Abstract
SYNOPSIS (for table of contents)
The identification of recurrent alterations in the melanoma genome has provided key insights into the biology of melanoma genesis and progression. These discoveries have come about as a result of the systematic deployment and integration of diverse genomic technologies, including DNA sequencing, chromosomal copy number analysis, and gene expression profiling. Here, we chronicle the discoveries of several key melanoma oncogenes affecting critical cell pathways and examine the role played by evolving genomics technologies in melanoma oncogene discovery. These advances are being exploited to improve prognosis and treatment of melanoma patients through the development of genome-based diagnostic and targeted therapeutic avenues.
Keywords: melanoma, oncogene, DNA sequencing, microarrays, gene amplification, targeted therapy
The notion of cancer as a genetic disease is, by now, well-established [1]. The accumulation of genomic mutations and epigenetic changes that dysregulate genes controlling key hallmarks of malignancy [2] (proliferation, survival, invasion, etc.) is considered to be the main cause of cancer. The last decade has witnessed the discovery of mutations in many genes driving the progression of a host of different cancers. These include activating mutations in growth-promoting oncogenes, inactivating mutations in growth-inhibiting tumor-suppressor genes, and alterations to stability genes. Collectively, somatic base pair mutations have been identified in almost 10% of genes in human cancers, although only a subset of these are believed to be driver events [3]. Ongoing efforts to elucidate these genes and their roles in promoting the growth of different tumor types constitute a major component of molecular cancer research.
Melanoma, a malignant tumor of melanocytes, is responsible for 80% of all deaths from skin cancer. An estimated 62,000 new melanoma cases were diagnosed in the United States in 2008 [4]. The poor prognosis of advanced melanoma and the relative ineffectiveness of conventional therapies for patients with metastatic disease underscore a dire need for improved therapeutic agents. Recent discoveries of recurrent mutations in the melanoma genome have provided an improved understanding of the biology of melanoma genesis and progression as well as an opportunity for the development of new classes of targeted drugs. In this review, we chronicle the discoveries of several critical melanoma oncogenes and highlight the role played by genomic technologies. Our goal is not to provide a comprehensive list of all genetic alterations observed in melanoma. Rather, we hope to illustrate through the lens of melanoma oncogene discovery how genomic analysis can lead to a better understanding of tumor biology and novel therapeutic avenues. We conclude with a discussion of emerging “omic” technologies and how these might lead to the discovery of additional oncogenes that articulate a path to improved prognosis and treatment of patients suffering from this deadly malignancy.
MELANOMA ONCOGENES
Genetic mutations involving numerous genes have been linked to melanoma genesis and progression. These genes encompass many signaling pathways, including the receptor tyrosine kinase (RTK), phosphtidylinositol-3-kinase (PI(3)K), retinoblastoma (RB), p53, Wnt, and NF-kB pathways. The supporting evidence implicating these oncogenes in melanoma ranges from positional cloning studies in familial melanoma to elevated frequencies of mutation or amplification in patient cohorts to functional studies in vtiro and in vivo. Below, we consider five well-studied melanoma oncogenes and their contributions to overall morbidity (Table 1). Each of these genes was identified by a different approach, highlighting alternate strategies for melanoma oncogene discovery and emphasizing the expanding importance of technology and integrative genomic analysis.
Table 1.
Summary of exemplary melanoma oncogenes
| Gene | Function | Alteration | Prevalence | Discovery Method |
|---|---|---|---|---|
| NRAS | Growth, Proliferation | Mutation | 15-30% | Transformation assay, genotyping |
| BRAF | Proliferation, Survival | Mutation | 50-70% | Systematic DNA sequencing |
| MITF | Lineage survival | Amplification | 10-20% | Copy number and gene expression profiling |
| NEDD9 | Invasion, Metastasis | Amplification | 36% | Copy number profiling and evolutionary conservation |
| KIT | Proliferation, Survival | Mutation, Amplification |
2-5% | DNA sequencing |
NRAS
One of the first genes shown to be specifically mutated in melanoma was NRAS, a gene encoding a member of the RAS family of small GTP-binding proteins. The RAS proteins lie at the “top” of the RAS/RAF/MEK/ERK MAP kinase pathway, which activates a large number of growth-promoting genes in response to signals transmitted from growth factors and cytokines. Regulation of RAS signaling is achieved by alternating between active (GTP-bound) and inactive (GDP-bound) forms of the RAS protein. The prototypic members of the RAS family, HRAS and KRAS, were originally discovered as the transforming oncoproteins in Harvey and Kirsten murine sarcoma viruses [5]. Human cellular versions of the HRAS and KRAS genes were subsequently identified, and single nucleotide mutations were shown to confer transforming capabilities on these genes upon transfection into the mouse embryonic fibroblast NIH 3T3 cell line [6-8]. NRAS was later discovered as the transforming gene of a neuroblastoma cell line, and sequence analysis revealed considerable homology with HRAS and KRAS [9, 10]. Mutations converting all three genes to active oncogenes nearly always occur in residues 12, 13, or 61 of the protein [11]. These mutations impair the GTPase catalytic activity of RAS, resulting in a constitutively GTP-bound and activated state. Altogether, approximately 20-30% of human tumors carry a mutation in one of the RAS genes.
NRAS was first shown to be mutated in melanoma in the mid-1980s when two studies demonstrated that NRAS DNA isolated from four human melanoma cell lies was sufficient to transform NIH 3T3 cells [12, 13]. Subsequent sequencing and genotyping efforts revealed that NRAS is mutated in 15-30% of melanomas, >10 times more frequently than HRAS or KRAS [14-18]. Notably, the mutations occur most often at residue 61. The spatial distribution of NRAS-mutated tumors on the skin and their proclivity for dipyrimidine mutations suggest a possible correlation with UV exposure [14, 17]. Transgenic mice with oncogenic NRAS targeted to the melanocytic compartment show hyperpigmented skin and develop cutaneous metastasizing melanoma [19]. In addition to its role in tumor formation, mutant RAS is also critical for tumor maintenance [20]. Knockdown of mutant NRAS (but not wild-type NRAS) by RNA interference reduces the viability of melanoma cell lines, suggesting a possible therapeutic avenue in patients whose tumors harbor this mutation [21]. However, the development of small molecules that target RAS proteins directly has proved exceedingly difficult.
As stated above, RAS operates in the MAP kinase pathway. This pathway is hyperactivated in up to 90% of human melanomas [22]. This underscores the contribution of NRAS mutations to the development of melanoma but also indicates the involvement of other genes in the pathway, discussed below. At the same time, RAS-activating mutations can influence the phosphoinositide-3-OH kinase (PI(3)K) pathway. The specificity of mutations in melanoma for NRAS compared to HRAS and KRAS is notable considering that all three isoforms are expressed in primary melanomas and melanoma-derived short-term cultures [23, 24], and that HRAS and KRAS are mutated in a variety of other human cancers. For example, KRAS mutations are found in 15-20% of lung cancers, ∼30% of colorectal cancers, and up to 90% of pancreatic adenocarcinomas [11]. HRAS mutations have been detected in Spitz nevi, yet they rarely occur in malignant melanoma [25]. Comparisons of the transformation efficiencies of different RAS isoforms suggest that the activity of different isoforms may depend on the cellular context [26].
In summary, the recurrence and high transforming potential of oncogenic NRAS mutations in human melanomas demonstrate the critical role of this gene and its downstream effector mechanisms in melanoma genesis and maintenance. As with many other “classic” oncogenes described in cancer, the original discovery and validation of NRAS as a melanoma oncogene required a laborious series of low-throughput experiments spanning many years and multiple research groups. The following examples illustrate how recent melanoma oncogene discoveries have emerged from powerful systematic approaches enabled by the massive technology revolution that accompanied the genomic era.
BRAF
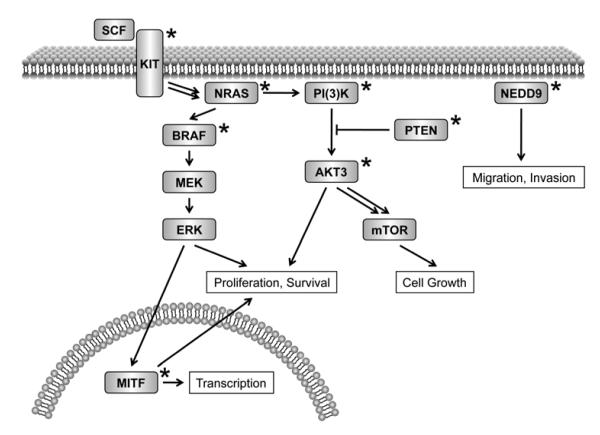
BRAF, a serine-threonine kinase, lies downstream of RAS in the MAP kinase signaling pathway Fig. 1) (BRAF induces MEK to phosphorylate ERK, which enhances cell growth and proliferation. In 2002, as part of a large-scale DNA sequencing effort to identify mutant oncogenes in the MAP kinase pathway, Davies et al. discovered highly recurrent oncogenic BRAF mutations in melanoma through systematic re-sequencing across 545 cancer cell lines [27]. The BRAF gene was observed to be mutated in 20 of 34 (59%) melanoma cell lines and an additional 18 of 24 (75%) patient-derived primary melanomas and short-term cultures, compared to an overall mutation frequency of 8% in all cancers. (After melanoma, BRAF was mutated in 15% of colon cancers, 11% of gliomas, and 10% of ovarian cancers, considering cell lines and primary tumors.) Notably, >90% of the BRAF mutations in melanoma involved a single substitution of valine to glutamic acid in the kinase domain (V600E) [27]. Subsequent studies have confirmed the specificity for V600E mutations and established the overall frequency of BRAF mutations at 50-70% in cutaneous melanomas [28-32]. Unlike the mutation patterns observed in NRAS, the T>A transversion associated with the V600E mutation is not suggestive of UV-induced DNA damage, although there is an apparent association between this mutation and intermittent exposure to sun [31]. Surprisingly, the V600E mutation was also observed in 39 of 44 (89%) benign melanocytic nevi, suggesting that mutation of BRAF represents an early event in melanocytic neoplasia but one that is insufficient by itself for tumorigenesis [29]. However, mutant BRAF does appear to induce transformation of NIH 3T3 cells and murine melanocytes and allow them to grow as tumors in nude mice, clearly implicating it as an oncogene [27, 33-35].
Figure 1.
Cell signaling pathways that undergo oncogenic dysregulation in melanoma. The receptor tyrosine kinase-driven MAP kinase and PI(3)K pathways are featured.. Genes mutated in melanoma are marked with an asterisk. These include KIT, NRAS, BRAF, MITF, PI(3)K, PTEN, AKT3, and NEDD9.
As noted above, the oncogenic potential of BRAF lies Nin its ability to phosphorylate MEK, leading to ERK activation and cell proliferation (Fig. 1). The kinase activity of BRAF is normally regulated by the phosphorylation of two amino acids, T599 and S602, which disrupts the Minteraction between the activation segment and P loop and causes a conformational change to the active state [36]. The V600E mutant mimics these phosphorylation events by inserting a negatively charged residue in the activation segment, resulting in constitutively active BRAF [27, 34]. The absence of comparable mutations from ARAF and CRAF (paralogs of BRAF that also function in MAP kinase signaling) is likely due to the further requirement of phosphorylation of an additional regulatory region (N region) in the kinase domain of these proteins. Unlike ARAF and CRAF, the N region of BRAF is constitutively phosphorylated such that BRAF is primed for activation [33]. Recent studies have identified several genes functioning downstream of BRAF in melanoma, including critical cell cycle regulators, tumor maintenance enzymes, and microphthalmia-associated transcription factor (MITF, discussed below) [37]. These findings underscore the key role of BRAF in melanoma genesis and progression.
Depletion of oncogenic BRAF (but not ARAF or CRAF) by RNA interference in cultured human melanoma cells and mouse xenograft models has been shown to inhibit proliferation and induce apoptosis [38, 39]. This suggests an essential role for the MAP kinase pathway in tumor maintenance. Consequently, inhibitors of BRAF and MEK are of considerable interest as potential targeted therapies for melanomas harboring BRAF mutations. To this end, several BRAF inhibitors (such as BAY 43-9006 and CHIR-265) and MEK inhibitors (such as PD0325901 and AZD6244) are currently in various stages of development and/or clinical trials [37]. One well-studied MEK inhibitor, CI-1040, clearly inhibits proliferation and soft-agar colony formation in vitro and causes rapid regression of BRAF mutant xenografts in mouse [40]. Given the additional role of RAS in MAP kinase signaling, it would stand to reason that MEK inhibition would produce a similar effect in NRAS mutant cells. However, human melanoma cell lines with BRAF mutations proved to be demonstrably more sensitive to CI-1040 and the related (but more potent) MEK inhibitor PD0325901 than did many NRAS mutant melanoma cell lines [41]. This suggests that activation of additional pathways by RAS, such as the PI(3)K/PTEN pathway, decreases the dependence of melanoma cells on MEK. Consistent with this, NRAS and BRAF mutations, as well as NRAS and PTEN mutations, are mutually exclusive in melanoma due to their shared pathway membership, whereas BRAF and PTEN mutations are coincident in up to 20% of melanomas [42]. BRAF and PTEN operate in different pathways downstream of RAS, such that separate mutations are required to activate both pathways. It thus remains uncertain as to why BRAF would be mutated so much more frequently than NRAS in melanoma.
MITF
In addition to nucleotide substitutions, DNA copy number alterations are frequently observed in cancer. More than just passive reflections of the genomic instability of cancer, such amplifications and deletions often function as driver events in tumorigenesis by altering the expression levels of important genes regulating cell growth, survival, etc. Changes in DNA copy number are readily profiled at high resolution by hybridizing tumor genomic DNA to microarrays comprised of oligonucleotides spanning the entire human genome [43]. As an illustration of the use of recurrent chromosomal aberrations to identify a novel melanoma oncogene, Garraway et al. profiled genomic copy gains and losses in the NCI-60 cancer cell line collection using high-density single nucleotide polymorphism (SNP) microarrays and observed a focal amplification at 3p14-3p13 shared among 6 out of 8 melanoma cell lines [44]. By integrating these copy number data with gene expression signatures for this cell line collection, the authors identified only one highly expressed gene located within the amplified region. This product of this gene, microphthalmia-associated transcription factor (MITF), was already known to be a master regulator of the melanocyte lineage and a sensitive marker for melanoma diagnosis [45, 46]. However, this integrative genomics approach provided the first evidence that MITF is an oncogene in human melanoma and the target of a specific somatic alteration in this malignancy.
Detailed characterization this locus by fluorescence in situ hybridization (FISH) established the frequency of MITF amplification in melanomas at 10-20%, with a higher incidence among advanced tumors and an associated decrease of 5 years in survival [44]. Interestingly, MITF amplifications are coincident with mutations in BRAF. Growth of NCI-60 cell lines harboring MITF amplification was inhibited following RNAi-mediated knockdown of MITF or introduction of dominant-negative MITF [44]. In an apparent contradiction, very high levels of MITF also seem to reduce tumorigenicity and promote melanocyte differentiation, perhaps by transcriptionally activating p16INK4A and p21, two cyclin-dependent kinase inhibitors [37]. Tight regulation of MITF levels may therefore be necessary for maximal proliferation.
As a master regulator of melanocyte development and differentiation, MITF represents an emerging class of “lineage-survival” oncogenes [47]. Unlike oncogenic NRAS and BRAF, which acquire new and tumor-specific cellular functions through nucleotide mutations, MITF becomes oncogenic via deregulation, affecting survival mechanisms that are also present in the normal melanocyte lineage. It is well established that wild-type MITF is essential for lineage survival and that deficiency of MITF results in the absence (or loss) of melanocytes during development [48]. The same survival mechanisms that govern melanocyte proliferation and development may subsequently persist or become deregulated during tumor progression. Additional lineage-survival oncogenes have been implicated in other cancers for which normal lineage programs become aberrantly regulated in cancer. For example, the androgen receptor (AR) and thyroid transcription factor 1 (TITF1) are targeted by genetic alterations in carcinomas of the prostate and lung, respectively [49, 50]. Like MITF, these genes encode transcription factors that are essential master regulators during development for their particular lineage, but whose functions provide key tumor survival roles in prostate and lung adenocarcinoma. These observations emphasize the importance of lineage in providing a context for additional oncogenic alterations and help to explain why certain mutations occur at different frequencies discrete tumor types.
NEDD9
The identification of MITF within a locus of recurrent amplification in cancer was enabled by the integration of copy number and gene expression data. Subsequently, Kim et al. used a related integrative genomics approach that incorporated evolutionary conservation to implicate NEDD9 as a melanoma metastasis gene from within a broad gain of chromosome 6p [51]. Amplification of 6p had been previously associated with 36% of human metastatic, but not primary, melanomas [52]. However, the shared amplified region spans >35 megabases, making recognition of the underlying driver gene(s) nearly impossible by analysis of copy number data alone. Using high-resolution microarray comparative genome hybridization, Kim et al. identified a recurrent focal amplification associated with metastatic potential in a genetically-engineered inducible mouse model of melanoma. By performing a cross-species comparison between the mouse and human genomes, the authors dramatically narrowed the minimal region of overlap to an 850-kilobase region on human chromosome 6p24-25. Further integration with expression data revealed NEDD9 (neural precursor cell expressed, developmentally downregulated 9) as the only resident gene that was overexpressed when compared to nontransformed primary melanocyte cultures. Accordingly, NEDD9 is amplified in 36% of human metastatic melanomas and overexpressed at both mRNA and protein levels in approximately 50%. NEDD9 protein expression was shown to be up-regulated in a manner correlated with tumor progression [51].
Functional studies provided further support of NEDD9 as a melanoma gene important for invasion and metastasis [51]. Knockdown of NEDD9 by RNAi reduced the number of distal metastases in vivo and limited cellular proliferation and invasion in vitro. Additionally, gain-of-function studies demonstrated that overexpression of NEDD9 enhances metastatic potential of primary melanocytes and nonmetastatic melanoma cells. This increased invasiveness is conferred by a functional and physical interaction between NEDD9 and focal adhesion kinase (FAK) at the cell periphery. These results demonstrate the utility of genome-wide cross-species comparisons for oncogene discovery; in particular, the integration of genomic data from both a mouse model and human tumors led to the identification of NEDD9. Genetically-engineered mouse models also enabled the design of appropriate functional assays and suggested the genetic context in which these assays would be most informative.
KIT
As discussed above, NRAS and BRAF mutations represent common mechanisms of upregulating the MAP kinase pathway in melanoma. In particular, BRAF mutations are prevalent in cutaneous melanomas without chronic sun-induced damage, while they almost never occur in melanomas of the palms and soles (acral melanomas), mucosal membranes (mucosal melanomas), and occur less frequently on skin with chronic sun-induced damage (CSD melanomas). In an attempt to discover oncogenic mutations in melanomas with wild-type NRAS and BRAF, Curtin et al. identified a shared region of amplification at 4q12 using comparative genomic hybridization (CGH) to microarrays [53]. This amplicon was specific to the acral and mucosal melanoma subtypes where BRAF mutations are rare, and it contained an attractive candidate oncogene, KIT. KIT encodes a receptor tyrosine kinase (RTK) for the stem cell factor (SCF) ligand and functions as an upstream activator of the MAP kinase signaling pathway (Fig. 1). Like MITF, KIT is an essential gene for melanocyte survival and development [54], although its expression is usually lost during cutaneous melanoma progression. Re-sequencing of KIT in melanomas with amplification of 4q12 revealed mutations in 3 of 7 cases. More extensive tumor sequencing confirmed that while they may occur in only 2–5% of all melanomas [55, 56], KIT mutations are much more prevalent in acral (12–23%), mucosal (16–25%), and CSD (8–20%) melanomas [53, 57]. Just as many additional cases exhibit amplifications without observed sequence mutations [53].
Of note, the KIT mutations that have been observed in melanoma nearly always occur in exon 11. This is similar to the spectrum of activating KIT mutations in patients with gastrointestinal stromal tumors (GISTs) [58]. GISTs have been successfully treated by the KIT inhibitor, imatinib mesylate [59]. (Imatinib, or Gleevec, also targets the BCR-ABL tyrosine kinase produced by the Philadelphia chromosome in chronic myelognous leukemia (CML) and leads to response in more than 95% of cases [60].) GIST patients whose tumors express an exon 11 mutant KIT protein exhibit an 84% partial response rate to imatinib [58]. These results have prompted multiple phase II clinical trials of imatinib in patients with metastatic melanoma. Results are inconclusive to date, but at least two melanoma patients with activating KIT mutations in exon 11 have demonstrated a marked response to imatinib treatment [61, 62]. These results speak to the emerging need for systematic cancer mutation profiling prior to enrolling patients in clinical trials for targeted agents, and suggest an immediate benefit of profiling melanoma patients for KIT mutations. Further, they anticipate a scenario that may ultimately typify personalized cancer medicine, in which a drug used to treat one type of cancer can be applied in a new cancer based on a shared genetic mechanism.
Additional Oncogenes
The genes described above account for only a fraction of the altered loci observed in melanoma. Oncogenes and tumor suppressor genes in many critical pathways have been implicated from a variety of experimental systems. The MAP kinase signaling pathway is commonly altered as evidenced by mutations in NRAS, BRAF, and KIT, but NRAS and receptor tyrosine kinases also contribute to the PI(3) kinase (PI(3)K) pathway. Oncogenic signaling through PI(3)K leads to the activation of AKT, a prominent downstream oncogenic effector in many tumor types. Interestingly, the AKT3 isoform is highly expressed in neural crest-derived cells such as melanocytes, and AKT3 has been found to undergo chromosomal copy gains and/or overexpression in up to 60% of melanomas [63]. Recently, activating point mutations in AKT3 have also been described in melanoma [64]. Hyperactivation of the PI(3)K/AKT pathway arises from a variety of additional perturbations as well, including activating PI(3)K mutations in 3% of metastatic melanomas [65], and deletions or loss-of-function mutations in PTEN in 40% of melanoma cell lines [66, 67]. The p16/RB pathway is another common target of melanoma genomic alterations; lesions affecting this pathway enable cells to avoid senescence and acquire extended proliferative potential. Germline mutations in the cyclin-dependent kinase inhibitor p16 (encoded by CDKN2A), as well as in the cyclin-dependent kinase CDK4, preventing binding by p16, are common in familial melanoma [68-71]. CDKN2A also encodes p14/ARF in an alternate reading frame, which serves to inhibit the p53 antagonist MDM2. Likewise, p53 is lost or mutated in 10–20% of melanomas [72, 73]. Finally, the Wnt pathway can provide additional proliferative and survival signals in melanoma through loss of adenomatous polyposis coli (APC) [74] and/or or mutation of beta-catenin (CTNNB1) [75, 76]. The complex interplay amongst each of these pathways warrants further study in order to determine their total contribution to melanoma genesis and progression.
MELANOMA ONCOGENES AND PERSONALIZED MEDICINE
The completion of the human genome paoject marked the beginning of the genomic era in biology. However, for this knowledge to truly transform medicine, accessible and affordable technologies are needed that can probe the genomes of many individuals in order to understand the genetic basis of disease and, ultimately, to stratify cancer patients in the clinical arena for optimal therapy. For cancer in particular, the ability to profile the genetic make-up of a patient’s tumor can illuminate which pathways are activated, thus enabling the prediction of disease outcome and the most appropriate course of treatment. The discovery of critical oncogenes, even if mutated in only a small percentage of tumors, can provide new putative targets for drugs tailored for specific patient subpopulations. DNA sequencing, copy number measurements, and gene expression profiling have already led to the identification of many key genes underlying melanoma progression. As described below, these technologies are likely to turn up many new candidate genes in the coming years and shed light on the dependencies operant among these genes and pathways in cancer subtypes defined by genomic or other molecular criteria (Table 2). Further, the emergence of “next-generation” DNA sequencing technologies is expected to dramatically accelerate the search for genes and the movement towards personalized medicine.
Table 2.
Genomic technologies for oncogene discovery
| Genomic Approach | Type of Tumor Data |
|---|---|
| SNP Microarrays, Comparative Genomic Hybridization (CGH) |
Chromosomal copy number |
| Loss of heterozygosity | |
| Gene Expression Microarrays | Gene expression profiles miRNA profiles |
| Sanger DNA Sequencing | Point mutations |
| Small insertions and deletions | |
| Mutation Profiling | Known and/or “druggable” mutations |
| Next-Generation DNA Sequencing | Point mutations |
| Small insertions and deletions | |
| Chromosomal rearrangements translocations | |
| Chromosomal copy number | |
| DNA methylation (with bisulphite treatment) | |
| Epigenetics (with chromatin immunoprecipitation) | |
| RNAi Screening | Essential genes |
| Suppressor genes |
Discovery of Additional Oncogenes
Despite the success of previous oncogene discovery efforts, a large part of the genetic basis of melanoma remains uncharacterized. As discussed above, the MAP kinase pathway is hyperactivated in 90% of melanomas [22]. This is frequently the result of BRAF mutations, but NRAS and KIT are often mutated as well. However, many melanomas harbor wild-type sequences for all three genes, suggesting that additional mutations may perturb this pathway. (The recent discovery of FGFR1 mutations in two melanoma lines with wild-type BRAF and NRAS may indicate a possible alternate mechanism for MAP kinase activation [23].) Further, perturbations to this pathway alone are not sufficient to drive melanoma progression. While BRAF is mutated in 50–70% of malignant melanomas, it is also mutated in as many as 90% of benign melanocytic nevi [29]. MITF overexpression and BRAF mutations together are sufficient to transform primary human melanocytes (in the context of deficient p53 and p16/RB pathways), but only ∼15% of BRAF mutated melanomas also exhibit MITF amplification [44]. Undoubtedly, additional melanoma oncogenes in many other pathways await discovery.
Large-scale, systematic DNA sequencing and copy number profiling efforts will undoubtedly aid the search for new oncogenes. Several groups have utilized large-scale Sanger sequencing to identify novel mutations in other tumor types. Greenman et al. sequenced the coding exons of 518 protein kinase genes in 210 diverse human cancers and found evidence for high-frequency “driver” mutations in more than 100 genes [77]. A separate, multi-institutional study identified 26 genes significantly mutated in a single tumor type upon sequencing 623 genes in 188 lung adenocarcinoma samples [78]. Vogelstein, Velculescu, Kinzler, and colleagues have impressively sequenced the coding exons of approximately 20,000 genes in breast, colorectal, and pancreatic cancers, as well as glioblastoma multiforme, albeit in relatively small discovery cohorts, which may have constrained the statistical power [79-82]. Genome-wide copy number profiling across many samples can also reveal regions of shared copy gain or loss, leading to the discovery of novel lineage-specific oncogenes and tumor suppressors [50]. To provide an organizational and scientific framework for systematically mapping genetic abnormalities that lead to cancer, the U.S. National Cancer Institute and National Human Genome Research Institute established The Cancer Genome Atlas (TCGA) initiative [83]. By integrating genomic data generated by multiple scientific teams using different platforms, including DNA sequence, gene expression, copy number alterations, and DNA methylation, TCGA will lead to the identification of genes and pathways significantly altered in different cancers [84]. Though the pilot stage of TCGA only includes glioblastoma, lung cancer, and ovarian cancer, it will serve as a model for similar large-scale efforts to comprehensively catalog and characterize the genomic changes involved in melanoma.
Most of the DNA mutations discussed to this point involve somatic changes that accumulate over the lifetime of an individual tumor. However, allelic variants of certain genes can be transmitted through the germline and result in an inherited predisposition to developing melanoma. Much remains to be understood about this heritable component of melanoma. Genetic linkage studies have demonstrated that rare mutations in CRCDKN2A on 9p21 segregate with melanoma susceptibility in large melanoma-prone families [68, 69]. More common physical characteristics, such as fair complexion and red hair, are also often accompanied by an increased risk of melanoma. To identify the common genetic variants underlying susceptibility to melanoma in large populations, genome-wide association studies will be particularly useful. Genome-wide association studies involve simultaneously genotyping hundreds of thousands of genetic polymorphisms in large sets of cases and controls. One recent study identified a susceptibility locus for melanoma on chromosome 20q11 by pooling 2,019 cases and 2,105 controls [85]. Additional genetic loci known to affect hair, eye, and skin MD pigmentation in Europeans show clear associations with cutaneous melanoma and basal cell carcinoma [86]. Further studies utilizing additional populations and larger cohorts will likely narrow these loci and reveal novel associations with melanoma susceptibility genes.
Further Characterization of Genetic Lesions
Even as more oncogenic mutations in melanoma become known, the precise mechanisms by which existing mutations confer tumorigenicity remain poorly understood. In vitro transformation assays monitoring proliferation, pathway activation, and anchorage-independent growth have been informative, yet these assays must be performed in the appropriate genetic background. Further, for certain types of mutations, the underlying oncogene may not always be apparent. Amplifications and deletions typically encompass many genes. To narrow down regions of copy gain or loss, one can incorporate additional genomic information. The examples of MITF and NEDD9 discussed above demonstrate how gene expression and evolutionary conservation can be successfully used to pinpoint the driver oncogene within a broader locus. However, many additional significant regions of copy number alteration remain uncharacterized. For example, a recent study identified 27 significant copy gains and losses across 101 melanoma cell lines and short-term cultures using high-density SNP arrays [23].
An additional strategy for the functional characterization of commonly amplified regions involves systematic gene knockdown by RNA interference. Though many regions harbor attractive candidate oncogenes, all constituent genes can be examined through systematic RNAi screens. For instance, one would expect RNAi knockdown of an oncogene to result in decreased proliferation of all cell lines harboring an amplification of that locus. Similarly, restoring the expression of a tumor suppressor gene that has been deleted should produce a similar anti-proliferative effect.
The high prevalence of particular mutations in melanoma underscores the critical roles played by the associated pathways. However, no single mutation is sufficient to lead to malignant melanoma. The interdependence among various mutations affecting different pathways needs to be further examined. From the distribution of mutations in NRAS, BRAF, and PTEN, it is clear that the MAP kinase pathway and the PI(3)K/PTEN pathway are often simultaneously targeted. The context-dependence of oncogenic mutations can be explored by RNAi screens in multiple cell lines with different genetic backgrounds [87]. Additional genes whose knockdown reduces proliferation in some, but not all, cell lines can be linked to common alterations through supervised analysis. The temporal order in which mutations occur can also be deduced from their distribution. BRAF is mutated in ∼90% of benign melanocytic nevi, whereas MITF amplification is restricted to malignant melanoma [29, 44]. This suggests that BRAF mutations occur early during melanoma progression, perhaps predisposing cells to hyperproliferation following additional genetic alterations.
Next-Generation Technologies for Oncogene Discovery
Looking forward, the discovery of new oncogenes in melanoma and other tumors will largely come about from genome-scale DNA sequencing efforts. Until recently, large-scale DNA sequencing in cancer has been conducted entirely by traditional capillary-based Sanger sequencing methods [77-82, 84]. New technologies have now emerged that promise to dramatically reduce the cost and increase the throughput of DNA sequencing [88]. These massively parallel “next-generation” sequencing platforms involve the clonal amplification of millions of individual template molecules without the need for multi-well plates and large quantities of costly reagents. The short sequence reads (ranging from 20-50 to a few hundred bases) that are generated can be mapped onto the reference human genome, and the nucleotide identity at each position is determined by counting discrete reads. This results in increased sensitivity for detecting rare sequence variants, which is especially important in cancer where confounding factors such as stromal contamination and genetic heterogeneity are common [89]. The first whole-genome sequence of a cancer patient’s tumor was recently completed using massively parallel sequencing [90]. In this study, 33-fold coverage was obtained for an acute myeloid leukemia genome, and 14-fold coverage was obtained for a normal skin sample from the sample patient, leading to the discovery and validation of several novel somatic mutations. For the purposes of oncogene discovery, it is currently more economical to sequence only protein-coding regions rather than sequence across the entire genome. Several groups have developed exon capture methods for targeted resequencing to overcome the cost limitations of parallel PCR reactions [91, 92]. Consequently, it will soon be feasible to sequence all human exons (or a subset thereof) in hundreds of tumors at reasonable cost.
The value of next-generation DNA sequencing technologies in cancer is not limited to genome sequencing applications. Microarray-based assays can be adapted to use DNA sequencing as a read-out, providing more sensitivity and a greater dynamic range. For instance, gene expression levels can be monitored by counting the number of sequence reads arising from different cDNAs. In doing so, novel transcription events and alternatively-spliced isoforms can be readily identified [93, 94]. To demonstrate its utility for cancer, Sugarbaker et al. used next-generation sequencing of cDNA to characterize RNA mutations and gene expression levels in malignant mesotheliomas [95]. In addition to expression, DNA methylation patterns can be evaluated in different samples using reduced representation bisulfite sequencing [96]. Chromatin modifications and transcription factor binding can be analyzed by DNA sequencing following chromatin immunoprecipitation (ChIP-Seq) [97, 98]. Further, copy number alterations can be identified in tumors at high-resolution from low-coverage shotgun sequencing of genomic DNA [99]. It is likely that integrative genomic characterization efforts such as The Cancer Genome Atlas will soon deal predominantly (if not exclusively) in various forms of DNA sequence data.
Targeted Therapies and Personalized Medicine
Ultimately, the key cancer-associated mutations uncovered through these discovery efforts can serve as important clinical biomarkers for the diagnosis and prognosis of melanoma, the prediction of a patient’s response to a particular treatment, and the development of novel rational therapeutics targeting specific subsets of patients. With regard to mutations as prognostic biomarkers, Curtin et al. used genome-wide copy number measurements and BRAF and NRAS mutational status to classify 126 melanomas into four clinically distinct groups with 70% percent accuracy [100]. Additionally, as discussed above, MITF amplification in melanoma correlates with a 5-year decrease in survival [44]. Global gene expression patterns have also revealed discernible subclasses of cutaneous melanoma [101]. The FDA has recently approved several anti-cancer drugs that specifically target the products of mutated genes in other malignancies. These include imatinib (BCR-ABL translocation in CML; KIT mutation in GIST) [59, 60], trastuzumab (ERBB2 amplification in breast cancer) [102], and gefitinib and erlotinib (EGFR mutation in non-small cell lung cancer) [103-105]. Imatinib represents a potential therapy for malignant melanomas harboring KIT mutations, as evidenced by the marked response of at least two patients to imatinib treatment [61, 62]. Given the large number of melanomas with activating mutations in the MAP kinase pathway (NRAS and BRAF), MEK inhibitors such as PD0325901 and AZD6244 are currently being developed and investigated in clinical trials [106].
The utility of these genetic biomarkers for the development and administration of targeted therapies hinges on being able to profile patients for mutations in an efficient, cost-effective manner. Although this goal has not yet been attempted categorically, recent advances suggest that there is room for optimism. Thomas et al. described a mass spectrometry-based strategy whereby they genotyped more than 200 high-frequency cancer-associated mutations in 1,000 human tumors [107]. Using this platform, termed OncoMap, the authors discovered that certain mutations commonly associated with particular cancers were also present at lower frequencies in many other cancers. This reinforced the notion that existing targeted treatments such as imatinib may be applied to additional cancers with shared genetic lesions. In its current embodiment, OncoMap relies on the observation that a relatively small number of recurrent base pair mutations accounts for the majority of somatic events known to drive tumor progression. However, extending this strategy to next-generation sequencing of whole exons will enable the rapid profiling of all mutations—both base pair mutations and copy number alterations—in cancer-associated genes. The ability to interrogate large panels of tumors will have a large impact on the discovery of targeted therapies, as pharmacological data from drug screens can be correlated with the mutational status of the associated tumor-derived cell lines [23, 108]. It will also enable efficient patient stratification for clinical trial design. Finally, such an approach, if applied universally, can transform patient care through improved diagnosis and prognosis in the movement towards personalized medicine.
SUMMARY
The identification of melanoma oncogenes, such as NRAS, BRAF, MITF, NEDD9, and KIT, have led to an improved understanding of the biology of melanoma. These discoveries have come about from a variety of genomic approaches, including systematic DNA sequencing, chromosomal copy number profiling, gene expression profiling evolutionary conservation analysis, and RNA interference. Despite these initial successes, the genetic basis of melanoma progression remains largely uncharacterized. It is clear that additional knowledge will come from the utilization of improved genomic technologies with increased throughput and sensitivity for mutation detection and discovery as well as analyses that integrate these diverse data types. These technologies (and the biological insight gained from their deployment) hold great promise for the development of improved diagnostics and targeted therapies and carry the potential to dramatically transform patient care.
Acknowledgments
This work was supported by Grant No. DP2OD002750 (NIH), the Burroughs-Wellcome Fund, and the Starr Cancer Consortium
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004 Aug;10(8):789–99. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000 Jan 7;100(1):57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 3.Futreal PA, Coin L, Marshall M, et al. A census of human cancer genes. Nat Rev Cancer. 2004 Mar;4(3):177–83. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cancer Facts and Figures: 2008. American Cancer Society; Atlanta: 2008. [Google Scholar]
- 5.Shih TY, Papageorge AG, Stokes PE, Weeks MO, Scolnick EM. Guanine nucleotide-binding and autophosphorylating activities associated with the p21src protein of Harvey murine sarcoma virus. Nature. 1980 Oct 23;287(5784):686–91. doi: 10.1038/287686a0. [DOI] [PubMed] [Google Scholar]
- 6.Parada LF, Tabin CJ, Shih C, Weinberg RA. Human EJ bladder carcinoma oncogene is homologue of Harvey sarcoma virus ras gene. Nature. 1982 Jun 10;297(5866):474–8. doi: 10.1038/297474a0. [DOI] [PubMed] [Google Scholar]
- 7.Santos E, Tronick SR, Aaronson SA, Pulciani S, Barbacid M. T24 human bladder carcinoma oncogene is an activated form of the normal human homologue of BALB- and Harvey-MSV transforming genes. Nature. 1982 Jul 22;298(5872):343–7. doi: 10.1038/298343a0. [DOI] [PubMed] [Google Scholar]
- 8.Der CJ, Krontiris TG, Cooper GM. Transforming genes of human bladder and lung carcinoma cell lines are homologous to the ras genes of Harvey and Kirsten sarcoma viruses. Proc Natl Acad Sci USA. 1982 Jun;79(11):3637–40. doi: 10.1073/pnas.79.11.3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shimizu K, Goldfarb M, Suard Y, et al. Three human transforming genes are related to the viral ras oncogenes. Proc Natl Acad Sci USA. 1983 Apr;80(8):2112–6. doi: 10.1073/pnas.80.8.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taparowsky E, Shimizu K, Goldfarb M, Wigler M. Structure and activation of the human N-ras gene. Cell. 1983 Sep;34(2):581–6. doi: 10.1016/0092-8674(83)90390-2. [DOI] [PubMed] [Google Scholar]
- 11.Bos JL. ras oncogenes in human cancer: a review. Cancer Res. 1989 Sep 1;49(17):4682–9. [PubMed] [Google Scholar]
- 12.Albino AP, Le Strange R, Oliff AI, Furth ME, Old LJ. Transforming ras genes from human melanoma: a manifestation of tumour heterogeneity? Nature. 1984 Mar 1-7;308(5954):69–72. doi: 10.1038/308069a0. [DOI] [PubMed] [Google Scholar]
- 13.Padua RA, Barrass NC, Currie GA. Activation of N-ras in a human melanoma cell line. Mol Cell Biol. 1985 Mar;5(3):582–5. doi: 10.1128/mcb.5.3.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van ’t Veer LJ, Burgering BM, Versteeg R, et al. N-ras mutations in human cutaneous melanoma from sun-exposed body sites. Mol Cell Biol. 1989 Jul;9(7):3114–6. doi: 10.1128/mcb.9.7.3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Albino AP, Nanus DM, Mentle IR, et al. Analysis of ras oncogenes in malignant melanoma and precursor lesions: correlation of point mutations with differentiation phenotype. Oncogene. 1989 Nov;4(11):1363–74. [PubMed] [Google Scholar]
- 16.Jafari M, Papp T, Kirchner S, et al. Analysis of ras mutations in human melanocytic lesions: activation of the ras gene seems to be associated with the nodular type of human malignant melanoma. J Cancer Res Clin Oncol. 1995;121(1):23–30. doi: 10.1007/BF01202725. [DOI] [PubMed] [Google Scholar]
- 17.van Elsas A, Zerp SF, van der Flier S, et al. Relevance of ultraviolet-induced N-ras oncogene point mutations in development of primary human cutaneous melanoma. Am J Pathol. 1996 Sep;149(3):883–93. [PMC free article] [PubMed] [Google Scholar]
- 18.Demunter A, Stas M, Degreef H, De Wolf-Peeters C, van den Oord JJ. Analysis of N- and K-ras mutations in the distinctive tumor progression phases of melanoma. J Invest Dermatol. 2001 Dec;117(6):1483–9. doi: 10.1046/j.0022-202x.2001.01601.x. [DOI] [PubMed] [Google Scholar]
- 19.Ackermann J, Frutschi M, Kaloulis K, McKee T, Trumpp A, Beermann F. Metastasizing melanoma formation caused by expression of activated N-RasQ61K on an INK4a-deficient background. Cancer Res. 2005 May 15;65(10):4005–11. doi: 10.1158/0008-5472.CAN-04-2970. [DOI] [PubMed] [Google Scholar]
- 20.Chin L, Tam A, Pomerantz J, et al. Essential role for oncogenic Ras in tumour maintenance. Nature. 1999 Jul 29;400(6743):468–72. doi: 10.1038/22788. [DOI] [PubMed] [Google Scholar]
- 21.Eskandarpour M, Kiaii S, Zhu C, Castro J, Sakko AJ, Hansson J. Suppression of oncogenic NRAS by RNA interference induces apoptosis of human melanoma cells. Int J Cancer. 2005 May 20;115(1):65–73. doi: 10.1002/ijc.20873. [DOI] [PubMed] [Google Scholar]
- 22.Cohen C, Zavala-Pompa A, Sequeira JH, et al. Mitogen-actived protein kinase activation is an early event in melanoma progression. Clin Cancer Res. 2002 Dec;8(12):3728–33. [PubMed] [Google Scholar]
- 23.Lin WM, Baker AC, Beroukhim R, et al. Modeling genomic diversity and tumor dependency in malignant melanoma. Cancer Res. 2008 Feb 1;68(3):664–73. doi: 10.1158/0008-5472.CAN-07-2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Winnepenninckx V, Lazar V, Michiels S, et al. Gene expression profiling of SC primary cutaneous melanoma and clinical outcome. J Natl Cancer Inst. 2006 Apr 5;98(7):472–82. doi: 10.1093/jnci/djj103. [DOI] [PubMed] [Google Scholar]
- 25.Bastian BC, LeBoit PE, Pinkel D. Mutations and copy number increase of HRAS in Spitz nevi with distinctive histopathological features. Am J Pathol. 2000 Sep;157(3):967–72. doi: 10.1016/S0002-9440(10)64609-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whitwam T, Vanbrocklin MW, Russo ME, et al. Differential oncogenic potential of activated RAS isoforms in melanocytes. Oncogene. 2007 Jul 5;26(31):4563–70. doi: 10.1038/sj.onc.1210239. [DOI] [PubMed] [Google Scholar]
- 27.Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature. 2002 Jun 27;417(6892):949–54. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 28.Brose MS, Volpe P, Feldman M, et al. BRAF and RAS mutations in human lung cancer and melanoma. Cancer Res. 2002 Dec 1;62(23):6997–7000. [PubMed] [Google Scholar]
- 29.Pollock PM, Harper UL, Hansen KS, et al. High frequency of BRAF mutations in nevi. Nat Genet. 2003 Jan;33(1):19–20. doi: 10.1038/ng1054. [DOI] [PubMed] [Google Scholar]
- 30.Gorden A, Osman I, Gai W, et al. Analysis of BRAF and N-RAS mutations in metastatic melanoma tissues. Cancer Res. 2003 Jul 15;63(14):3955–7. [PubMed] [Google Scholar]
- 31.Maldonado JL, Fridlyand J, Patel H, et al. Determinants of BRAF mutations in primary melanomas. J Natl Cancer Inst. 2003 Dec 17;95(24):1878–90. doi: 10.1093/jnci/djg123. [DOI] [PubMed] [Google Scholar]
- 32.Shinozaki M, Fujimoto A, Morton DL, Hoon DS. Incidence of BRAF oncogene mutation and clinical relevance for primary cutaneous melanomas. Clin Cancer Res. 2004 Mar 1;10(5):1753–7. doi: 10.1158/1078-0432.ccr-1169-3. [DOI] [PubMed] [Google Scholar]
- 33.Garnett MJ, Marais R. Guilty as charged: B-RAF is a human oncogene. Cancer Cell. 2004 Oct;6(4):313–9. doi: 10.1016/j.ccr.2004.09.022. [DOI] [PubMed] [Google Scholar]
- 34.Wan PT, Garnett MJ, Roe SM, et al. Mechanism of C activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell. 2004 Mar 19;116(6):855–67. doi: 10.1016/s0092-8674(04)00215-6. [DOI] [PubMed] [Google Scholar]
- 35.Wellbrock C, Ogilvie L, Hedley D, et al. V599EB-RAF is an oncogene in melanocytes. Cancer Res. 2004 Apr 1;64(7):2338–42. doi: 10.1158/0008-5472.can-03-3433. [DOI] [PubMed] [Google Scholar]
- 36.Zhang BH, Guan KL. Activation of B-Raf kinase requires phosphorylation of the conserved residues Thr598 and Ser601. EMBO J. 2000 Oct 16;19(20):5429–39. doi: 10.1093/emboj/19.20.5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gray-Schopfer V, Wellbrock C, Marais R. Melanoma biology and new targeted therapy. Nature. 2007 Feb 22;445(7130):851–7. doi: 10.1038/nature05661. [DOI] [PubMed] [Google Scholar]
- 38.Hingorani SR, Jacobetz MA, Robertson GP, Herlyn M, Tuveson DA. Suppression of BRAF(V599E) in human melanoma abrogates transformation. Cancer Res. 2003 Sep 1;63(17):5198–202. [PubMed] [Google Scholar]
- 39.Hoeflich KP, Gray DC, Eby MT, et al. Oncogenic BRAF is required for tumor growth and maintenance in melanoma models. Cancer Res. 2006 Jan 15;66(2):999–1006. doi: 10.1158/0008-5472.CAN-05-2720. [DOI] [PubMed] [Google Scholar]
- 40.Collisson EA, De A, Suzuki H, Gambhir SS, Kolodney MS. Treatment of metastatic melanoma with an orally available inhibitor of the Ras-Raf-MAPK cascade. Cancer Res. 2003 Sep 15;63(18):5669–73. [PubMed] [Google Scholar]
- 41.Solit DB, Garraway LA, Pratilas CA, et al. BRAF mutation predicts sensitivity to MEK inhibition. Nature. 2006 Jan 19;439(7074):358–62. doi: 10.1038/nature04304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsao H, Goel V, Wu H, Yang G, Haluska FG. Genetic interaction between NRAS and BRAF mutations and PTEN/MMAC1 inactivation in melanoma. J Invest Dermatol. 2004 Feb;122(2):337–41. doi: 10.1046/j.0022-202X.2004.22243.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hoheisel JD. Microarray technology: beyond transcript profiling and genotype analysis. Nat Rev Genet. 2006 Mar;7(3):200–10. doi: 10.1038/nrg1809. [DOI] [PubMed] [Google Scholar]
- 44.Garraway LA, Widlund HR, Rubin MA, et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005 Jul 7;436(7047):117–22. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- 45.King R, Weilbaecher KN, McGill G, Cooley E, Mihm M, Fisher DE. Microphthalmia transcription factor. A sensitive and specific melanocyte marker for MelanomaDiagnosis. Am J Pathol. 1999 Sep;155(3):731–8. doi: 10.1016/S0002-9440(10)65172-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Levy C, Khaled M, Fisher DE. MITF: master regulator of melanocyte development and melanoma oncogene. Trends Mol Med. 2006 Sep;12(9):406–14. doi: 10.1016/j.molmed.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 47.Garraway LA, Sellers WR. Lineage dependency and lineage-survival oncogenes in human cancer. Nat Rev Cancer. 2006 Aug;6(8):593–602. doi: 10.1038/nrc1947. [DOI] [PubMed] [Google Scholar]
- 48.Chin L, Garraway LA, Fisher DE. Malignant melanoma: genetics and therapeutics in the genomic era. Genes Dev. 2006 Aug 15;20(16):2149–82. doi: 10.1101/gad.1437206. [DOI] [PubMed] [Google Scholar]
- 49.Heinlein CA, Chang C. Androgen receptor in prostate cancer. Endocr Rev. 2004 Apr;25(2):276–308. doi: 10.1210/er.2002-0032. [DOI] [PubMed] [Google Scholar]
- 50.Weir BA, Woo MS, Getz G, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007 Dec 6;450(7171):893–8. doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim M, Gans JD, Nogueira C, et al. ComparPative oncogenomics identifies NEDD9 as a melanoma metastasis gene. Cell. 2006 Jun 30;125(7):1269–81. doi: 10.1016/j.cell.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 52.Bastian BC, LeBoit PE, Hamm H, Brocker EB, Pinkel D. Chromosomal gains and losses in primary cutaneous melanomas detected by comparative genomic hybridization. Cancer Res. 1998 May 15;58(10):2170–5. [PubMed] [Google Scholar]
- 53.Curtin JA, Busam K, Pinkel D, Bastian BC. Somatic activation of KIT in distinct subtypes of melanoma. J Clin Oncol. 2006 Sep 10;24(26):4340–6. doi: 10.1200/JCO.2006.06.2984. [DOI] [PubMed] [Google Scholar]
- 54.Geissler EN, Ryan MA, Housman DE. The dominant-white spotting (W) locus of the mouse encodes the c-kit proto-oncogene. Cell. 1988 Oct 7;55(1):185–92. doi: 10.1016/0092-8674(88)90020-7. [DOI] [PubMed] [Google Scholar]
- 55.Willmore-Payne C, Holden JA, Tripp S, Layfield LJ. Human malignant melanoma: detection of BRAF- and c-kit-activating mutations by high-resolution amplicon melting analysis. Hum Pathol. 2005 May;36(5):486–93. doi: 10.1016/j.humpath.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 56.Willmore-Payne C, Holden JA, Hirschowitz S, Layfield LJ. BRAF and c-kit gene copy number in mutation-positive malignant melanoma. Hum Pathol. 2006 May;37(5):520–7. doi: 10.1016/j.humpath.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 57.Beadling C, Jacobson-Dunlop E, Hodi FS, et al. KIT gene mutations and copy number in melanoma subtypes. Clin Cancer Res. 2008 Nov 1;14(21):6821–8. doi: 10.1158/1078-0432.CCR-08-0575. [DOI] [PubMed] [Google Scholar]
- 58.Heinrich MC, Corless CL, Demetri GD, et al. Kinase mutations and imatinib response in patients with metastatic gastrointestinal stromal tumor. J Clin Oncol. 2003 Dec 1;21(23):4342–9. doi: 10.1200/JCO.2003.04.190. [DOI] [PubMed] [Google Scholar]
- 59.Demetri GD, von Mehren M, Blanke CD, et al. Efficacy and safety of imatinib mesylate in advanced gastrointestinal stromal tumors. N Engl J Med. 2002 Aug 15;347(7):472–80. doi: 10.1056/NEJMoa020461. [DOI] [PubMed] [Google Scholar]
- 60.Kantarjian H, Sawyers C, Hochhaus A, et al. Hematologic and cytogenetic responses to imatinib mesylate in chronic myelogenous leukemia. N Engl J Med. 2002 Feb 28;346(9):645–52. doi: 10.1056/NEJMoa011573. [DOI] [PubMed] [Google Scholar]
- 61.Kim KB, Eton O, Davis DW, et al. Phase II trial of imatinib mesylate in patients with metastatic melanoma. Br J Cancer. 2008 Sep 2;99(5):734–40. doi: 10.1038/sj.bjc.6604482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hodi FS, Friedlander P, Corless CL, et al. Major response to imatinib mesylate in KIT-mutated melanoma. J Clin Oncol. 2008 Apr 20;26(12):2046–51. doi: 10.1200/JCO.2007.14.0707. [DOI] [PubMed] [Google Scholar]
- 63.Stahl JM, Sharma A, Cheung M, et al. Deregulated Akt3 activity promotes development of malignant melanoma. Cancer Res. 2004 Oct 1;64(19):7002–10. doi: 10.1158/0008-5472.CAN-04-1399. [DOI] [PubMed] [Google Scholar]
- 64.Davies MA, Stemke-Hale K, Tellez C, et al. A novel AKT3 mutation in melanoma tumours and cell lines. Br J Cancer. 2008 Oct 21;99(8):1265–8. doi: 10.1038/sj.bjc.6604637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Omholt K, Krockel D, Ringborg U, Hansson J. Mutations of PIK3CA are rare in cutaneous melanoma. Melanoma Res. 2006 Apr;16(2):197–200. doi: 10.1097/01.cmr.0000200488.77970.e3. [DOI] [PubMed] [Google Scholar]
- 66.Wu H, Goel V, Haluska FG. PTEN signaling pathways in melanoma. Oncogene. 2003 May 19;22(20):3113–22. doi: 10.1038/sj.onc.1206451. [DOI] [PubMed] [Google Scholar]
- 67.Guldberg P, Straten P thor, Birck A, Ahrenkiel V, Kirkin AF, Zeuthen J. Disruption of the MMAC1/PTEN gene by deletion or mutation is a frequent event in malignant melanoma. Cancer Res. 1997 Sep 1;57(17):3660–3. [PubMed] [Google Scholar]
- 68.Hussussian CJ, Struewing JP, Goldstein AM, et al. Germline p16 mutations in familial melanoma. Nat Genet. 1994 Sep;8(1):15–21. doi: 10.1038/ng0994-15. [DOI] [PubMed] [Google Scholar]
- 69.Kamb A, Shattuck-Eidens D, Eeles R, et al. Analysis of the p16 gene (CDKN2) as a candidate for the chromosome 9p melanoma susceptibility locus. Nat Genet. 1994 Sep;8(1):23–6. doi: 10.1038/ng0994-22. [DOI] [PubMed] [Google Scholar]
- 70.Wolfel T, Hauer M, Schneider J, et al. A p16INK4a-insensitive CDK4 mutant targeted by cytolytic T lymphocytes in a human melanoma. Science. 1995 Sep 1;269(5228):1281–4. doi: 10.1126/science.7652577. [DOI] [PubMed] [Google Scholar]
- 71.Zuo L, Weger J, Yang Q, et al. Germline mutations in the p16INK4a binding domain of CDK4 in familial melanoma. Nat Genet. 1996 Jan;12(1):97–9. doi: 10.1038/ng0196-97. [DOI] [PubMed] [Google Scholar]
- 72.Straume O, Akslen LA. Alterations and prognostic significance of p16 and p53 protein expression in subgroups of cutaneous melanoma. Int J Cancer. 1997 Oct 21;74(5):535–9. doi: 10.1002/(sici)1097-0215(19971021)74:5<535::aid-ijc10>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 73.Ragnarsson-Olding BK, Karsberg S, Platz A, Ringborg UK. Mutations in the TP53 gene in human malignant melanomas derived from sun-exposed skin and unexposed mucosal membranes. Melanoma Res. 2002 Oct;12(5):453–63. doi: 10.1097/00008390-200209000-00007. [DOI] [PubMed] [Google Scholar]
- 74.Worm J, Christensen C, Gronbaek K, Tulchinsky E, Guldberg P. Genetic and epigenetic alterations of the APC gene in malignant melanoma. Oncogene. 2004 Jul 1;23(30):5215–26. doi: 10.1038/sj.onc.1207647. [DOI] [PubMed] [Google Scholar]
- 75.Omholt K, Platz A, Ringborg U, Hansson J. Cytoplasmic and nuclear accumulation of beta-catenin is rarely caused by CTNNB1 exon 3 mutations in cutaneous malignant melanoma. Int J Cancer. 2001 Jun 15;92(6):839–42. doi: 10.1002/ijc.1270. [DOI] [PubMed] [Google Scholar]
- 76.Rubinfeld B, Robbins P, El-Gamil M, Albert I, Porfiri E, Polakis P. Stabilization of beta-catenin by genetic defects in melanoma cell lines. Science. 1997 Mar 21;275(5307):1790–2. doi: 10.1126/science.275.5307.1790. [DOI] [PubMed] [Google Scholar]
- 77.Greenman C, Stephens P, Smith R, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007 Mar 8;446(7132):153–8. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ding L, Getz G, Wheeler DA, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008 Oct 23;455(7216):1069–75. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jones S, Zhang X, Parsons DW, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008 Sep 26;321(5897):1801–6. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008 Sep 26;321(5897):1807–12. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sjoblom T, Jones S, Wood LD, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006 Oct 13;314(5797):268–74. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 82.Wood LD, Parsons DW, Jones S, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007 Nov 16;318(5853):1108–13. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 83.Collins FS, Barker AD. Mapping the cancer genome. Pinpointing the genes involved in cancer will help chart a new course across the complex landscape of human malignancies. Sci Am. 2007 Mar;296(3):50–7. [PubMed] [Google Scholar]
- 84.Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008 Oct 23;455(7216):1061–8. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Brown KM, Macgregor S, Montgomery GW, et al. Common sequence variants on 20q11.22 confer melanoma susceptibility. Nat Genet. 2008 Jul;40(7):838–40. doi: 10.1038/ng.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gudbjartsson DF, Sulem P, Stacey SN, et al. ASIP and TYR pigmentation variants associate with cutaneous melanoma and basal cell carcinoma. Nat Genet. 2008 Jul;40(7):886–91. doi: 10.1038/ng.161. [DOI] [PubMed] [Google Scholar]
- 87.Luo B, Cheung HW, Subramanian A, et al. Highly parallel identification of essential genes in cancer cells. Proc Natl Acad Sci USA. 2008 Dec 23;105(51):20380–5. doi: 10.1073/pnas.0810485105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shendure J, Ji H. Next-generation DNA sequencing. Nat Biotechnol. 2008 Oct;26(10):1135–45. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- 89.Thomas RK, Nickerson E, Simons JF, et al. Sensitive mutation detection in heterogeneous cancer specimens by massively parallel picoliter reactor sequencing. Nat Med. 2006 Jul;12(7):852–5. doi: 10.1038/nm1437. [DOI] [PubMed] [Google Scholar]
- 90.Ley TJ, Mardis ER, Ding L, et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008 Nov 6;456(7218):66–72. doi: 10.1038/nature07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hodges E, Xuan Z, Balija V, et al. Genome-wide in situ exon capture for selective resequencing. Nat Genet. 2007 Dec;39(12):1522–7. doi: 10.1038/ng.2007.42. [DOI] [PubMed] [Google Scholar]
- 92.Olson M. Enrichment of super-sized resequencing targets from the human genome. Nat Methods. 2007 Nov;4(11):891–2. doi: 10.1038/nmeth1107-891. [DOI] [PubMed] [Google Scholar]
- 93.Sultan M, Schulz MH, Richard H, et al. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science. 2008 Aug 15;321(5891):956–60. doi: 10.1126/science.1160342. [DOI] [PubMed] [Google Scholar]
- 94.Wang ET, Sandberg R, Luo S, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008 Nov 27;456(7221):470–6. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sugarbaker DJ, Richards WG, Gordon GJ, et al. Transcriptome sequencing of malignant pleural mesothelioma tumors. Proc Natl Acad Sci USA. 2008 Mar 4;105(9):3521–6. doi: 10.1073/pnas.0712399105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Meissner A, Mikkelsen TS, Gu H, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008 Aug 7;454(7205):766–70. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mikkelsen TS, Ku M, Jaffe DB, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007 Aug 2;448(7153):553–60. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007 Jun 8;316(5830):1497–502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 99.Chiang DY, Getz G, Jaffe DB, et al. High-resolution mapping of copy-number alterations with massively parallel sequencing. Nat Methods. 2008 Nov 30; doi: 10.1038/nmeth.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Curtin JA, Fridlyand J, Kageshita T, et al. Distinct sets of genetic alterations in melanoma. N Engl J Med. 2005 Nov 17;353(20):2135–47. doi: 10.1056/NEJMoa050092. [DOI] [PubMed] [Google Scholar]
- 101.Bittner M, Meltzer P, Chen Y, et al. Molecular classification of cutaneous malignant melanoma by gene expression profiling. Nature. 2000 Aug 3;406(6795):536–40. doi: 10.1038/35020115. [DOI] [PubMed] [Google Scholar]
- 102.Slamon DJ, Leyland-Jones B, Shak S, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001 Mar 15;344(11):783–92. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- 103.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004 May 20;350(21):2129–39. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 104.Paez JG, Janne PA, Lee JC, et al. EGFR mutations in lung cancer: corr Uelation with clinical response to gefitinib therapy. Science. 2004 Jun 4;304(5676):1497–500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 105.Pao W, Miller V, Zakowski M, et al. EGF receptor gene mutations AN are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci USA. 2004 Sep 7;101(36):13306–11. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Adjei AA, Cohen RB, Franklin W, et al. Phase I pharmacokinetic and pharmacodynamic study of the oral, small-molecule mitogen-activated protein kinase kinase 1/2 inhibitor AZD6244 (ARRY-142886) in patients with advanced cancers. J Clin Oncol. 2008 May 1;26(13):2139–46. doi: 10.1200/JCO.2007.14.4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Thomas RK, Baker AC, Debiasi RM, et al. Hight throughput oncogene mutation profiling in human cancer. Nat Genet. 2007 Mar;39(3):347–51. doi: 10.1038/ng1975. [DOI] [PubMed] [Google Scholar]
- 108.McDermott U, Sharma SV, Dowell L, et al. Identification of genotype-correlated sensitivity to selective kinase inhibitors by using high-throughput tumor cell line profiling. Proc Natl Acad Sci USA. 2007 Dec 11;104(50):19936–41. doi: 10.1073/pnas.0707498104. [DOI] [PMC free article] [PubMed] [Google Scholar]