Abstract
The allosteric regulation of human liver pyruvate kinase (hL-PYK) by fructose-1,6-bisphosphate (Fru-1,6-BP; activator), ATP (inhibitor) and alanine (Ala; inhibitor) was monitored over a pH range from 6.5 to 8.0 at 37°C. As a function of increasing pH, hL-PYK's affinity for the substrate phosphoenolpyruvate (PEP), and for Fru-1,6-BP decreases, while affinities for ATP and Ala slightly increases. At pH 6.5, Fru-1,6-BP and ATP elicit only small allosteric impacts on PEP affinity. As pH increases, Fru-1,6-BP and ATP elicit greater allosteric responses, but the response to Ala is relatively constant. Since the magnitudes of the allosteric coupling for ATP and for Ala inhibition are different and the pH dependences of these magnitudes are not similar, these inhibitors likely elicit their responses using different molecular mechanisms. In addition, our results fail to support a general correlation between pH dependent changes in effector affinity and pH dependent changes in the corresponding allosteric response.
Keywords: pyruvate kinase, liver form, allosteric coupling, allostery, pH dependence
In mammals, approximately 90% of blood glucose produced by gluconeogenesis is generated in the liver. During periods of gluconeogenesis, glycolysis in this tissue must be regulated to prevent futile cycling of gluconeogenic substrates. Pyruvate kinase (PYK) catalyzes the conversion of phosphoenolpyruvate (PEP) and MgADP to pyruvate and MgATP as the last step of glycolysis. Since the liver PYK isozyme (L-PYK) catalyzes a reaction that opposes the first enzymatic step of gluconeogenesis, several mechanisms regulate L-PYK activity. The regulatory features of L-PYK embody many of the classic examples of regulatory mechanisms used to control metabolic and signal transduction pathways: 1) phosphorylation of L-PYK by cAMP dependent protein kinase (cAMP-PK) and the countering dephosphorylation (both in response to hormonal signals) coordinate the balance of glycolysis and gluconeogenesis with whole body energy needs and supply; 2) the Fru-1,6-BP regulation of L-PYK is a textbook example of an intermediate from a metabolic pathway acting as a feed forward activator in the same pathway; 3) Ala inhibits L-PYK during periods of protein breakdown (starvation) when Ala is used as the primary carbon source for gluconeogenesis; 4) allosteric inhibition of L-PYK by ATP offers examples both of a feedback inhibition by the final product of the metabolic pathway and of the sensitivity of energy producing catalytic reactions to the intracellular energy supply. Allostery and phosphorylation appear to be the primary mechanisms of rapidly controlling PYK activity since, in contrast to mRNA levels [1], enzyme concentrations are held within narrow limits [2-4].
Allosteric ATP inhibition and regulation by phosphorylation on the N-terminus are unique to the pyruvate kinase isozymes from mammalian liver and red blood cell. These two isozymes are products of a single gene due to the use of different translation start sites and differ only by the additional 31 amino acids located at the N-terminus of the red blood cell isozyme (R-PYK) [5]. Amino acid inhibition of both L-PYK and R-PYK is common with the other two mammalian PYK isozymes, the muscle form (M1-PYK) and the “fetal” form (M2-PYK). However, the regulatory amino acid ligand is different. M1-PYK is inhibited by hydrophobic amino acids. Ala binds to M1-PYK competitively with Phe, but elicits a negligible allosteric response [6]. Fru-1,6-BP activation is shared amongst not only L-PYK, R-PYK, and M2-PYK, but also isozymes from a variety of prokaryotic and eukaryotic sources [2, 7-10]. It should be noted that these general regulatory features of L-PYK have been summarized from studies using a variety of mammalian model systems and/or the similar mammalian R-PYK isozymes; there are only a limited number of publications which have focused on the purified human L-PYK (hL-PYK) isozyme [11-16] and much of the work reported in those publications are techniques to purify the enzyme from human liver tissue.
Binding of allosteric effectors to L-PYK alters the affinity of the enzyme for PEP without altering the catalytic rate (kcat) or the affinity of the enzyme for MgADP [2, 10, 17]. We find the best definition for this “K-type” allostery is a comparison of how one ligand binds in the absence vs. in the presence of a second ligand [18]. Consistent with this definition, the extent to which the affinity of a protein for one ligand is modified when the second ligand is present in saturating concentrations gives rise to the magnitude of the allosteric coupling. This magnitude is distinguished from a protein's affinity for the effector using a thermodynamic linked-function analysis [19, 20]. Multiple pathways of interaction (linked-reactions) [18] within the protein contribute to the observed allosteric coupling. The contributions of these multiple linked-reactions to the observed magnitude can vary with changes in solution conditions (e.g. pH). Multiple linked-reactions are expected throughout the protein even if a single chemical moiety on the effector elicits the allosteric response. Considering such an example (although likely over-simplified), if two effectors formed the exact same regulatory ligand-protein interaction (and no additional regulatory interactions), the observed magnitude of the allosteric response would be expected to be equivalent. In addition, the pH dependences of these magnitudes (in the absence of a protonation event on either effector) would be equivalent. However, due to the very restrictive limitations of this theoretical example, similar coupling magnitudes and/or pH dependencies of these magnitudes for two different effectors seem just as likely to result from contributions of completely or partially different sets of linked-reactions. Therefore, dissimilar magnitudes and pH dependencies of allosteric functions associated with two effectors are consistent with two different allosteric mechanisms (defined here as the sum of all linked-reactions involved), but similarities in these responses do not necessitate similar allosteric mechanisms. In the case of hL-PYK, a comparative evaluation of allosteric functions is of particular interest since the binding site for the allosteric nucleotide remains unknown (the allosteric amino acid and Fru-1,6-BP binding sites have been determined by co-crystallization [6, 21, 22]). As determined in the current work, hL-PYK inhibition by ATP and inhibition by Ala do not have similar magnitudes of allosteric coupling, nor are the dependencies of these magnitudes on changes in pH similar. This result is consistent with different ATP and Ala allosteric mechanisms functioning in hL-PYK.
In addition to insights into the allosteric regulation of hL-PYK, we are interested in potential general correlations associated with allostery. As one example, we reasoned that there might be a physiological benefit if a change in solution conditions simultaneously increased effector affinity and the corresponding allosteric coupling. This type of coordinated response could amplify the regulation impact elicited by an allosteric effector. Since hL-PYK is regulated by multiple allosteric effectors that collectively elicit inhibiting and activating responses, this system is ideal to test for a general correlation associated with allostery. However, our results do not support this general correlation.
Materials and Methods
Materials
Materials for gene disruption were as previously reported [23]. The pLC1 and pLC11 plasmids encoding human R-PYK and hL-PYK, respectively, were obtained as a gift from Drs. Andrea Mattevi and Giovanna Valentini [22]. The potassium salt of PEP was purchased from Fluka. Fast-Flow/Fibrous-Form DEAE cellulose, NADH, L-lactic dehydrogenase (Type III bovine heart), glycerol, and the potassium salt of ADP were purchased from Sigma. Other buffer components were from Fisher Scientific.
E. coli expression system development
E. coli has two genes (pykA and pykF) that encode two pyruvate kinase isozymes (EcPYK-A and EcPYK-F) [24, 25]. Deleting endogenous pyruvate genes, rather than interrupting these genes by inserting antibiotic resistance markers [26-31], eliminates the possibility of the E. coli strain reverting to express endogenous pyruvate kinase activity. The procedure to delete genes as overviewed here has previously been detailed in Lovingshimer et al. [23]. Steps include the 1) development of a linear DNA cassette including a selection marker flanked by segments of the target gene, 2) electroporation of this cassette into an E. coli engineered to facilitate recombination (DY329), 3) selection of colonies that incorporated the selection marker, 4) transduction of the disrupted gene/marker into the desired E. coli strain, and 5) removal of the selection marker using the FLP recombinase system.
E. coli gene deletions were performed in the laboratory of Dr. Gregory D. Reinhart (Texas A&M University) using methods described by Yu et al. [32] and Datsenko and Wanner [33]. We selected the MQ E. coli strain as a parent cell strain based on the benefits previously reviewed [23] and the fact that this strain possesses the lac operon; the latter property enables the use of lactose as an inducer of gene expression using lac and tac promoters. The genotypes of the bacterial strains used in this study are listed in Table 1. The KanR-cassettes that contained homology to pykA or pykF were made by PCR using FastStart Taq polymerase (Roche Applied Sciences), the primers listed in Supplemental Table 1, and the pRATT04 plasmid as a template for the KanR gene. The product size after PCR was confirmed by electrophoresis. Electroporation of these cassettes (500-700 ng of PCR product) into electrocompetent DY329 cells (50 μL) was done using a MicroPulser Electroporator and 0.1-cm electroporation cuvettes (BioRad). One mL of ice-cold SOC was added and cells were allowed to recover at 30°C for 1 hr. Cells were plated on LB agar containing 30 μg/mL kanamycin. Single colonies were selected for further steps. P1 lysates of the modified DY329 strains were made and these lysates were used to transduce the KanR insertion into MQ. Cells were plated on LB agar containing kanamycin and single colonies were selected. The KanR-cassette was deleted by transforming pCP20 into the modified MQ cell strain. Transformed cells were selected by ampicillin-resistance when cells were grown at 30°C. The pCP20 plasmid encodes FLP recombinase under a thermally induced promoter. In addition, the plasmid has temperature sensitive-replication. Therefore, growth at 42°C results in the removal of the KanR-cassette and the loss of plasmid replication. Upon removal of the KanR-cassettes, a FRT scar (FRT) remains. Cells were screened for the loss of both antibiotic resistances. The presence or absence of the KanR-cassettes into pykA was verified by PCR according to Lovingshimer et al. [23]. The primers designed to the pykF gene did not produce a PCR product; no further effort to identify new primers was made after the enzymatic verification of the removal of PYK activity in knockout strains (Supplemental Figure 1). For PCR verification of the deletion of pykA, the forward primer was CTTATACGACATCCGAATGAG and the reverse primer was GAGAGGCCTTCGCCTGATG. Primers designed internal to the KanR gene were as previously reported [23]. Created cell strains are summarized in Table 1.
Table 1.
Strains used and constructed
| Name | Description | Ref. |
|---|---|---|
| DY329 | W3110 ΔlacU169 nadA∷Tn10 gal490 λ-cI857 Δ(cro-bioA) | [32] |
| FF01 | DY329 ΔpykA∷kan | — |
| FF02 | DY329 ΔpykF∷kan | — |
| MQ | F- λ- lac+ lacIq araD139 rpsL150 ptsF25 flhD5301 rpsR deoC relA1 | [23] |
| FF11 | MQ ΔpykA∷kan | — |
| FF12 | MQ ΔpykF∷kan | — |
| FF20 | MQ ΔpykA∷FRT | — |
| FF30 | MQ ΔpykF∷FRT | — |
| FF31 | MQ ΔpykF∷FRT ΔpykA∷kan | — |
| FF50 | MQ ΔpykF∷FRT ΔpykA∷FRT | — |
A comparison of pyruvate kinase activity recovered from MQ, FF20, and FF30 cultures (Supplement Figure 1) reveals that the removal of pykA (FF20) results in the loss of 36% of the activity and deletion of the pykF gene (FF30) causes a 72% reduction in activity. The sum of activities recovered from FF20 and FF30 cells accounts for 92% of the activity from MQ cells. Finally, simultaneously removing both the pykA and the pykF genes removes all detectable pyruvate kinase activity in the resulting FF50 strain. FF50 cells retain the ability to grow on glucose as the only carbon source. An E. coli strain with the two endogenous pyruvate kinase genes interrupted by antibiotic resistance markers also retain the ability to grow on glucose, an ability that has been attributed to the generation of pyruvate by the Entner-Doudoroff pathway and the conversion of PEP to pyruvate by the phosphotransferase transport system [26, 34].
FF50 cells were transformed with pLC11; this plasmid is constructed identical to pLC1, but encodes hL-PYK [22]. Optimum expression of hL-PYK in FF50 cells transformed with pLC11 was obtained using 5 mM lactose added at the time of inoculation and grown at 37°C for 24 hours (Supplemental Figure 2). Using this optimum condition, FF50 cells expressing hL-PYK or human R-PYK were lysed by sonication. Clarified cell extracts were analyzed by Western blot analysis. The polyclonal antibody made to rat L-PYK used for detection was a gift from Dr. James B. Blair (Virginia Tech). hL-PYK is expressed in FF50 cells as a single band on an SDS gel (Supplemental Figure 3). This result was further confirmed by measurement of the mass of the purified protein using MALDI-TOF mass spectrometry (Supplemental Figure 4). The homogeneous expression of hL-PYK is a result that is of particular interest since human R-PYK purified from E. coli has been reported to contain two different sizes of subunits [35]. Consistent with our finding, rat L-PYK purified from an E. coli expression system also does not show signs of proteolysis [36].
Protein Purification of hL-PYK
All purification steps were performed on ice or at 4°C and using pre-chilled buffers. FF50 cells expressing hL-PYK from the pLC11 plasmid were lysed in buffer A (10 mM MES-pH 6.0, 2 mM MgCl2, 25% glycerol, 2 mM DTT, and 1 mM PMSF). Recovery of PYK activity in soluble cell extract was comparable whether cells were lysed with a French-press or using sonication (data not shown); sonication was used in this study. Cell debris was removed by centrifugation and solid ammonium sulfate was added to the clarified cell extract to a final of 26% w/v (0.15 g of ammonium sulfate/mL of cell extract) at 4°C. After centrifugation, solid ammonium sulfate was added to the supernatant to a final 46% ammonium sulfate (an additional 0.12 g of ammonium sulfate/mL of the initial cell extract) at 4°C. The 26%/46% protein pellet was collected by centrifugation. The protein pellet was first suspended in ice cold buffer B (10 mM MES at pH 6.8, 5 mM MgCl2, 10 mM KCl, and 2 mM DTT) and then dialyzed against the same buffer. The dialyzed protein sample was loaded onto a column packed with a minimum of 30 g pre-washed DEAE-cellulose per liter of the initial cell culture and pre-equilibrated with buffer B. Buffer B was passed through the column at a rate of 2.0 mL/min. 2 mL samples were collected. Pyruvate kinase activity and A280 were determined for each fraction and those with the highest specific activity were pooled. The pooled protein was concentrated by adding 0.35 g of ammonium sulfate followed by centrifugation. The protein pellet was suspended in less than 5 mLs of buffer C (50 mM MES and 50 mM HEPES at pH 6.5, 10 mM MgCl2, 100 mM KCl, 0.1 mM EDTA, and 2 mM DTT). Using a flow rate of 1.3 mL/min, this sample was further fractionated with a S-200 pre-packed size exclusion column (Amersham Biosciences) pre-equilibrated with buffer C. Active fractions were pooled and stored in buffer C at 4°C. After the DEAE-cellulose column, hL-PYK is greater than 95% pure as analyzed by SDS-PAGE (Supplemental Figure 5). This enzyme fraction had a specific activity of 121 U/mg (Supplemental Table 2). A final size-exclusion column results in greater than 99% purity, but had a reduced specific activity. The specific activity of the DEAE fraction is within the range reported for other mammalian L-PYK isozymes [2]. hL-PYK purified with the size-exclusion column was used in this work, however protein purified with the DEAE-cellulose column, but not the size-exclusion column gives similar results (Supplemental Figure 6).
Determination of the Molar Absorption Coefficient for hL-PYK
Pace et al. reported that the best approach for determining the extinction coefficient of a protein is the Edelhoch method [37]. Following this method, the protein concentration of size-exclusion purified hL-PYK was determined in 6 M guanidine-HCl using the molar absorption coefficients of Trp (5,685 M-1 cm-1) and Tyr (1,285 M-1 cm-1) in 6 M guanidine-HCl (note the absence of cystine in hL-PYK). The absorbance of the protein in buffer was then determined in a 1 cm cuvette to calculate the molar absorption coefficient of hL-PYK in buffer. The subunit molar coefficient at 280 nm for size-exclusion purified hL-PYK is 29400±300 M-1cm-1 (0.502±0.006 mL*mg-1cm-1), a result that is only 1.8% different from that predicted by generalized equation of Pace et al [37].
Kinetic Assays
Activity measurements were carried out at 37°C using a lactate dehydrogenase coupled assay. Reactions contained 100 mM HEPES, 10 mM MgCl2, 2 mM ADP (10 times the K0.5-ADP; Supplemental Figure 7), 0.1 mM EDTA, 0.18 mM NADH, and 19.6 U/mL lactate dehydrogenase. PEP and effector concentrations were varied as indicated. Stock solutions of PEP and effectors were adjusted to the proper pH before addition, and dilutions were in KCl to maintain constant K+ concentration. To prevent competition between MgADP and MgATP in the active site, ATP was added as a solution of equal molar MgADP/MgATP. (Effectors, including ATP, elicit K-type effects on PEP affinity without altering maximal velocity [2, 10, 17]. Therefore, a decrease in maximal velocity upon the addition of ATP indicates competitive binding between ADP and ATP at the active site. As can be seen in Figure 1 below, the addition of an equal molar MgADP/MgATP solution elicits an allosteric response on PEP affinity without causing changes on the maximal velocity. Therefore, this equal molar concentration of MgADP prevents competitive binding of ATP in the active site.) The concentration of cations varies considerably between an assay at pH 6.0, low PEP, and in the absence of effectors versus an assay at pH 8.0, at 10 mM PEP and at the highest concentrations of effectors. Therefore, to maintain a constant ionic strength, K+ and Na+ concentrations from additions of KOH and from counter ions of ligands were summed and KCl and NaCl were supplemented to a total K+ concentration of 500 mM and a total Na+ concentration of 150 mM in all assays. The enzymatic reaction was initiated with PEP and monitored at 340 nm over time in a UV-Star flat bottom 96-well plate (Greiner bio-one) containing a total reaction volume of 350 μL. All activity readings were collected using a Molecular Devices Spectramax Plus384 spectrophotometer.
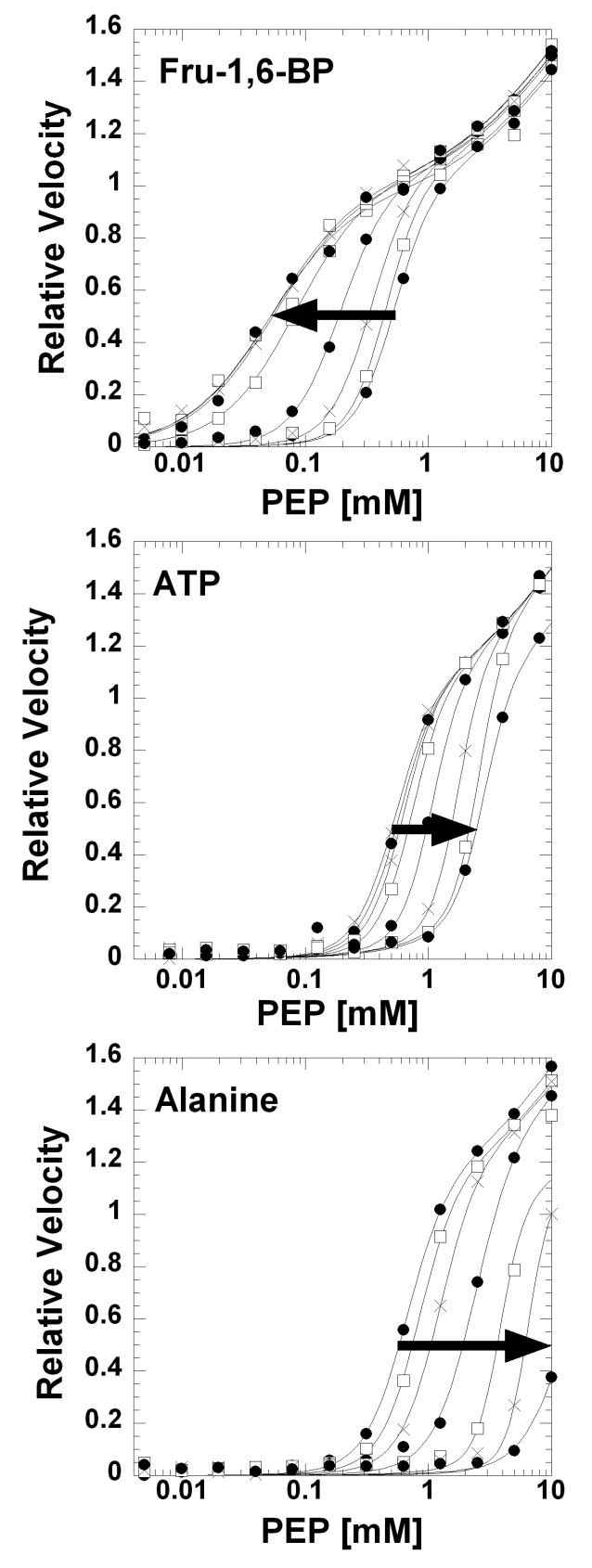
Figure 1.
Effects of Fru-1,6-BP, ATP, and Ala on the response of initial velocity to varying PEP concentration at pH 7.5 and 37°C. Lines represent the best fits to Equation 1 with the restrictions described in Material and Methods. Arrows indicate increasing effector concentration of Fru-1,6-BP (0, 0.000070, 0.00021, 0.0063, 0.0019, 0.0057, 0.017, and 0.051 mM), ATP (0, 0.51, 1.01, 2.02, 4.05, 8.1, 16.2, and 32.4 mM), or Ala (0, 0.782, 2.34, 7.03, 21, 63, and 570 mM).
Data fitting
Data were fit to appropriate equations using the nonlinear least-squares fitting analysis of Kaleidagraph (Synergy) software. Kapp-PEP-1 values were obtained by fitting initial rates obtained from kinetic assay to:
| Equation 1 |
Where Vmax-1 is the maximum velocity associated with the low PEP phase, Vmax-2 is the maximum velocity associated with the high PEP phase, Kapp-PEP-1 is the concentration of substrate that yields a rate equal to one-half the Vmax-1, Kapp-PEP-2 is the equivalent of Kapp-PEP-1 associated with the high PEP phase, and nH is the Hill coefficient associated with the low PEP phase. As detailed in Results and Discussion, fits using Equation 1 were performed with Vmax-2 set to equal Vmax-1 and Kapp-PEP-2 fixed at either 10 mM (when varying ATP or Fru-1,6-BP) or 15 times the Kapp-PEP-1 (when varying Ala). Only Kapp-PEP-1 fit values were considered further (without the “-1” distinction). Kapp-PEP values were plotted as a function of effector concentration and fit to:
| Equation 2 |
where Ka = Kapp-PEP when [Effector]=0, Kx = the dissociation constant for effector when [PEP]=0 and Qax=the coupling constant between PEP and the effector. Qax=1 defines no allosteric response, Qax>1 defines allosteric activation, and Qax<1 defines allosteric inhibition. Qax is related to the coupling free energy, ΔGax between PEP and effector by [38]:
| Equation 3 |
Results and Discussion
PEP Elicits a Biphasic Activity Response
The response of hL-PYK activity to varying concentrations of PEP is biphasic (Figure 1). This finding was a surprise to us since a biphasic response has not previously been reported. The two phases will be referred to as the low PEP phase and the high PEP phase. When assaying activity at 37°C, very little of the high PEP phase could be determined using PEP concentrations up to 10 mM. Reducing temperature to 5°C and reducing total ionic strength in the assay did not increase apparent affinity sufficiently to allow full characterization of the high PEP phase, again using PEP concentration up to 10 mM. At the 5°C condition, concentrations greater than 10 mM are inhibiting. We rule out pyruvate contamination in PEP and/or non-catalyzed dephosphorylation of PEP, since activity is negligible in the absence of enzyme, even at greater than 10 mM PEP (see Supplemental Figure 8 for 5°C data). Completely ruling out the presence of a contaminating enzymatic activity that gives rise to the high PEP phase is difficult. However, the biphasic response is observed at all steps during protein purification (Supplemental Figure 6). In addition, the activity in both PEP phases is dependent on the presence of both Mg and ADP; no activity is observed when either Mg or ADP is removed from the assay, at all PEP concentrations up to 10 mM and in the absence of effector or in the presence of 0.51 mM Fru-1,6-BP (data not shown). A biphasic response might result if some fraction of hL-PYK is covalently modified, resulting in mixed population of modified and unmodified proteins. Unlike E. coli expressed R-PYK, hL-PYK expressed in E. coli is homogeneous (Supplemental Figures 3 and 4). In addition, phosphorylation (at Ser12 [39-43]) or oxidation of the unmodified protein purified from E. coli does not remove the biphasic response to PEP (data not shown).
If the biphasic response is a property of the unmodified enzyme, we can speculate why the biphasic response to PEP has not previously been reported. First, unless covalent modifications were enzymatically modulated after purification, L-PYK previously purified from liver was a mixture of phosphorylated and non-phosphorlated enzyme. Next, the concentration of PEP used in the current work spans a much greater range than has previously been employed. Finally, even with the expanded substrate range used here, the only reason that the biphasic nature is apparent is the use of a logarithmic x-axis to plot data, necessary to compare data collected over a large concentration range. When a linear x-axis is used, the biphasic nature is not obvious (Supplemental Figure 9).
If the biphasic response is a property of the unmodified protein, it may provide an insight into understanding mechanistic aspects of the enzyme; however, this property is not likely physiologically consequential since it only occurs at PEP concentrations above a physiological range [44]. One potential mechanism that would give rise to a biphasic response comes from the fact that the PYK tetramer is constructed as a dimer of dimers. One structure of a M1-PYK tetramer contains subunits in two different conformations [6]. The two subunits diagonal to one another in the tetramer share a common conformation. This may indicate that the PYK tetramer functions as a dimer of dimers. As an additional consideration, previously reported nH-PEP values likely reflect data from the low PEP phase. Therefore, the fact that these nH-PEP values for L-PYK isozymes is often near 2 (see below and [2]) may be consistent with a functioning dimer of dimers; two dimers may act cooperatively to generate the low PEP phase and the second two monomers may be responsible for the high PEP phase. Taken together, the PYK tetramer might not only be constructed as a dimer of dimers, but it may also function as a dimer of dimers.
Fit Evaluations Reveal the Robustness of Qax Determination
To quantify the magnitude of the allosteric response, PEP affinity must be determined over a concentration range of effector [18, 20, 45]. Only the low PEP phase of the response curve appears to be responsive to Fru-1,6-BP and ATP (Figure 1). Whether the high PEP phase is responsive to Ala is less certain. To address if Ala causes the two phases to become a single phase, a PEP titration of activity was determined in the presence of Ala at 5°C (Supplemental Figure 8). This data appears to indicate that the response to PEP is a single phase in the presence of a saturating concentration of Ala, but does not satisfactorily address if the high PEP phase is responsive to Ala.
The more challenging problem was determining the best approach for data fitting to obtain Kapp-PEP values. Unfortunately, even the 5°C data do not provide clues to this best fitting approach. Therefore, based on the speculation that the biphasic response is due to a functioning dimer of dimers, the high PEP phase was forced to have a Vmax activity that is equal to the Vmax activity from the low PEP phase. Data in the low PEP phase were fit to a Hill equation; data in the high PEP phase were fit to a Michaelis-Menten equation (Equation 1). Additionally, in the presence of Ala concentrations greater than 15 mM, the Vmax for the low PEP phase was assumed to be unmodified by Ala and therefore was set to equal the Vmax for the low PEP phase in the absence of Ala. Allosteric regulation focuses on a comparison of affinity values in the absence vs. presence of effector [18, 20]; therefore, even if this fitting approach was not capable of accurately evaluate Kapp-PEP, it facilitates an evaluation of allosteric coupling of hL-PYK (further justified below).
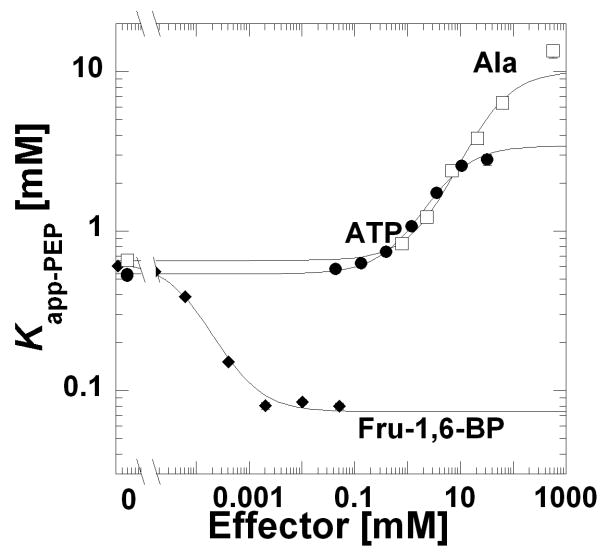
Using this fitting strategy, Kapp-PEP for the low PEP phase was determined over a concentration range of allosteric effector. In Figure 2, these Kapp-PEP values were plotted as a function of the effector concentration. With the format used in Figure 2, Ka-PEP is the left y-intercept. Due to the use of a logarithmic y-axis in Figure 2, the magnitude of allosteric coupling is the distance between the lower plateau in the absence of effector and the upper plateau obtained at high effector concentration [18, 20]. Although Kix-effector is determined by fitting this data to Equation 2, identifying this parameter by visual inspection of Figure 3 is less intuitive. Kix-effector is reflected in the horizontal position of the transition between the upper and lower plateaus; however, this parameter is not the midpoint of the curve. Quantitative values for these three parameters (Ka-PEP, Kix-Effector, and Qax) are obtained when fitting this data to Equation 2 and are reported in Table 2. In a similar case in which saturation of substrate was unobtainable, Vmax/Km values were employed to fit the primary data [46]. For data in Figure 1, when the Vmax/Km region of each PEP response curve was fit according to the previous description [46], the Kix-Effector and Qax parameters were similar to those obtained by the fitting strategy outlined above (Table 2 and Supplemental Figure 10). Qax-FBP shows some variation depending on the fitting strategy. As can be seen in Figure 1, the Fru-1,6-BP dependent shift in K0.5-PEP is accompanied by a decrease in the cooperativity of PEP binding (a decrease in nH-PEP from near 2.6 to 1.5). The Vmax/Km fitting approach does not account for homotropic cooperativity. Therefore, the greater Qax-FBP determined by the Vmax/Km fitting strategy here (Table 2) is consistent with the increase in the observed allosteric response upon the loss of homotropic cooperativity previously detailed in phosphofructokinase from E. coli [46]. However, the change in homotropic cooperativity represented here is not as severe as that observed in phosphofructokinase, the apparent reason why the two Qax-FBP values reported in Table 2 remain in reasonable agreement with each other. The apparent overall robustness of Qax determination, independent of the fitting strategy used to analyze primary data is due to the fact that Qax is a ratio [18, 20] that can be estimated even in the absence of completely accurate evaluations of Kapp-PEP.
Figure 2.
Kapp-PEP as a function of Fru-1,6-BP (◆), ATP (●), or Ala (□) at pH 7.5 and 37°C. Kapp-PEP were obtained from fits to Equation 1. Lines represent the best fits of data as exemplified in Figure 1 to Equation 2. When error bars are not apparent, they are smaller than the data point symbols.
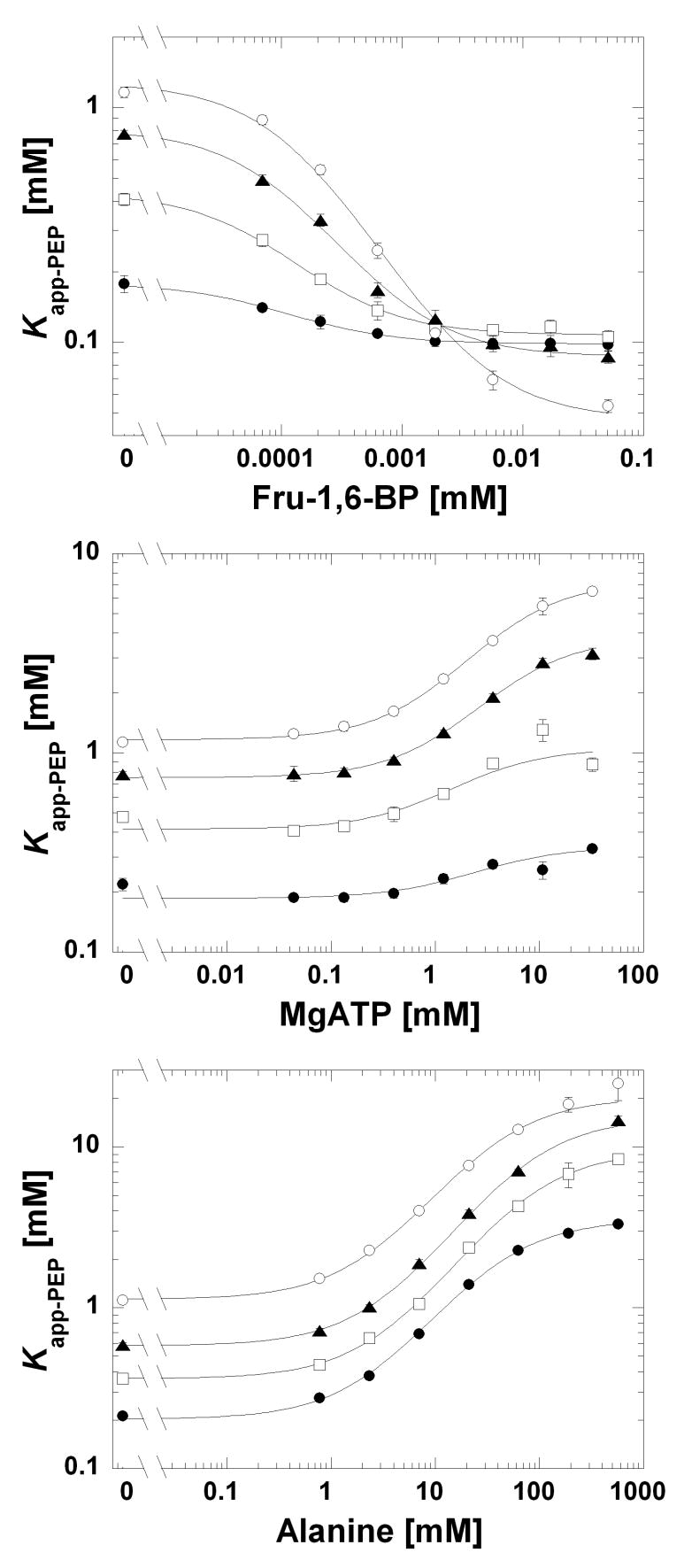
Figure 3.
Kapp-PEP as a function of Fru-1,6-BP, ATP, or Ala at and 37°C and pH 6.0 (●), 7.0 (□), 7.5 (▲), and 8.0 (○). The range of the y-axis is not consistent between graphs. Lines represent the best fits to Equation 2. When error bars are not apparent, they are smaller than the data point symbols.
Table 2.
Parameters from Linkage Analysis Obtained at pH 7.5, 37°C
| Fit strategy of PEP response | ||
|---|---|---|
| Parameters from Equation 2 | as in text (Figure 2) | Vmax/Km (Supplemental Figure 10) |
| Ka-PEP (mM) | 0.57±0.01 | NA |
| Kix-FBP (mM) | 0.00064±0.00005 | 0.00040±0.00006 |
| Kix-ATP (mM) | 0.81±0.06 | 1.5±0.3 |
| Kix-Ala (mM) | 2.3±0.2 | 2.5±0.3 |
| Qax-FBP (ΔGax-FBP (kcal/mol)) | 8.7±0.2 (-1.33±0.01) | 18±1 (-1.78±0.03) |
| Qax-ATP (ΔGax-ATP (kcal/mol)) | 0.16±0.01 (1.13±0.04) | 0.09±0.04 (1.5±0.3) |
| Qax-Ala (ΔGax-Ala (kcal/mol)) | 0.064±0.004 (1.69±0.04) | 0.033±0.003 (2.10±0.06) |
A comparison of fit parameters with those previously reported for purified hL-PYK [11, 47] can also be used to evaluate our fitting approach. Our Ka-PEP 0.57±0.01 mM (Table 2) agrees well with the previously reported Kapp-PEP value of 0.4 mM obtained in the absence of effectors. Likewise, the previously reported Kapp-PEP values of 1.2 mM and 1.05 mM in the presence of 3 mM Ala or 1 mM ATP, respectively, are in very good agreement with an estimate of the Kapp-PEP at the same effector concentrations derived from the data in Figure 2. The Kapp-PEP value of 0.04 mM in the presence of 0.5 mM Fru-1,6-BP is in reasonable agreement with the approximately 7.5 mM value estimated from Figure 2. The assumptions incorporated into fitting the biphasic PEP response are likely to impact the obtained nH-PEP values (especially as Kapp-PEP increases), therefore we have not made an extensive evaluation of this parameter. In the absence of effectors, an nH-PEP value of 2.6 was associated with the response of initial velocity to varying concentrations of PEP. This value is near the nH-PEP=1.5 to 2.5 range obtained from purified L-PYK isozymes from several mammalian species [2]. Overall, the parameters associated with the low PEP phase obtained here are in good agreement with previously obtained values for purified hL-PYK [11, 47]. Therefore, even if the high PEP phase is an artifact that cannot be attributed to unmodified protein, our fitting approach allows for an estimate of the Kapp-PEP associated with the low PEP phase that can, in turn, be used to evaluate allosteric properties of hL-PYK.
The pH Dependence Inhibition by Ala and ATP are not Coincidental
Although the binding site for amino acid effectors has been determined [6], the allosteric nucleotide binding site has not been determined for any PYK isozyme. Therefore, it is currently unknown if ATP and Ala bind to the same binding site and/or if these effectors elicit inhibition through a common allosteric mechanism. Unfortunately, the potential that the two inhibitors bind at distinct sites on a protein with mutual antagonism due to an allosteric mechanism is not easily distinguished from competitive binding of the two effectors to the same site on the protein. Therefore, competitive binding studies between ATP and Ala would not likely be instructive to understanding the mechanism of ATP binding and/or allosteric mechanism. As outlined in the introduction, differences in the magnitudes of Qax-ATP and Qax-Ala (Table 2) are consistent with the two inhibitors functioning through two different allosteric mechanisms. In other systems, modification of the allosteric effector influences the magnitude of the allosteric coupling (i.e. changing which set of linked-reactions are elicited) [6] and in one extreme example, an allosteric activator and an allosteric inhibitor bind competitively to the same allosteric site of bacterial phosphofructokinse [48]. Therefore, even though differences in the magnitudes of Qax-ATP and Qax-Ala support two different allosteric mechanisms, these differences cannot be extended to infer that the two inhibitors do not bind to the same site on the protein.
The pH-dependence of Qax-ATP and Qax-Ala were also determined to give additional insights into whether these two inhibitors elicit their responses through different allosteric mechanisms. In Figure 3, the Kapp-PEP vs. effector concentration was determined over a moderate pH range from pH 6.5 to 8.0. To appreciate the dependence of Qax on pH, these parameters are plotted as a function of pH in Figures 4 and 5. Recall that Qax=1 defines the absence of allosteric regulation. Therefore, over this pH range the smallest allosteric response to ATP occurs at pH 6.5. As pH increases, ATP elicits more inhibition (a decrease in Qax-ATP). In contrast, the Qax-Ala is relatively insensitive to changes in this pH range. Ka-PEP and Kix-effector values from the same data set are plotted as a function of pH in Figure 5. In contrast to Ka-PEP, Kix-ATP and Kix-Ala both decrease (increased affinity) with increasing pH.
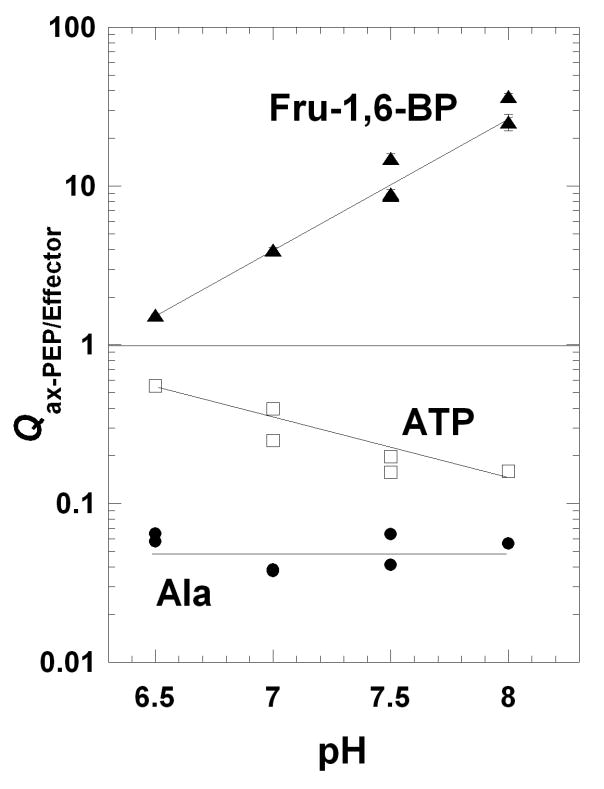
Figure 4.
Qax for the regulation by Fru-1,6-BP (▲), ATP (□), and Ala (●) as a function of pH. Qax values were obtained from fits of data as exemplified in Figure 3 to Equation 2. Lines represent data trends. When error bars are not apparent, they are smaller than the data point symbols.
Figure 5.
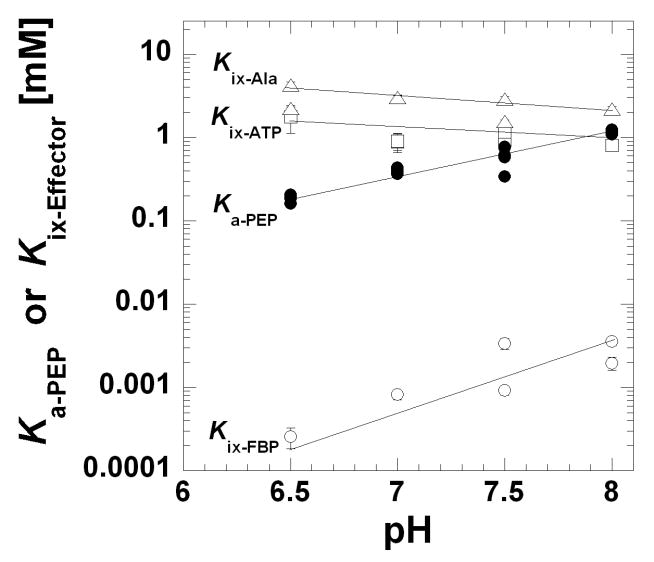
Ka-PEP (●), Kix-FBP (○), Kix-ATP (□), and Kix-Ala (△) as a function of pH. Affinity values were obtained from fits of the data in as exemplified in Figure 3 to Equation 2. Lines represent data trends. When error bars are not apparent, they are smaller than the data point symbols.
In general, the apparent influence of pH may be a result of any combination of multiple protonation events on the ligand and/or multiple protonation events on the protein. Ala has pKa values of 2.34 and 9.69 [49]. The pKa values of ATP in buffers with greater than 100 mM KCl are 3.97 and near 6 [50]. These pKa values for the two inhibitors are outside of the pH range considered in this study. Therefore, in as much as the relevant protonation events can be assigned to changes on the protein, a comparison of the regulations by ATP and Ala is possible. Primarily, if two effectors elicit their allosteric effects through different allosteric mechanisms, we would expect protonation of different parts of the protein would impact the two inhibitions and could result in different pH-dependences of the resulting allosteric couplings (the moieties in the protein that undergo protonation do not necessarily reside in the protein's binding site for the effector). Therefore, the fact Qax-ATP and Qax-Ala demonstrate different dependences on pH is consistent with two different allosteric mechanisms.
pH Dependence of Qax and Kix-effector are not Correlated
The pH-dependence of the allosteric regulation of hL-PYK can also be used to test if a change in solution conditions simultaneously increases effector affinity and the magnitude of the corresponding allosteric coupling, a potential general correlation to allosteric functions. Each of the parameters in Equation 2 can vary independently of the others [18]; however, we can envision a potential physiological benefit for a coordinated response between the two properties.
In consideration of the potential correlation between Kix and Qax for any one effector, the pH dependence of the Fru-1,6-BP activation of hL-PYK can be considered in addition to the pH dependence of the inhibition by Ala and ATP (Figures 4 and 5). Like inhibition by ATP, the smallest allosteric response to Fru-1,6-BP occurs at pH 6.5 (Figure 4) and increases with increasing pH (an increase in Qax-FBP). If the increased Fru-1,6-BP activation (increased Qax-FBP) of hL-PYK at high pH is interpreted as being physiologically beneficial, Fru-1,6-BP might also be expected to bind the enzyme more tightly at higher pH. However, in direct contrast to this expectation, Fru-1,6-BP affinity decreases (i.e. increased Kix-FBP) with increased pH (Figure 5).
More consistent with the expectation, the affinity of hL-PYK for ATP slightly increases (decreased Kix-ATP) as pH rises, corresponding with the increased ability of ATP to inhibit (decreased Qax-ATP). However, Qax-Ala is relatively insensitive to the pH range studied while the Kix-Ala decreases over the same range.
These results do not support a general correlation between changes in effector affinity and allosteric coupling. Our results have not ruled out that simultaneously increasing effector affinity and increasing the allosteric response might be important to the regulation of a protein by any one effector. A physiological relevance may also be rationalized in cases in which opposite responses in effector affinity vs. the magnitude of the allostery are elicited by pH changes; at a set concentration of effector, compensating changes between effector affinity and the allosteric response may result in an equivalent allosteric response over a broad pH range (i.e. an apparent pH insensitivity). Our results also do not rule out that some modulation other than changes in pH might elicit a coordinated effect on effector affinity and the magnitude of the allosteric response. However, upon considering the examples from the current study, it may not be reasonable to expect a general correlation between Qax and Kix-effector values.
Supplementary Material
Supporting Information Available: 1) Primers used in gene deletion from E. coli, 2) Enzymatic activity in knock-out cell strains, 3) Optimization of hL-PYK Expression, 4) Western blot for expression of homogeneous hL-PYK, 5) MALDI-TOF determination of protein mass, 6) Protein purification table, 7) SDS-PAGE of purification steps, 8) Effects of level of protein purification, 9) ADP response 10) PEP response curve at 5°C, 11) Data presentation on a linear x-axis, 12) Alternative evaluation of primary data.
Acknowledgments
The knockout E. coli strains were constructed in the lab of Dr. Gregory D. Reinhart with the advice of Dr. Michelle R. Lovingshimer and the aid of Lin Fan. We are grateful for their willingness to share facilities and expertise. The assistance of Rachel Williams, Luke Vierthaler, Doanh Tran and Mathews Athiyal in characterizing the knockout E. coli strains and the Mass Spectrometry/Proteomics Core at the University of Kansas Medical Center directed by Dr. Antonio Artigues in determining whole protein mass was appreciated. We would like to thank Drs. Andrea Mattevi and Giovanna Valentini for their willingness to share the pLC1 and pLC11 plasmids. Work in the laboratory of the author is supported by NIH grant DK78076.
Abbreviations
- PYK
pyruvate kinase
- L-PYK
PYK from mammalian liver
- hL-PYK
human liver pyruvate kinase
- R-PYK
PYK from mammalian erythrocyte
- M1-PYK
the PYK found in mammal brain and muscle
- EcPYK-A and EcPYK-F
the two E. coli pyruvate kinase isozymes
- PEP
phospho(enol)pyruvate
- DTT
dithiothreitol
- PMSF
phenylmethylsulfonyl fluoride
- FRT
FRT scare
- Fru-1,6-BP
FBP denotes fructose-1,6-bisphosphate in text and parameters, respectively
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Yamada K, Noguchi T. Biochem J. 1999;337(Pt 1):1–11. [PMC free article] [PubMed] [Google Scholar]
- 2.Blair JB. In: The Regulation of Carbohydrate Formation and Utilization in Mammals. Veneziale CM, editor. University Park Press; Baltimore: 1980. pp. 121–151. [Google Scholar]
- 3.Johnson ML, Veneziale CM. Biochemistry. 1980;19:2191–2195. doi: 10.1021/bi00551a030. [DOI] [PubMed] [Google Scholar]
- 4.Tanaka T, Harano Y, Sue F, Morimura H. J Biochem (Tokyo) 1967;62:71–91. doi: 10.1093/oxfordjournals.jbchem.a128639. [DOI] [PubMed] [Google Scholar]
- 5.Noguchi T, Yamada K, Inoue H, Matsuda T, Tanaka T. J Biol Chem. 1987;262:14366–14371. [PubMed] [Google Scholar]
- 6.Williams R, Holyoak T, McDonald G, Gui C, Fenton AW. Biochemistry. 2006;45:5421–5429. doi: 10.1021/bi0524262. [DOI] [PubMed] [Google Scholar]
- 7.Dombrauckas JD, Santarsiero BD, Mesecar AD. Biochemistry. 2005;44:9417–9429. doi: 10.1021/bi0474923. [DOI] [PubMed] [Google Scholar]
- 8.Mesecar AD, Nowak T. Biochemistry. 1997;36:6803–6813. doi: 10.1021/bi962870s. [DOI] [PubMed] [Google Scholar]
- 9.Mattevi A, Valentini G, Rizzi M, Speranza ML, Bolognesi M, Coda A. Structure. 1995;3:729–741. doi: 10.1016/s0969-2126(01)00207-6. [DOI] [PubMed] [Google Scholar]
- 10.Munoz ME, Ponce E. Comp Biochem Physiol B Biochem Mol Biol. 2003;135:197–218. doi: 10.1016/s1096-4959(03)00081-2. [DOI] [PubMed] [Google Scholar]
- 11.Kahn A, Marie J. Methods in Enzymology. 1982;90:131–140. doi: 10.1016/s0076-6879(82)90119-7. [DOI] [PubMed] [Google Scholar]
- 12.Marie J, Kahn A, Boivin P. Biochim Biophys Acta. 1977;481:96–104. doi: 10.1016/0005-2744(77)90141-3. [DOI] [PubMed] [Google Scholar]
- 13.Marie J, Kahn A. Enzyme. 1977;22:407–411. doi: 10.1159/000458826. [DOI] [PubMed] [Google Scholar]
- 14.Marie J, Kahn A, Boivin P. Biochim Biophys Acta. 1976;438:393–406. doi: 10.1016/0005-2744(76)90256-4. [DOI] [PubMed] [Google Scholar]
- 15.Marie J, Kahn A, Boivin P. Hum Genet. 1976;31:35–45. doi: 10.1007/BF00270397. [DOI] [PubMed] [Google Scholar]
- 16.Sprengers ED, Staal GE. FEBS Lett. 1979;101:287–288. doi: 10.1016/0014-5793(79)81027-3. [DOI] [PubMed] [Google Scholar]
- 17.Hall ER, Cottam GL. Int J Biochem. 1978;9:785–793. doi: 10.1016/0020-711x(78)90027-7. [DOI] [PubMed] [Google Scholar]
- 18.Fenton AW. Trends Biochem Sci. 2008;33:420–425. doi: 10.1016/j.tibs.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weber G. Biochemistry. 1972;11:864–878. doi: 10.1021/bi00755a028. [DOI] [PubMed] [Google Scholar]
- 20.Reinhart GD. Methods Enzymol. 2004;380:187–203. doi: 10.1016/S0076-6879(04)80009-0. [DOI] [PubMed] [Google Scholar]
- 21.Jurica MS, Mesecar A, Heath PJ, Shi W, Nowak T, Stoddard BL. Structure. 1998;6:195–210. doi: 10.1016/s0969-2126(98)00021-5. [DOI] [PubMed] [Google Scholar]
- 22.Valentini G, Chiarelli LR, Fortin R, Dolzan M, Galizzi A, Abraham DJ, Wang C, Bianchi P, Zanella A, Mattevi A. J Biol Chem. 2002;277:23807–23814. doi: 10.1074/jbc.M202107200. [DOI] [PubMed] [Google Scholar]
- 23.Lovingshimer MR, Siegele D, Reinhart GD. Protein Expr Purif. 2006;46:475–482. doi: 10.1016/j.pep.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 24.Valentini G, Iadarola P, Somani BL, Malcovati M. Biochim Biophys Acta. 1979;570:248–258. doi: 10.1016/0005-2744(79)90145-1. [DOI] [PubMed] [Google Scholar]
- 25.Malcovati M, Valentini G, Kornberg HL. Acta Vitaminol Enzymol. 1973;27:96–111. [PubMed] [Google Scholar]
- 26.Ponce E, Flores N, Martinez A, Valle F, Bolivar F. J Bacteriol. 1995;177:5719–5722. doi: 10.1128/jb.177.19.5719-5722.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gosset G, Yong-Xiao J, Berry A. J Ind Microbiol. 1996;17:47–52. doi: 10.1007/BF01570148. [DOI] [PubMed] [Google Scholar]
- 28.Garrido-Pertierra A, Cooper RA. FEBS Lett. 1983;162:420–422. doi: 10.1016/0014-5793(83)80799-6. [DOI] [PubMed] [Google Scholar]
- 29.Pertierra AG, Cooper RA. J Bacteriol. 1977;129:1208–1214. doi: 10.1128/jb.129.3.1208-1214.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Siddiquee KA, Arauzo-Bravo MJ, Shimizu K. FEMS Microbiol Lett. 2004;235:25–33. doi: 10.1016/j.femsle.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 31.Al Zaid Siddiquee K, Arauzo-Bravo MJ, Shimizu K. Appl Microbiol Biotechnol. 2004;63:407–417. doi: 10.1007/s00253-003-1357-9. [DOI] [PubMed] [Google Scholar]
- 32.Yu D, Ellis HM, Lee EC, Jenkins NA, Copeland NG, Court DL. Proc Natl Acad Sci U S A. 2000;97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Datsenko KA, Wanner BL. Proc Natl Acad Sci U S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Emmerling M, Dauner M, Ponti A, Fiaux J, Hochuli M, Szyperski T, Wuthrich K, Bailey JE, Sauer U. J Bacteriol. 2002;184:152–164. doi: 10.1128/JB.184.1.152-164.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang C, Chiarelli LR, Bianchi P, Abraham DJ, Galizzi A, Mattevi A, Zanella A, Valentini G. Blood. 2001;98:3113–3120. doi: 10.1182/blood.v98.10.3113. [DOI] [PubMed] [Google Scholar]
- 36.Loog M, Oskolkov N, O'Farrell F, Ek P, Jarv J. Biochim Biophys Acta. 2005;1747:261–266. doi: 10.1016/j.bbapap.2004.11.016. [DOI] [PubMed] [Google Scholar]
- 37.Pace CN, Vajdos F, Fee L, Grimsley G, Gray T. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson JL, Reinhart GD. Biochemistry. 1994;33:2635–2643. doi: 10.1021/bi00175a036. [DOI] [PubMed] [Google Scholar]
- 39.Inoue H, Noguchi T, Tanaka T. Eur J Biochem. 1986;154:465–469. doi: 10.1111/j.1432-1033.1986.tb09420.x. [DOI] [PubMed] [Google Scholar]
- 40.Humble E. Biochim Biophys Acta. 1980;626:179–187. doi: 10.1016/0005-2795(80)90209-3. [DOI] [PubMed] [Google Scholar]
- 41.Pilkis SJ, El-Maghrabi MR, Coven B, Claus TH, Tager HS, Steiner DF, Keim PS, Heinrikson RL. J Biol Chem. 1980;255:2770–2775. [PubMed] [Google Scholar]
- 42.Hjelmquist G, Andersson J, Edlund B, Engstroom L. Biochem Biophys Res Commun. 1974;61:559–563. doi: 10.1016/0006-291x(74)90993-0. [DOI] [PubMed] [Google Scholar]
- 43.Edlund B, Andersson J, Titanji V, Dahlqvist U, Ekman P, Zetterqvist O, Engstoom L. Biochem Biophys Res Commun. 1975;67:1516–1521. doi: 10.1016/0006-291x(75)90198-9. [DOI] [PubMed] [Google Scholar]
- 44.Flory W, Peczon BD, Koeppe RE, Spivey HO. Biochem J. 1974;141:127–131. doi: 10.1042/bj1410127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reinhart GD. Arch Biochem Biophys. 1983;224:389–401. doi: 10.1016/0003-9861(83)90225-4. [DOI] [PubMed] [Google Scholar]
- 46.Fenton AW, Reinhart GD. Biochemistry. 2002;41:13410–13416. doi: 10.1021/bi026450g. [DOI] [PubMed] [Google Scholar]
- 47.Marie J, Buc H, Simon MP, Kahn A. Eur J Biochem. 1980;108:251–260. doi: 10.1111/j.1432-1033.1980.tb04718.x. [DOI] [PubMed] [Google Scholar]
- 48.Evans PR, Hudson PJ. Nature. 1979;279:500–504. doi: 10.1038/279500a0. [DOI] [PubMed] [Google Scholar]
- 49.Lide DR, editor. CRC Handbook of Chemistry and Physics. CRC Press, Inc.; Ann Arbor: 1992. [Google Scholar]
- 50.De Stefano C, Milea D, Pettignano A, Sammartano S. Biophys Chem. 2006;121:121–130. doi: 10.1016/j.bpc.2005.12.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information Available: 1) Primers used in gene deletion from E. coli, 2) Enzymatic activity in knock-out cell strains, 3) Optimization of hL-PYK Expression, 4) Western blot for expression of homogeneous hL-PYK, 5) MALDI-TOF determination of protein mass, 6) Protein purification table, 7) SDS-PAGE of purification steps, 8) Effects of level of protein purification, 9) ADP response 10) PEP response curve at 5°C, 11) Data presentation on a linear x-axis, 12) Alternative evaluation of primary data.