Sigma. Rabbit polyclonal antibody to human α-globin chain was kindly provided by L. Bernini (Leiden University, Medical Genetics Center, The Netherlands). Untreated sections were incubated overnight on a drop of polyclonal antibody to either human hemoglobin or human α-globin chain, undiluted or diluted 1:10, 1:100, or 1:200. After washing in buffer, sections were incubated with a secondary antibody diluted in PBS (1:5 or 1:10) for 4 h; the secondary antibody consisted of anti-rabbit IgG conjugated to 10 nm colloidal gold (British Biocell, Cardiff, United Kingdom). Sections were counterstained with aqueous uranyl acetate and viewed on a Philips EM400 electron microscope. All incubations were carried out at room temperature. Control incubations, in which the primary antibody was omitted or replaced with normal serum, were carried out in parallel with incubations with the primary antibody. All solutions used in the immunocytochemical incubations contained 1% BSA (Sigma) and 0.l% gelatin (British Biocell).
RESULTS AND DISCUSSION
In an effort to identify what we suspected were the unlinked modifying genes responsible for the severity of α-thalassemia (12), we explored several inbred mouse strains for their ability to transmit the milder phenotype in a dominant fashion. The inbred C57BL/6 strain proved suitable. Crosses between normal C57BL/6 and 129sv/ev α1/α1-deficient mice generated F1 offspring heterozygous for the α1-null mutation. These mice were backcrossed to 129sv/ev mice carrying the α1 mutation, and mice homozygous for the α1−/− mutation were analyzed for phenotype severity (Table 1).
Table 1.
A comparison of hematologic parameters in phenotypically severe and mild α-thalassemia in the offspring of F1 129sv/ev/C57BL mice backcrossed to 129sv/ev
| Group | Genotype | n | Hb, g/dl | MCV, μm3 | MCH, pg | RDW, % | Retics, % |
|---|---|---|---|---|---|---|---|
| I (Severe) | |||||||
| α1−/− homozygotes | α1−α2/α1−α2 | 14 | 8.0 ± 0.6 | 31 ± 4.3 | 13.7 ± 1 | 41.3 ± 1.6 | 32 ± 5.5 |
| II (Mild) | |||||||
| α1−/− homozygotes | α1−α2/α1−α2 | 19 | 10.1 ± 0.8 | 33 ± 2.4 | 14.6 ± 1 | 34 ± 1.9 | 9.0 ± 4.0 |
| Wild type | |||||||
| 129 | α1α2/α1α2 | 4 | 15.3 ± 0.1 | 45 ± 0.3 | 15.2 ± 1 | 15 ± 2 | 1.9 ± 0.9 |
| Wild type | |||||||
| B6 | α1α2/α1α2 | 4 | 15.6 ± 0.5 | 46 ± 0.4 | 15.6 ± 0.3 | 12.6 ± 0.4 | 1.5 ± 0.1 |
MCV, mean corpuscular volume; MCH, mean corpuscular hemoglobin; RDW, red cell distribution width; Retics, reticulocytes in peripheral blood; 129, 129sv/ev inbred mice; B-6, C57B1-6 inbred mice; backcrossed mice: product of α1−/+/(129/B6) × α1− 129/129. Values given are mean ± SD.
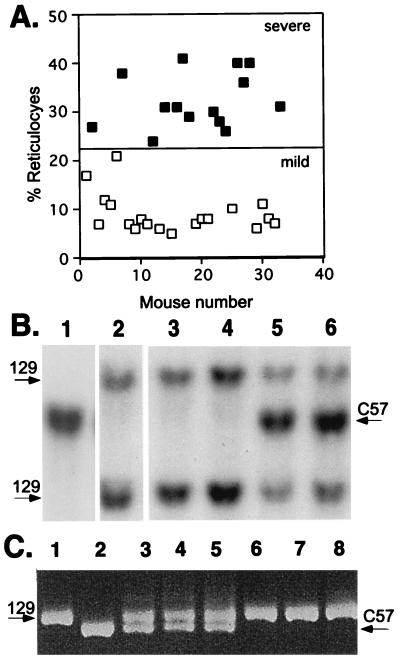
Blood analyses of over 30 backcrossed α1-deficient mice (α1−α2/α1−α2) (Table 1) revealed that all suffered from microcytic anemia with red cell parameters characteristic of human thalassemia. As can be seen in the scattergram shown in Fig. 1A, the level of reticulocytes in the peripheral blood varied from 6% to 37% among the backcrossed α1−α2/α1−α2 mice. This reticulocytosis provided a quantitative measure of phenotypic severity. Indeed, two relatively evenly divided and distinguishable groups could be ascertained among the backcrossed mice by arbitrarily selecting a 22% reticulocyte value to distinguish between the severe and mild phenotypes. A t value of 13.7 and a P value < 0.001 confirmed a statistically significant difference between the two groups. This quantitative variation among backcrossed α-thalassemic mice could—in theory at least—be accounted for by assuming that offspring displaying the milder phenotype inherit a dominant C57BL/6 modifying gene(s) from the F1 parent, whereas those displaying the severe phenotype inherit a recessive 129sv/ev modifying gene(s). That these offspring fall roughly into two relatively equal groups as regards the severity of their disease (Fig. 1A) further suggests that there might be few, possibly even one, modifying gene(s).
Figure 1.
Analysis of the severity of α-thalassemia in mice backcrossed as F1 129sv/ev/C57BL hybrids into 129sv/ev genetic backgrounds. (A) F1 129sv/ev/C57BL α1− heterozygotic mice were backcrossed to 129sv/ev α1− heterozygotes to produce α1−/− mice in the backcrossed background. The severity of the α-thalassemia was judged by the level of reticulocytosis with 22% reticulocytes chosen to arbitrarily separate severe from mild disease (see Table 1). These mice were individually plotted in a scattergram against their blood reticulocyte levels. Their β-globin genotype was determined by using the polymorphic markers shown in B and C. ■, mice homozygous for 129sv/ev β-globin; □, mice with one 129sv/ev allele and one C57BL/6 allele. (B) Southern blot of tail DNAs from controls and backcrossed mice restricted with EcoRI and hybridized with β-globin cDNA. Lane 1, C57BL/6 control; lane 2, 129sv/ev control; lanes 3 and 4, backcrossed mice with two 129sv/ev β-globin alleles; lanes 5 and 6, backcrossed mice with one 129sv/ev globin allele and one C57BL6 β-globin allele (129/B6). (C) PCR analysis of tail DNAs from control and backcrossed mice using D7MIT-40 oligonucleotides as primers. D7MIT-40 is a marker adjacent to the β-globin locus (see Materials and Methods). Lane 1, 129sv/ev control; lane 2, C57BL/6 control; lanes 3–5, backcrossed mice with both 129sv/ev and C57BL/6 β-globin alleles; lanes 6–8, backcrossed mice with two 129sv/ev β-globin alleles. A 208-bp fragment is the amplified band of C57BL/6 genomic DNA, and approximately 230 bp is the amplified band of 129sv/ev genomic DNA.
Whereas any of a number of candidate genes could be responsible for the modifying effects we observe, our attention was initially drawn to the β-globin locus. Unbalanced β-globin chains contribute to the pathology of α-thalassemia because the product of the excess β-chain, hemoglobin β4, in addition to being a poor O2 transporter, has detrimental effects on the integrity and survival of the red blood cell (13–15).
β-globin linkage to the modified phenotype in backcrossed 129/C57BL mice was tested by making use of two unambiguously polymorphic genetic markers for the two β-globin alleles (Fig. 1 B and C). A clear correlation between the severity of the α-thalassemia and the inherited β-globin genotype emerged. Mice belonging to the severely affected group (Fig. 1A, ▪), carried two 129sv/ev β-globin alleles, whereas mice in the mildly affected group carried one C57BL/6 and one 129sv/ev allele (Fig. 1A, □). This correlation between the genotype and the quantitative data was tested further by using the map manager qt program version 26 (16), which calculates the statistical likelihood of any association between a genotype and a quantitative trait (in this case, the reticulocyte count). By using these 30 meioses (Fig. 2A), this yields a Lod score of 13.3 and calculates that the genotype at this locus accounts for 87% of the variation in the reticulocyte count, arguing that the β-locus is the major correlating locus and that further genomewide analysis is unlikely to reveal any other significant association.
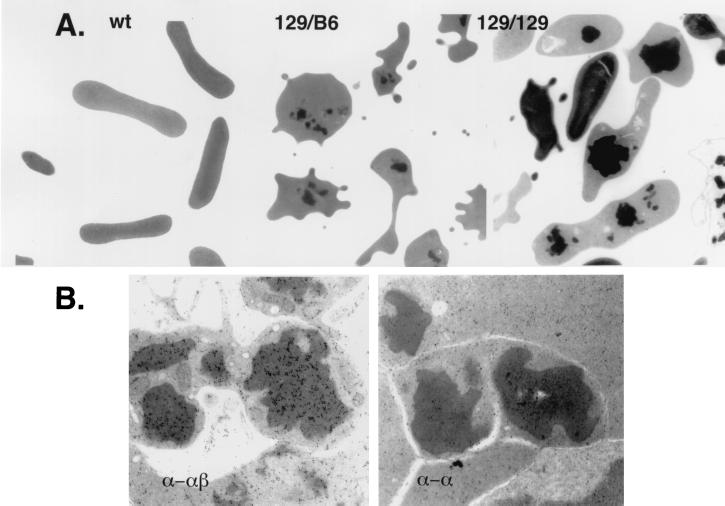
Figure 2.
Electron microscopic and immunoelectron microscopic analyses of red blood cell inclusion bodies. (A) Left to Right, wild-type (wt) and thalassemic red blood cells derived from mice bearing the indicated β-globin allele, 129/B6 (129sv/ev/C57BL/6) and 129/129 (129sv/ev/129sv/ev). The inclusion bodies are the readily visible electron-dense structures. Magnification ×7,000 (Left), ×6,000 (Center), and ×6000 (Right). (B) Shown are inclusions from sections of marrow from an α-thalassemic mouse bearing the 129sv/ev/129sv/ev β-globin alleles. The sections were immunogold-labeled with polyclonal antibody directed against hemoglobin (both α- and β-chains) (α-αβ) and antibody directed only against purified α-chain (α-α). Magnification is ×26,000. Inclusions show a positive reaction with the antibody against hemoglobin but are not reactive with the antibody against α-globin chains.
Given the linkage between β-globin loci and the severity of α-thalassemia, one might predict that other mouse strains that carry 129sv/ev-like β-globin alleles would also be permissive for the severe phenotype. To test this possibility, we took advantage of the fact that both 129sv/ev and BALB/c mice carry the Hbbd hemoglobin haplotype at their β-loci (17). We cloned and sequenced both the β-globin major and minor genes from the 129sv/ev mouse and found, perhaps not surprisingly, that their amino acid sequences were identical to those of BALB/c (18). [A comparison of these sequences revealed one silent substitution in β-globin major (CTT in BALB/c to CTG in 129sv/ev at amino acid 118) and one silent substitution in β-globin minor (CTA to CTG at amino acid 106 of 129sv/ev).] Consistent with this prediction, we found that backcrossed α-thalassemic mice bearing the 129sv/ev/129sv/ev β-globin genotype and those bearing the 129sv/ev/BALB/c β-globin genotype all developed severe disease with reticulocyte values averaging 35% and 39%, respectively (Table 2).
Table 2.
β-Globin and hematologic parameters in backcrossed α1-deficient mice of the 129sv/ev/BALB/c genetic background
| α1 mutation | β-globin alleles | n | Hb, g/dl | MCV, μm3 | MCH, pg | RDW, % | Retic, % |
|---|---|---|---|---|---|---|---|
| α1−/− homozygotes | 129/129 | 4 | 8.6 ± 0.9 | 34 ± 2.4 | 12.5 ± 1.3 | 41 ± 4.3 | 35 ± 4.1 |
| α1−α2/α1−α2) | |||||||
| α1−/− homozygotes | 129/BALB | 4 | 8.3 ± 0.5 | 37 ± 4.5 | 13 ± 0.8 | 40 ± 1 | 39 ± 2.5 |
| α1−α2/α1−α2 | |||||||
| Wild-type 129 | 129/129 | 4 | 15.6 ± 0.5 | 46 ± 0.4 | 15.6 ± 0.3 | 12.6 ± 0.4 | 1.5 ± 0.2 |
| α1α2/α1α2 | |||||||
| Wild-type BALB | BALB/BALB | 4 | 16.4 ± 0.3 | 49.5 ± 1.3 | 16.8 ± 0.3 | 15.9 ± 0.6 | 3.1 ± 0.8 |
| α1α2/α1α2 |
Abbreviations are as in Table 1. BALB, BALB/c inbred mice. Backcrossed mice: product of α1+/− (129/BALB) × α1+/− (129/129). The β-globin alleles were distinguished using PCR and D7Mit-40 oligonucleotides as primers. Values are mean ± SD.
As noted above, a major cause of erythroid cell destruction in α-thalassemia is the excess of unpaired β-globin chains that aggregate within inclusion bodies and exert a harmful effect on the cell membrane (13–15). We wondered whether the two groups of mice showing different degrees of erythrocyte destruction—indicated by the degree of reticulocytosis—were dissimilar as to the frequency and size of such inclusions. Hence, erythroid cells in the bone marrow and blood was examined by using the electron microscope. In both groups of mice, a considerable proportion of bone marrow reticulocytes and blood erythrocytes showed electron-dense inclusions (Fig. 2A). These were elongated, branching, stellate or rounded in appearance, had distinct edges, and varied considerably in size. Only few such inclusions were seen within erythroblasts (data not shown). The percentages of bone marrow reticulocytes and blood erythrocytes that contained inclusions were only slightly higher in the 129sv/ev/129sv/ev than in the 129sv/ev/C57BL/6 mice. However, between the two groups of animals, there was a striking difference in the size of the inclusions (Fig. 2A). As shown in Table 3, the percentages of bone marrow reticulocytes and blood erythrocytes that showed large inclusions in the 129sv/ev/129sv/ev mice were markedly and significantly higher than those in the 129sv/ev/C57BL/6 animals.
Table 3.
Large inclusion bodies in erythroid cells of group I and group II α-thalassemic mice
| β-globin alleles | n | Cells with large inclusions, %
|
|
|---|---|---|---|
| Reticulocytes, bone marrow | Erythrocytes, blood | ||
| 129/129 | 4 | 53 ± 6 | 28 ± 8 |
| 129/BL-6 | 4 | 19 ± 8 | 8 ± 3 |
| p < 0.001 | p < 0.01 | ||
That these inclusion bodies contained globin chains was demonstrated by using an immunogold anti-hemoglobin antibody to stain bone marrow reticulocytes. As shown in Fig. 2B, greater density of gold particles is seen over the inclusions than over the surrounding cytoplasm. By contrast, after the application of an anti-α-globin chain antibody, inclusions and cytoplasm exhibited similar labeling intensity (Fig. 2B), suggesting that the inclusions contain β-, but not α-globin chains. Normal rabbit serum used as a negative control failed to stain either inclusions or cytoplasm.
The correlation between the severity of the disease, the inheritance of a specific β-globin allele, and the large size of the inclusion bodies in the severely affected red blood cells prompted us to compare the amino acid sequences of the 129sv/ev and C57BL alleles for clues as to the basis of this phenotypic difference. A comparison of the β-globin amino acid sequences of the 129sv/ev and the C57BL strains (19–20) is shown in Table 4. Of the three amino acid substitutions between C57BL and the 129sv/ev β-globin major sequences (β-globin major is the predominantly expressed gene in these mice), the substitution of Gly-13 in C57BL for Cys-13 in 129sv/ev is particularly interesting. The same glycine-to-cysteine substitution, along with other amino acid substitutions, is found for β-globin minor. This Cys-13, likely to be located on the solvent surface of hemoglobin β4 (21), could provide for extensive and efficient aggregation of hemoglobin β4 via disulfide bridges. The elimination of this Cys-13 and its replacement by glycine would be expected to reduce this aggregation and thus account for the milder phenotype. In any case, our studies provide a framework for considering mechanisms of genetic modification in thalassemia in which the nature of the chain in excess, even when it is a normal polymorphic form of a globin gene, determines the severity of the thalassemic disorder.
Table 4.
Amino acid changes between 129sv/ev β-globin and C57BL-10 β-globin
| β-globin amino acid change | C57 BL-10 residue | 129sv/ev residue |
|---|---|---|
| Major | ||
| 13 | Gly | Cys |
| 20 | Ala | Ser |
| 139 | Ala | Thr |
| Minor | ||
| 9 | Ala | Ser |
| 13 | Gly | Cys |
| 16 | Gly | Ala |
| 20 | Ala | Pro |
| 58 | Ala | Pro |
| 73 | Asp | Glu |
| 76 | Asn | Lys |
| 77 | His | Asn |
| 80 | Ser | Asn |
| 109 | Met | Ala |
| 139 | Ala | Thr |
Although having a normal gene in the role of a modifier is not entirely unexpected, it certainly could make its identification much more difficult. We were aided in this particular effort by the vast knowledge of the hemoglobin molecule and the numerous mutations that have been described for its α- and β-chains and the well studied pathophysiology of the disease. In less well studied cases in which the underlying mutations and their ramifications for the organism are not well understood, tracking a normal gene as a modifier could be extremely difficult.
Acknowledgments
We are very grateful to Drs. Frank Bunn, David Beier, Jon Seidman, and William Dietrich for their helpful advice and discussion. We are also most grateful to Anne Harrington for her expertise in the management of the mouse colony and mating protocols used in these studies and to Terri Broderick for her editorial assistance and advice.
References
- 1.Nathan D, Orkin S H. In: Hematology of Infancy and Childhood. 5th Ed. Nathan D, Oski F A, editors. Philadelphia: Saunders; 1998. pp. 811–886. [Google Scholar]
- 2.Nathan D G, Gunn R B. Am J Med. 1966;41:815–830. doi: 10.1016/0002-9343(66)90039-8. [DOI] [PubMed] [Google Scholar]
- 3.Kan Y W, Nathan D G. J Clin Invest. 1970;49:635–642. doi: 10.1172/JCI106274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Knox-Macaulay H H, Weatherall D J, Clegg J B, Bradley J, Brown M J. Br J Haematol. 1972;22:497–512. doi: 10.1111/j.1365-2141.1972.tb05695.x. [DOI] [PubMed] [Google Scholar]
- 5.Bate C M, Humphries G. Lancet. 1977;1:1031–1034. doi: 10.1016/s0140-6736(77)91261-2. [DOI] [PubMed] [Google Scholar]
- 6.Furbetta M, Galanello R, Ximenes A, Angius A, Melis M A, Serra P, Cao A. Br J Haematol. 1979;41:203–210. doi: 10.1111/j.1365-2141.1979.tb05849.x. [DOI] [PubMed] [Google Scholar]
- 7.Weatherall D. Mol Med Today. 1995;1:1357–4310. doi: 10.1016/1357-4310(95)80014-x. [DOI] [PubMed] [Google Scholar]
- 8.Higgs D R. Clin Haematol. 1993;6:117–150. doi: 10.1016/s0950-3536(05)80068-x. [DOI] [PubMed] [Google Scholar]
- 9.Ho P J, Hall G W, Luo L Y, Weatherall D J, Thein S L. Br J Haematol. 1998;100:70–78. doi: 10.1046/j.1365-2141.1998.00519.x. [DOI] [PubMed] [Google Scholar]
- 10.Thein S L, Wood W G, Wickkramasinghe S N, Galvin M C. Blood. 1993;82:961–967. [PubMed] [Google Scholar]
- 11.Wilkie A O, Zeitlin H C, Lindenbaum R H, Buckle V J, Fischel-Ghodsian N, Chui D H, Gardner-Medwin D, MacGillivray M H, Weatherall D J, Higgs D R. Am J Hum Genet. 1990;46:1127–1140. [PMC free article] [PubMed] [Google Scholar]
- 12.Leder A, Daugherty C, Whitney B, Leder P. Blood. 1997;90:1275–1282. [PubMed] [Google Scholar]
- 13.Nathan D G, Stossel T B, Gunn R B, Zarkowsky H S, Laforet M T. J Clin Invest. 1969;48:33–41. doi: 10.1172/JCI105972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rigas D A, Koler R D. Blood. 1961;18:1–17. [PubMed] [Google Scholar]
- 15.Rachmilewitz E A. Ann N Y Acad Sci. 1969;165:171–184. doi: 10.1111/j.1749-6632.1969.tb27787.x. [DOI] [PubMed] [Google Scholar]
- 16.Manly K F, Olson J M. Mamm Genome. 1999;10:327–334. doi: 10.1007/s003359900997. [DOI] [PubMed] [Google Scholar]
- 17.Russel E S, Bernstein S E. In: Biology of the Laboratory Mouse. 2nd Ed. Green E R, editor. New York: Dover; 1975. pp. 351–372. [Google Scholar]
- 18.Konkel D A, Maizel J V, Jr, Leder P. Cell. 1979;18:865–873. doi: 10.1016/0092-8674(79)90138-7. [DOI] [PubMed] [Google Scholar]
- 19.Erhart M A, Simons K-S, Weaver S. Mol Biol Evol. 1985;2:304–320. doi: 10.1093/oxfordjournals.molbev.a040353. [DOI] [PubMed] [Google Scholar]
- 20.Holdener-Kenny B, Weaver S. Proc Natl Acad Sci USA. 1986;83:4374–4378. doi: 10.1073/pnas.83.12.4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dickerson R E, Geis I. Hemoglobin: Structure, Function, Evolution and Pathology. Menlo Park, CA: Benjamin/Cummings; 1983. [Google Scholar]