Abstract
Hyaluronan (HA), a large glycosaminoglycan abundant in the extracellular matrix, is important in cell migration during embryonic development, cellular proliferation, and differentiation and has a structural role in connective tissues. The turnover of HA requires endoglycosidic breakdown by lysosomal hyaluronidase, and a congenital deficiency of hyaluronidase has been thought to be incompatible with life. However, a patient with a deficiency of serum hyaluronidase, now designated as mucopolysaccharidosis IX, was recently described. This patient had a surprisingly mild clinical phenotype, including notable periarticular soft tissue masses, mild short stature, an absence of neurological or visceral involvement, and histological and ultrastructural evidence of a lysosomal storage disease. To determine the molecular basis of mucopolysaccharidosis IX, we analyzed two candidate genes tandemly distributed on human chromosome 3p21.3 and encoding proteins with homology to a sperm protein with hyaluronidase activity. These genes, HYAL1 and HYAL2, encode two distinct lysosomal hyaluronidases with different substrate specificities. We identified two mutations in the HYAL1 alleles of the patient, a 1412G → A mutation that introduces a nonconservative amino acid substitution (Glu268Lys) in a putative active site residue and a complex intragenic rearrangement, 1361del37ins14, that results in a premature termination codon. We further show that these two hyaluronidase genes, as well as a third recently discovered adjacent hyaluronidase gene, HYAL3, have markedly different tissue expression patterns, consistent with differing roles in HA metabolism. These data provide an explanation for the unexpectedly mild phenotype in mucopolysaccharidosis IX and predict the existence of other hyaluronidase deficiency disorders.
Hyaluronidase (reviewed in refs. 1 and 2) is required for the breakdown of hyaluronan (HA), a high molecular mass glycosaminoglycan that is abundant in the extracellular matrix of connective tissues. HA is recognized as an important structural and functional component of the extracellular matrix. Changes in its turnover are associated with many processes involving cell proliferation, migration, and differentiation—including embryogenesis, inflammation, wound healing, and metastasis (reviewed in refs. 3 and 4). Because hyaluronidase is involved in such key physiologic processes, a deficiency of hyaluronidase has been thought to be incompatible with life (5).
In 1996, a deficiency of serum hyaluronidase activity was described in a patient with a surprisingly mild clinical phenotype (6); this lysosomal storage disorder has now been designated mucopolysaccharidosis IX (Online Mendelian Inheritance in Man, The Johns Hopkins University, Baltimore; http://www.ncbi.nlm.nih.gov/omim/). This patient had periarticular soft tissue masses, mild short stature and acetabular erosions, and an absence of neurological or visceral involvement. Histological examination of affected tissues revealed macrophages with HA-filled lysosomes and lesser accumulations of HA in the lysosomes of fibroblasts. Serum hyaluronidase activity was noted to be deficient, and the concentration of HA in the serum was elevated 38- 90-fold over normal (6).
Historically, two forms of hyaluronidase, one with a neutral pH optimum and one with an acid-pH optimum, were described (7). A sperm-specific protein, PH-20, was identified as the hyaluronidase with a neutral pH optimum (8). Acid-active hyaluronidase has been detected in many tissues and, until recently, was believed to represent a single lysosomal enzyme. It has become apparent through the study described herein, and others, that multiple hyaluronidase genes exist, although their protein products have not been fully characterized.
Human plasma hyaluronidase was recently purified, and its cDNA was cloned (9). The gene encoding this enzyme, now termed HYAL1, maps to human chromosome 3p21.3 (10) and had previously been isolated as a candidate gene LuCa1 in the search for a lung cancer tumor suppressor in the 3p21 region (11). The product of the HYAL1 gene was found to have an an acidic pH optimum and had properties that suggested it was membrane associated (9, 12).
A second hyaluronidase gene, HYAL2, was identified based on homology to the previously described PH-20 protein (13). Like HYAL1, this gene had previously been identified as a candidate lung cancer tumor suppressor LuCa2 in the 3p21.3 region (11). HYAL2 was found to localize to the lysosome and to have an acidic pH optimum and, in contrast to HYAL1, was only able to degrade high molecular mass HA to intermediately sized products of ≈20 kDa (13).
Two additional hyaluronidase genes have been defined. One of these, a third gene in the 3p21.3 region, LuCa3, also was defined on the basis of homology to PH-20 and is designated HYAL3; its protein product is uncharacterized. Another gene, MGEA5, recently cloned and mapped to human chromosome 10q24.1-q24.3, was noted to have hyaluronidase activity but is otherwise not well characterized (14).
In a search for the gene causing mucopolysaccharidosis IX in our patient, we evaluated two candidate genes with homology to the sperm protein PH-20, HYAL1 and HYAL2. Two HYAL1 mutations, a nonconservative amino acid substitution on one allele and a complex intragenic rearrangement on the other allele, were identified in the patient and were shown to be inherited in an autosomal recessive fashion. Furthermore, analysis of the expression pattern of HYAL1, HYAL2, and HYAL3 in human tissues revealed markedly different tissue distributions of the three hyaluronidase gene products. The expression patterns of these genes and the tissue distribution of HA is consistent with the relatively mild phenotype of our hyaluronidase deficient patient. This explains the nonlethality of “hyaluronidase deficiency” and predicts the existence of other hyaluronidase deficient conditions.
MATERIALS AND METHODS
Substrate Gel Electrophoresis (Zymography).
Serum samples (2 μl) were diluted in Laemmli’s sample buffer (15) without reducing agent or SDS present. Sample electrophoresis and hyaluronidase activity detection were based on the method of Miura et al. (16). In brief, samples were separated in a 7.5% native polyacrylamide gel containing 170 μg/ml HA (Sigma human umbilical cord). The gels were incubated in sodium formate buffer (pH 3.5) for 20 hours at 37°C and then were incubated for an additional 20 hours at 37°C in 20 mM Tris⋅HCl (pH 8) (with or without 200 μg/ml pronase and 100 μg/ml trypsin) before staining with Alcian blue. Hyaluronidase activity was indicated by a band or zone of absent staining. A small amount of residual hyaluronidase activity appeared to be present in the proband if the gels were not protease-treated before staining because of interference by abundant proteins with the staining of HA.
Sequence Analysis.
Homology searches using the PH-20 protein sequence, the HYAL1 cDNA sequence, or the HYAL2 cDNA sequence against nonredundant and expressed sequence tag (EST) databases were performed by using blast (17). Multiple alignments of the hyaluronidase protein sequences were done with clustal w 1.74 (18).
Mutation Detection/Confirmation.
Genomic DNA was prepared from fibroblasts and leukocytes of the patient and her extended family as described (19). The PCR products of the HYAL1 and HYAL2 genes were generated and analyzed by single strand conformational polymorphism analysis at room temperature and at 4°C as described (20). The sense and antisense primers used for amplification of fragments were, for A, 5′-GTCTGAGAGGCAACTCGGATGTG-3′ and 5′-ATCAGGCTGGCATTCTGGGGCAG-3′ (62°C annealing); for B, 5′-GGAGCCTGTGTTTGGTGGTCTGC-3′ and 5′-GCTGGTCATTTTGGGCACGGCTG-3′ (60°C annealing); for C, 5′-CCCTGACTGCTACAACTATGACTT-3′ and 5′-AAAGCTAAAGTACCCCAAGGCTGG-3′ (60°C annealing); for D, 5′-GTCCCATGGCCAGAGCAGCCCCA-3′ and 5′-ACCTGCTGGTCAGCCAGGACTTT-3′ (64°C annealing); for E, 5′-AAAGTCCTGGCTGACCAGCAGGT-3′ and 5′-AGGTTCTCAATATGTGCAACTCAG-3′ (62°C annealing); for F, 5′-ATGGGCTTTGGGAGCATAGATG-3′ and 5′-CACAGACCTTCCGGCAGAATC-3′ (58°C annealing); for G, 5′-ACCGCGACCGTCTAGGCCTGTAT-3′ and 5′-AGAGGTAGAAGCCCCAGAGGTGC-3′ (62°C annealing); for H, 5′-ACCGGGACCGTCTAGGCCTGTAT-3′ and 5′-TAACCCAAATGTGCAGTGGATC-3′ (62°C annealing); for I, 5′-GGTTGAGCTGGGAGTTCAGCAGG-3′ and 5′-ACCTGAGGTTGGTAGCCAAAGGC-3′ (58°C annealing); for J, 5′-CTGTGGGCTGAGGCAGCTGACC-3′ and 5′-AGGCCAGCTTGCTAGGCAGCTA-3′ (62°C annealing). Fragment C was subcloned into pCRII by using a TA Cloning Kit (Invitrogen), and clones were sequenced by using a T7 Sequencing Kit (Amersham Pharmacia). For confirmation of the mutations, 15-μl aliquots of the fragment C PCR products were incubated with 1 μl of MscI at 37°C for 2 hours and then were separated by electrophoresis on a 10% polyacrylamide gel.
Northern Blot Analysis.
The adult human multiple-tissue Northern blot containing 2 μg of polyA+-RNA from each of 12 tissues was purchased from CLONTECH. The hybridizations were carried out in Express Hyb solution (CLONTECH) according to the manufacturer’s instructions. The HYAL1 cDNA probe was prepared from the clone 650884 insert by digestion with PvuII to generate a 340-bp fragment. The HYAL2 cDNA probe was prepared from the clone 428219 insert by digestion with CfoI/BsrFI to generate a 508-bp fragment, and the HYAL3 cDNA probe was prepared from clone 547239 by digestion with StuI to generate a 312-bp fragment. The fragments were gel purified by using a Geneclean II Kit (BIO/CAN, Montreal), and the β-actin cDNA probe was supplied with the CLONTECH blot. All DNA fragments were random primer-labeled with α32P-dATP to generate a probe with a specific activity of at least 1 × 109 cpm/μg. The blot was hybridized sequentially with the following probes: HYAL3 (two times), HYAL1, HYAL2, and β-actin. With the exception of the initial HYAL3-probed blot, which was exposed to XOMAT-AR film (Kodak), the hybridized blots were exposed to BioMax AS (Kodak) film with two intensifying screens at −80°C.
GenBank Accession Numbers.
GenBank accession numbers are as follows: human HYAL1 cDNA, U96078, gi2314820; human HYAL2 cDNA, AJ000099, gi2370092; LUCA13, AC002455; LUCA14, U731667; human HYAL3 or LuCa-3 cDNA, AF040710, gi2935328; bee venom hyaluronidase gi155680; Caenorhabditis elegans hyaluronidase, gi3879998; clone 650884, AA223264; clone 428219, AA001817; and clone 547239, AA085250.
RESULTS AND DISCUSSION
Confirmation of Serum Hyaluronidase Deficiency.
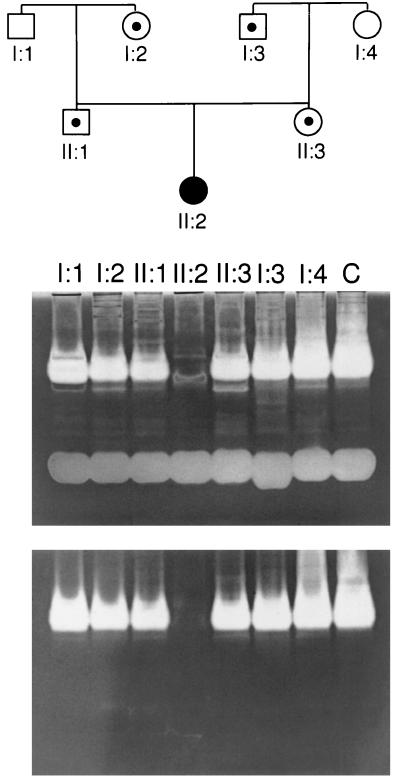
To confirm the complete absence of serum hyaluronidase activity in the patient described by Natowicz et al. (6), we used a substrate-gel based assay (16). This assay is at least 10-fold more sensitive than the aqueous assay (21) previously used to characterize the patient’s deficiency and allows inhibitors of the enzyme to be separated from the enzyme before the assay. The disadvantage of the gel-based assay is that abundant proteins can interfere with the staining of HA, appearing as an unstained region representing hyaluronidase activity. Such interference was observed in the proband sample (II:2) in the upper panel of Fig. 1. This nonspecific banding can be readily eliminated by treating the gel with proteases after the activity assay but before staining for HA. As shown in the bottom panel of Fig. 1, under these conditions, there is no detectable hyaluronidase activity in the serum of the proband. The heterozygous parents and grandparents were defined in the pedigree in Fig. 1 on the basis of the original aqueous assay (6). These individuals could be differentiated from normal controls by using the gel-based assay when less serum (1 μl) was loaded on the gel (data not shown).
Figure 1.
Serum hyaluronidase activity. Serum hyaluronidase degraded the HA that had been incorporated in the gels. The gels were counterstained with Alcian blue before (Upper) or after (Lower) pretreatment with proteases. The regions in which the HA is degraded appear as clear bands against a blue background (white against gray in the photo). Lanes were overloaded with sample (2 μl serum) to demonstrate the complete deficiency of hyaluronidase activity in the proband in this assay system. Use of 0.2–1 μl of serum provides optimal discrimination of heterozygotes. Lanes are labeled with the numbers that correspond to the pedigree above.
Identification of Candidate Genes.
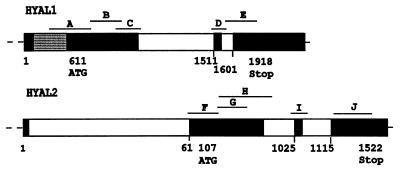
To identify the gene encoding the hyaluronidase that is deficient in our patient, we searched the EST and nonredundant databases for sequences with homology to PH-20, a hyaluronidase with a neutral pH optimum (8). Two cDNAs, LuCa-1 and LuCa-2 (now designated HYAL1 and HYAL2, respectively), as well as many ESTs representing these two cDNAs, were identified. Subsequent blast searches with the HYAL1 and HYAL2 cDNAs identified the sequences of two overlapping cosmids that form part of a contig spanning a region on human chromosome 3p21.3 that contains a tumor suppressor for small cell lung cancer (11). By comparing the sequences of the HYAL1 and HYAL2 cDNAs to the sequences of the cosmids LUCA13 and LUCA14, the genes were found to be separated by ≈14 kilobases, and the protein coding sequence of each gene was restricted to three exons (Fig. 2). There was evidence for alternative splicing for HYAL1 and an upstream exon for HYAL2 (Fig. 2). Csoka et al. (10) localized HYAL1 to 3p21.2-p21.3 by using other methods. The broad distribution of the ESTs representing both genes suggested that they could both be candidates for the deficiency of the lysosomal hyaluronidase in our patient.
Figure 2.
Partial HYAL1 and HYAL2 gene structures and mutation screening strategy. Exons are indicated by black boxes, introns are indicated by white boxes, and alternatively spliced exons are indicated by hatched boxes. Nucleotide numbering below the genes is based on the cDNA sequences. Fragments that were PCR-amplified for mutation analysis are labeled A through J and represent all of the transcribed regions of the gene that are protein encoding.
Identification of a Mutation in the HYAL1 Gene.
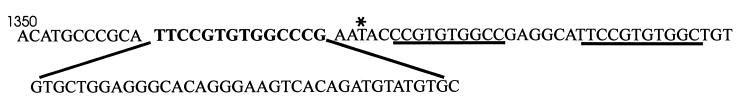
The coding regions of the patient’s HYAL1 and HYAL2 genes were PCR-amplified as 10 overlapping fragments (Fig. 2) and were examined for mutations by using single strand conformational polymorphism analysis. A normal and a deleted PCR product, as well as multiple single strand conformational polymorphism shifts, were observed for fragment C of the HYAL1 gene; no shifts were detected in other fragments (data not shown). Fragment C was PCR-amplified from patient DNA and was cloned, and five clones were fully sequenced. Three of the clones contained both a 37-bp deletion starting at nucleotide 1,361 and a 14-bp insertion of 5′-TTCCGTGTGGCCCG-3′ (1361del37ins14); two of the clones contained a 1412G → A mutation that results in the substitution of Lys for Glu at position 268. The 14-bp sequence that is inserted in one allele is very similar to two different sequences, each downstream of the deletion (Fig. 3), and suggests that the deletion/insertion mutation may have been generated by some form of slippage and misalignment (22).
Figure 3.
Nucleotide sequence around and including the 1361del37ins14 mutation. The number of the first nucleotide is given, and the inserted 14 nucleotides are in bold. The deleted 37 nucleotides are shown below. As asterisk indicates the 1400C → T mutation that is also present on the 1361del37ins14 allele. The underlined sequences are homologous to parts of the 14-nt insertion.
Both the 1361del37ins14 and 1412G → A mutations are expected to destroy the activity of HYAL1. The 1361del37ins14 mutation results in a frameshift beginning at amino acid 250 and terminating 19 amino acids downstream, 185 amino acids premature of the termination codon. The three clones that contained the del/ins mutation also had a 1400C → T substitution that would change His264 to Tyr if it were translated in the normal reading frame. The second mutation substitutes the negatively charged Glu268 with the a positively charged Lys. Clustal W multiple alignments revealed that Glu268 is conserved in all of the related hyaluronidases, including bee venom and C. elegans hyaluronidases (Fig. 4). Further, when the equivalent residue was mutated to a Gln in either human sperm (PH-20) or bee venom hyaluronidase, it completely disrupted enzyme activity (23). Glycosyl hydrolases typically have two critical acidic residues in the active site (24), and Arming et al. (23) predict that Glu268 is one of them. Both mutations would prevent the correct translation of the critical Glu268 in the HYAL1 protein product.
Figure 4.
Alignment of hyaluronidase protein sequences in the region of the Glu268Lys mutation. Related human hyaluronidases, Hyal 1, Hyal 2, Hyal 3, and PH-20, as well as bee venom and C. elegans hyaluronidase sequences, were aligned by using clustal w 1.74. A small portion of the alignment, including the E268 residue that is mutated in the Hyal 1-deficient patient, is shown. The asterisks indicate residues that are identical in all of the proteins. The numbers to the right of the sequences indicate the amino acid number of the protein sequences used in the alignment.
Confirmation of the HYAL1 Mutations.
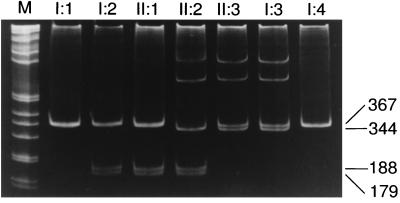
PCR-based methods were used to confirm the presence of both mutations in the genomic DNA of the proband and to examine the segregation of the mutations in the family. The smaller PCR product (344 bp) and the heteroduplexes in the proband resulted from the presence of the 1361del37ins14 mutation (Fig. 5). A similar pattern in the mother and maternal grandfather indicates that this mutation was inherited from the maternal side of the family. The 1412G → A mutation, which introduces a MscI site, was inherited from the father (Fig. 5). The segregation of the mutations was consistent with results of the enzyme-based analysis (6). Furthermore, neither mutation was detected in 100 randomly selected alleles that were analyzed by using the same PCR-based strategies (data not shown).
Figure 5.
Confirmation and segregation of HYAL1 mutations. Fragment C (Fig. 2) was PCR-amplified from genomic DNA, was digested with MscI, and was separated by PAGE. The 1412G → A mutation creates an MscI site that generates 188- and 179-bp fragments from the 367-bp PCR product, as seen in the patient (II:2), her father (II:1), and her paternal grandmother (I:2). The two slower-migrating bands in lanes II:2, II:3, and I:3 are heteroduplexes formed between the normal (367 bp) and 1361del37ins14 (344 bp) PCR products. The sizes of the bands are shown to the right of the gel. The numbering refers to the pedigree in Fig. 1.
Identification of HYAL3.
Recently, two ESTs corresponding to a third PH-20 related hyaluronidase, HYAL3, were identified in a blast search of the EST database. The corresponding gene had been previously identified as LuCa3 in the search for a lung cancer tumor suppressor on chromosome 3p21.3. The gene structure of HYAL3 is similar to that of the other two genes, having its coding sequence restricted to three exons (data not shown). The order of the genes is HYAL2-HYAL1-HYAL3 (data not shown). Only two ESTs from HYAL3 have been isolated, one from a colon library and one from a neuronal library, suggesting that the HYAL3 gene is weakly expressed. The identity between the protein sequences of HYAL1, HYAL2, and HYAL3 is very similar, ≈37%.
Tissue Expression Profiles of HYAL1, HYAL2, and HYAL3.
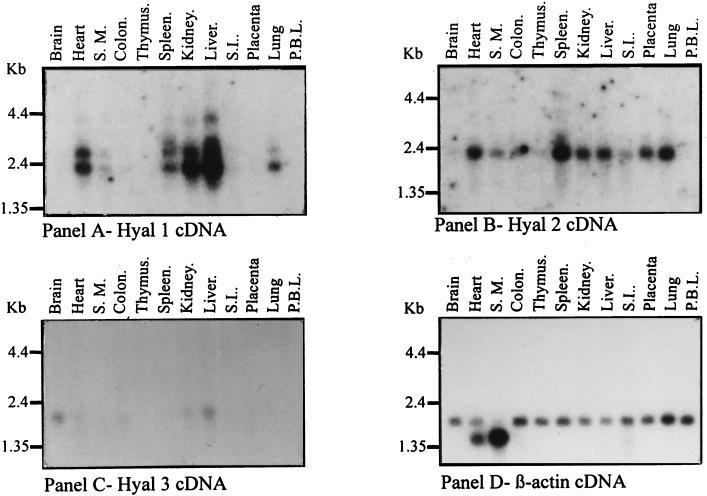
In contrast to most lysosomal storage disorders, our patient has a mild clinical phenotype in which the pathology is limited to specific cell types despite the broad distribution of HA in the body. This suggested to us that there may be more than one lysosomal hyaluronidase. The biochemical characteristics of human plasma hyaluronidase (9, 21) and the histological findings in our patient (6), together with the data presented, indicate that HYAL1 encodes a lysosomal enzyme. The studies of Lepperdinger et al. (14) show that HYAL2 also encodes a lysosomal enzyme whereas the subcellular localization of HYAL3 has not yet been examined. Because these enzymes may have differing roles in HA turnover, we examined the expression profile of HYAL1, HYAL2, and HYAL3 genes in human tissues (Fig. 6). Striking differences were noted. HYAL2 is the most broadly expressed gene; HYAL1 expression, although limited, is particularly high in the liver, a major site of HA degradation. The level of HYAL3 expression is very low and limited to a few tissues. Of interest, HYAL3 expression is highest in the brain, where neither HYAL1 nor HYAL2 is expressed. The restriction of HYAL1 expression to a few tissues and the notable absence of HYAL1 expression in the brain are consistent with the mild phenotype of our patient, including the absence of neurological abnormalities that characterize most other lysosomal enzyme deficiencies. The fact that HA is so abundant in synovial fluid and skeletal tissues is also consistent with the phenotype of our patient.
Figure 6.
Expression of HYAL1, HYAL2, and HYAL3 mRNA in adult human tissues. The probe used is shown below each panel. The exposure time was 48 hours for A, 89 hours for B, 48 hours for C, and 3 hours for D. S.M., skeletal muscle; S.I., small intestine; P.B.L., peripheral blood leukocytes.
Northern blot analysis revealed up to four HYAL1 mRNA species in human tissues. The precise identity of these mRNA species is unknown. The identification of an alternatively spliced exon by comparison of ESTs suggests that the two smaller species (2.4 and 2.7 kilobases) may be accounted for by alternative splicing.
The lysosomal hyaluronidase genes comprise a unique, tandemly distributed, multigene family, unlike previously reported lysosomal genes or gene families. It had previously been thought that lysosomal hyaluronidase was a single enzyme and that a genetic deficiency had not been identified because its importance in embryonic morphogenesis made its deficiency lethal in early development (5). The presence of at least three presumed lysosomal hyaluronidase genes, with distinct patterns of expression and different substrate specificities, reveals heretofore unexpected complexities in HA catabolism and suggests the possibility of additional and as yet undescribed phenotypes caused by hyaluronidase deficiencies.
Acknowledgments
This article is dedicated to the memory of a dear friend and colleague, Dr. Phyllis J. McAlpine. This work was supported by Medical Research Council Grant MT-11708 to B.T.-R.
ABBREVIATIONS
- HA
hyaluronan
- EST
expressed sequence tag
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Kreil G. Protein Sci. 1995;4:1666–1669. doi: 10.1002/pro.5560040902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frost G I, Csoka T, Stern R. Trends Glycosci Glycobiol. 1996;8:419–434. [Google Scholar]
- 3.Fraser J R E, Laurent T C. J Intern Med. 1997;242:23–27. doi: 10.1046/j.1365-2796.1997.00170.x. [DOI] [PubMed] [Google Scholar]
- 4.Toole B P. J Intern Med. 1997;242:35–40. doi: 10.1046/j.1365-2796.1997.00171.x. [DOI] [PubMed] [Google Scholar]
- 5.Roden L, Campbell P, Fraser J R E, Laurent T C, Pertoft H, Thompson J N. Ciba Found Symp. 1989;143:60–86. doi: 10.1002/9780470513774.ch5. [DOI] [PubMed] [Google Scholar]
- 6.Natowicz M R, Short M P, Wang Y, Dickersin G R, Gebhardt M C, Rosenthal D I, Sims K B, Rosenberg A E. N Engl J Med. 1996;335:1029–1033. doi: 10.1056/NEJM199610033351405. [DOI] [PubMed] [Google Scholar]
- 7.Bowness J M, Tan Y H. Biochim Biophys Acta. 1968;151:288–290. [PubMed] [Google Scholar]
- 8.Gmachl M, Sagan S, Ketter S, Kreil G. FEBS Lett. 1993;336:545–548. doi: 10.1016/0014-5793(93)80873-s. [DOI] [PubMed] [Google Scholar]
- 9.Frost G I, Csoka T B, Wong T, Stern R. Biochem Biophys Res Commun. 1997;236:10–15. doi: 10.1006/bbrc.1997.6773. [DOI] [PubMed] [Google Scholar]
- 10.Csoka T, Frost G I, Heng H H, Scherer S W, Mohapatra G, Stern R. Genomics. 1998;48:63–70. doi: 10.1006/geno.1997.5158. [DOI] [PubMed] [Google Scholar]
- 11.Wei M, Latif F, Bader S, Kashuba V, Chen J Y, Duh F-M, Sekido Y, Lee C-C, Geil L, Kuzmin I, et al. Cancer Res. 1996;56:1487–1492. [PubMed] [Google Scholar]
- 12.Csoka T, Frost G I, Wong T, Stern R. FEBS Lett. 1997;417:307–310. doi: 10.1016/s0014-5793(97)01309-4. [DOI] [PubMed] [Google Scholar]
- 13.Lepperdinger G, Strobl B, Kreil G. J Biol Chem. 1998;273:22466–22470. doi: 10.1074/jbc.273.35.22466. [DOI] [PubMed] [Google Scholar]
- 14.Heckel D, Comtesse N, Brass N, Blin N, Zang K D, Meese E. Hum Mol Genet. 1998;7:1859–1872. doi: 10.1093/hmg/7.12.1859. [DOI] [PubMed] [Google Scholar]
- 15.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 16.Miura R O, Yamagata S, Miura Y, Harada T, Yamagata T. Anal Biochem. 1995;225:333–340. doi: 10.1006/abio.1995.1163. [DOI] [PubMed] [Google Scholar]
- 17.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 18.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoar D I, Haslam D B, Starozik D M. Prenatal Diagn. 1984;4:241–247. doi: 10.1002/pd.1970040402. [DOI] [PubMed] [Google Scholar]
- 20.Triggs-Raine B L, Akerman B R, Clarke J T, Gravel R A. Am J Hum Genet. 1991;49:1041–1054. [PMC free article] [PubMed] [Google Scholar]
- 21.Natowicz M R, Wang Y. Clin Chim Acta. 1996;245:1–6. doi: 10.1016/0009-8981(95)06182-7. [DOI] [PubMed] [Google Scholar]
- 22.Greenblatt M S, Grollman A P, Harris C C. Cancer Res. 1996;56:2130–2136. [PubMed] [Google Scholar]
- 23.Arming S, Strobl B, Wechselberger C, Kreil G. Eur J Biochem. 1997;247:810–814. doi: 10.1111/j.1432-1033.1997.t01-1-00810.x. [DOI] [PubMed] [Google Scholar]
- 24.Davies G, Henrissat B. Curr Biol. 1995;3:853–859. doi: 10.1016/S0969-2126(01)00220-9. [DOI] [PubMed] [Google Scholar]