Abstract
Diversity and colonization levels of naturally occurring arbuscular mycorrhizal fungi (AMF) in onion roots were studied to compare organic and conventional farming systems in the Netherlands. In 2004, 20 onion fields were sampled in a balanced survey between farming systems and between two regions, namely, Zeeland and Flevoland. In 2005, nine conventional and ten organic fields were additionally surveyed in Flevoland. AMF phylotypes were identified by rDNA sequencing. All plants were colonized, with 60% for arbuscular colonization and 84% for hyphal colonization as grand means. In Zeeland, onion roots from organic fields had higher fractional colonization levels than those from conventional fields. Onion yields in conventional farming were positively correlated with colonization level. Overall, 14 AMF phylotypes were identified. The number of phylotypes per field ranged from one to six. Two phylotypes associated with the Glomus mosseae–coronatum and the G. caledonium–geosporum species complexes were the most abundant, whereas other phylotypes were infrequently found. Organic and conventional farming systems had similar number of phylotypes per field and Shannon diversity indices. A few organic and conventional fields had larger number of phylotypes, including phylotypes associated with the genera Glomus-B, Archaeospora, and Paraglomus. This suggests that farming systems as such did not influence AMF diversity, but rather specific environmental conditions or agricultural practices.
Electronic supplementary material
The online version of this article (doi:10.1007/s00572-009-0237-2) contains supplementary material, which is available to authorized users.
Keywords: Allium cepa, Glomus, Organic farming, Mycorrhizal colonization, Phylotype
Introduction
Onion (Allium cepa L.) has a sparse rooting system without root hairs which makes the crop dependent for water and nutrient acquisition on arbuscular mycorrhizal fungi (AMF) (De Melo 2003; Stribley 1990). This dependency is especially true in case of cultivation under nutrient-poor soil conditions as is frequently the case in low-input and organic agriculture. AMF enlarge the soil volume from which nutrients can be taken up, via an extensive mycelium network, enabling host plants to access more resources (Finlay 2004). As a consequence, AMF enhance uptake of nutrients, particularly phosphorus (Hayman and Mosse 1971), and may allow for a reduction of the amount of fertilizers applied (Linderman and Davis 2004). Furthermore, AMF can protect the plant against biotic (diseases) and abiotic (drought) stress, and improve soil aggregation (Gosling et al. 2006).
Research on Allium species and their interactions with AMF has a long history that dates back to 1884, when Mollberg described in roots of Allium scorodoprasum what we currently know as AMF (Koide and Mosse 2004). Allium species, and in particular onion, are excellent models for mycorrhizal research because they have a simple rooting system, slow growth, and high response to AMF. The knowledge on Allium–AMF interactions benefited greatly from the work of Mosse and co-workers, who presented detailed analyses of AMF functioning under field conditions (Hayman and Mosse 1971; Mosse and Hayman 1971; Mosse 1973; Owusu-Bennoah and Mosse 1979).
Onion, with a total annual acreage of 16,000–19,000 ha and an organically managed acreage of 600 ha, is an important crop in the Netherlands and a good model to monitor and compare the AMF status of agricultural soils. Numerous studies have shown that agricultural soils have low AMF species richness in comparison to natural ecosystems, such as woodlands and grasslands. The difference in AMF diversity is thought to be due to tilling-induced disruption of hyphal networks, rotation with non-mycorrhizal crop species, the occurrence of fallow periods, and the use of fertilizers and fungicides (Helgason et al. 1998; Daniell et al. 2001; Merryweather 2001; Jansa et al. 2002a). However, agricultural soils can differ in species richness and composition of AMF because their management systems differ significantly. This is the case for example in low- and high-input input farming systems (Ryan et al. 2000). In low-input and organic farming systems, synthetic fungicides and soluble phosphate fertilizers are limited or excluded. This may increase AMF inoculum potential and colonization levels compared to conventional farming systems, as has been observed for wheat (Douds et al. 1993; Ryan et al. 1994) and clover–ryegrass pastures (Eason et al. 1999; Ryan et al. 2000). Furthermore, recent research showed that AMF biodiversity was higher in low-input systems compared to high-input systems (Oehl et al. 2003, 2004), although the relationship is not always straightforward (Hijri et al. 2006).
The contribution of AMF to crop-production increase in high-input agriculture is low because phosphorus is amply available. In contrast, AMF might have a significant role in increasing crop production in low-input and organic agricultural systems. However, this concept is far from being practically applied due to the lack of understanding of the functioning of AMF species (Scullion et al. 1998).
The present research aimed to study AMF species richness and composition in onion fields in the Netherlands, by comparing organic and conventional cultivation systems. In this way, we investigate if the adoption of organic practices on formerly conventionally managed farmlands leads to higher AMF diversity. Unlike previous studies, this research was carried out in a large number of sites. The primer sets developed by Redecker (2000) and Redecker et al. (2003) were used to identify AMF phylotypes that colonize onion plant roots, and therefore, only the AMF assemblage of the target host species was analyzed rather than the complete diversity in the soil.
Materials and methods
Survey of onion fields and root colonization
Two traditional onion-growing regions in the Netherlands were sampled, namely, Zeeland in the southwest and Flevoland (the Flevopolder and the Noordoostpoder) in the center of the country. In Zeeland, onion cultivation takes place already for centuries, whereas in Flevoland, on the land recently reclaimed from the sea, onion cultivation only takes place since the second half of the twentieth century. In both regions, the soils are classified as clay to loess–clay soils. Clay content ranged from 23% to 40% for the soils investigated in Zeeland, and 7% to 55% in Flevoland (Table 1). Seed-onions in the Netherlands are cultivated in rotation with other field crops. The soils are ploughed every year, either before or after the winter, and seedbeds are prepared consisting of fine soil aggregates. Sowing date is at the end of March and early April, and harvest takes place in the second half of August (early cultivars) and September (late cultivars). In both years, samplings were done in the second half of June. Plants were in leaf development phase, with four to six expanded leaves.
Table 1.
Average soil chemical parameters and soil history of onion fields surveyed in 2004 and 2005, by cultivation system and region in the Netherlands
| Cultivation system | Region | Number of fields | Years under organic cultivationa | Onion yield (ton/ha) | Soil properties | ||||
|---|---|---|---|---|---|---|---|---|---|
| OMb | pH | Pwc | Cab | Kd | |||||
| Organic | 20 | 12.1 | 32 | 3.1 | 7.4 | 32 | 7.2 | 29 | |
| Flevoland | 15 | 13.8 (4–32) | 33 | 3.1 | 7.4 | 32 | 6.5 | 30 | |
| Zeeland | 5 | 5.7 (1–12) | 28 | 2.9 | 7.2 | 30 | 9.2 | 20 | |
| Conventional | 19 | – | 70 | 3.0 | 7.3 | 44 | 5.1 | 26 | |
| Flevoland | 14 | – | 76 | 3.2 | 7.3 | 45 | 5.5 | 23 | |
| Zeeland | 5 | – | 59 | 2.5 | 7.5 | 42 | 2.8 | 33 | |
aAverage values. The range of values is indicated between brackets
bOM organic matter (%), Ca CaCO3 (%)
cmg P2O5 l−1 of dry soil. The difference in P content between cultivation systems was almost significant (REML analysis, p = 0.071)
dmg K kg−1 soil
Twenty onion fields were sampled in a balanced survey between cultivation systems and regions: five organic and five conventional fields in Zeeland, plus five organic and five conventional fields in Flevoland. Ten plants per field were randomly sampled. The surrounding soil was excavated in order to take out the rooting system as intact as possible. A new survey was done in June 2005, only in Flevoland, because the 2004 study suggested that management practices in this region could have an influence on AMF diversity. The 2005 study comprised ten organic and nine conventional onion fields. All of them were different from fields sampled in 2004, although in some cases, they were located within the same farm. Organic farm fields followed ecological or biodynamic practices, and fulfilled the basic standards for organic production (available from IFOAM 2007) as certified by SKAL (www.skal.com). Information on chemical characteristics of the soils (pH, organic matter, content of phosphorus, and other nutrients) and onion yields were obtained from the farmers for 28 of the 39 sites, and the number of years under organic management queried for all 20 organic fields (Table 1).
AMF colonization was estimated only for samples collected in 2004. Staining of fungal structures was done using trypan blue, and colonization was quantified following the magnified intersections method (McGonigle et al. 1990). Hyphal (HC), arbuscular (AC), and vesicular colonization (VC) were quantified separately. Data analysis was carried out via analysis of variance using Genstat 9.2 (Lawes Agricultural Trust, Rothamsted Exp. St., UK, 2006). The relationships between colonization parameters and environmental variables were studied using the Pearson correlation coefficient and linear regression analysis.
Molecular diversity analysis
AMF species colonizing onion roots were identified by sequencing the partial 18S-ITS1–5.8S-ITS2 rDNA region, as described by Redecker (2000), with minor modifications. DNA was isolated from 1-cm root pieces randomly taken from each onion sample. rDNA was amplified using a nested polymerase chain reaction (PCR) approach. The primers NS5/ITS4i were used in the first step (Redecker et al. 2003) with an annealing temperature of 51°C. For the second amplification, PCR products were diluted 1:100. This second step was performed using the primers ACAU1660, ARCH1311, GLOM1310, and LETC1670 in combination with ITS4i in separate reactions. Primer GIGA5.8R was used only in combination with NS5. Primer sequences and protocols for PCR amplifications are available from Redecker (2000) and Redecker et al. (2003).
PCR products having the expected size were cloned by the pGEM-T vector system (Promega, Madison, WI, USA). An aliquot of 8 μl of the successful clones were digested with restriction enzymes MboI, HinfI, and AluI in 15 μl at 37°C for 6 h. Restriction fragment patterns were run on a 3% gel made from RESponse agarose (Byozim group, Landgraaf, The Netherlands) and analyzed by the Phoretix 1D software (Nonlinear Dynamics, Durham, NC, USA). A representative clone for each distinct restriction profile was sequenced by the dideoxynucleotide chain termination method using BigDyeTM Terminator Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA) and an automatic sequencer (ABI PRISM 3700 DNA Analyser, Applied Biosystems). After removing chimerical results, a set of 65 sequences were obtained from the 5.8S-ITS2 rDNA region and, when available, the partial 18S region (each region 400–500 bps in length). Sequences are deposited in the European Molecular Biology Laboratory (EMBL and NCBI databases) under the accession numbers AM992799 to AM992864. These sequences were used for identification of AMF phylotypes after phylogeny analysis. In order to obtain that, sequences were submitted to the BLAST query tool (Altschul et al. 1997) and database www.ncbi.nih.gov/blast/ for an initial similarity analysis. Sequence alignment was performed manually. Finally, a phylogenetic analysis was carried out by distance analysis using the neighbor-joining method in PAUP 4b10 (Swofford 2003) with the Kimura two-parameter model, and a gamma shape parameter of 0.5. Bootstrap analyses were done with 1,000 replications. Sequences selected from public databases belonging to known AMF species were included in the phylogenetic analysis.
Phylotypes were determined from cladograms as clearly distinct monophyletic taxa which were also present in the respective maximum likelihood trees. AMF genera or morphospecies associated to each phylotype were assigned on the basis of the position of already known sequences from databases (Table 3). Phylotypes were associated either to a group of related species (e.g., Glomus caledonium–geosporum) or a genus (e.g., Paraglomus), and therefore, the number of phylotypes is a conservative estimate for the number of morphospecies. Code names were assigned to phylotypes, as an acronym for the genus followed by a correlative number. Glomus-A and Glomus-B groups were distinguished according to Schwarzott et al. (2001).
Table 3.
Number of fields containing each AMF phylotype, by cultivation system and region in the Netherlands
| Phylotypes a | Cultivation systems | Regions | Total nr of fields | ||
|---|---|---|---|---|---|
| Organic | Conventional | Flevoland | Zeeland | ||
| GloA1 (Glomus-A) | 1 | 1 | 1 | ||
| GloA2 (Glomus-A) | 1 | 1 | 1 | ||
| GloA3 (G. intraradices) | 3 | 1 | 2 | 2 | 4 |
| GloA4 (Glomus-A) | 2 | 2 | 2 | ||
| GloA5 (G. caledonium-geosporum) | 16 | 15 | 22 | 9 | 31 |
| GloA6 (G. mosseae-coronatum) | 19 | 19 | 28 | 10 | 38 |
| GloB1 (G. walkeri) | 2 | 2 | 2 | ||
| GloB2 (Glomus-B) | 1 | 1 | 1 | ||
| GloB3 (Glomus-B) | 1 | 1 | 1 | ||
| GloB4 (Glomus-B) | 2 | 2 | 2 | ||
| GloB5 (G. claroideum) | 1 | 1 | 1 | ||
| Par1 (Paraglomus sp.) | 5 | 5 | 5 | ||
| Arch1 (Archaeospora sp.) | 1 | 4 | 5 | 5 | |
| Arch2 (Archaeospora sp.) | 2 | 2 | 2 | ||
| Total number of surveyed fields | 20 | 19 | 29 | 10 | 39 |
aDistinct monophyletic taxa after neighbor-joining analysis of rDNA sequences. Whenever known, species names associated with a phylotype are indicated. Glomus-A and B, as defined by Schwarzott et al. (2001)
AMF diversity was analyzed by integrating the data from 2004 and 2005, using the number of phylotypes found per onion field. In addition, Shannon–Weaver diversity indices (H′) were calculated based on the relative abundance of phylotypes per field, as 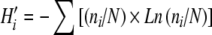 , being ni the number of observation for the i phylotype, whereas N is the total number of observations recorded (Mueller et al. 2004). Differences in AMF diversity indices between cultivation systems or between regions were tested by residual maximum likelihood analysis (REML) using Genstat 9.2. Besides, the associations between AMF diversity with chemical soil parameters and with the number of years under organic agriculture were studied by Pearson correlation coefficient and linear regression analysis.
, being ni the number of observation for the i phylotype, whereas N is the total number of observations recorded (Mueller et al. 2004). Differences in AMF diversity indices between cultivation systems or between regions were tested by residual maximum likelihood analysis (REML) using Genstat 9.2. Besides, the associations between AMF diversity with chemical soil parameters and with the number of years under organic agriculture were studied by Pearson correlation coefficient and linear regression analysis.
Correspondence analysis (CA) was performed in CANOCO 4.53 (Ter Braak and Smilauer 2004). CA allows to study the contribution of each sampled site (n = 39) to the total variation based on their AMF species composition, as well as the contribution from each phylotype (n = 14) based on their abundance along sampled sites (Jongman et al. 1995).
Results
AMF colonization in onions
In 2004, all sampled onion plants were colonized. The average colonization in both sampled regions and cultivation systems was 60% for arbuscular colonization (AC) and 84% for hyphal colonization (HC) as grand means (Table 2). The presence of vesicles was much lower, namely, 7% on average. While neither region nor cultivation system was a significant source of variation, the interaction between regions and cultivation systems was significant for AC (p = 0.004) and HC (p = 0.005). This was due to the fact that conventional cultivation in Zeeland had a mean AC of 46%, a mean HC of 72%, and both means differed significantly from the means of the other three combinations of region and cultivation system. Furthermore, onions grown in conventional fields in Zeeland had a significantly lower AC/HC ratio.
Table 2.
Arbuscular mycorrhizal colonization parameters by cultivation systems and regions in the Netherlands (Survey 2004)
| Cultivation system | Region | Arbuscular colonization (AC) (%) | Hyphal colonization (HC) (%) | Vesicular colonization (%) | Ratio AC/HC |
|---|---|---|---|---|---|
| Organic | Flevoland | 62 a | 89 a | 9.2 | 0.69 a |
| Zeeland | 65 a | 85 a | 6.5 | 0.76 a | |
| Conventional | Flevoland | 67 a | 91 a | 7.7 | 0.73 a |
| Zeeland | 46 b | 72 b | 3.6 | 0.62 b |
Means within each column followed by the same letter do not differ statistically (Fischer LSD test, p > 0.05)
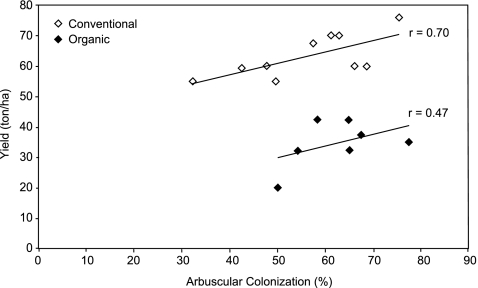
No correlation was found between average AC and HC and soil parameters, on the one hand, and the number of years under organic cultivation, on the other hand, except for the correlation between content of calcium in the soil and AC (r = 0.55; p < 0.05). In addition, a significant correlation was found between AC and onion yield (OY) under conventional management (r = 0.70, p < 0.05; Fig. 1), as well as between HC and OY (r = 0.85, p < 0.05). In case of conventional management (n = 10), the linear regression was estimated as 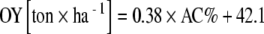 . With a smaller range of variation in AMF colonization level (AC, HC) and less data points available (n = 7), in case of organic management, the regression was estimated as
. With a smaller range of variation in AMF colonization level (AC, HC) and less data points available (n = 7), in case of organic management, the regression was estimated as 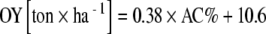 , though this regression was not significant.
, though this regression was not significant.
Fig. 1.
The correlation between arbuscular colonization (%) and onion yield (tons/ha) for conventional (n = 10) and organic (n = 7) management systems (survey 2004)
AMF diversity in onion fields
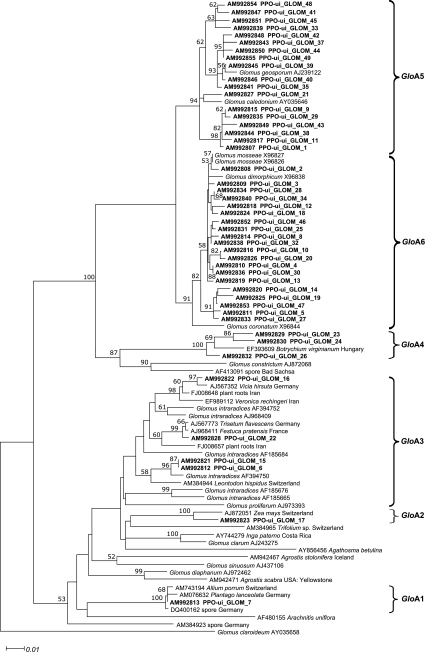
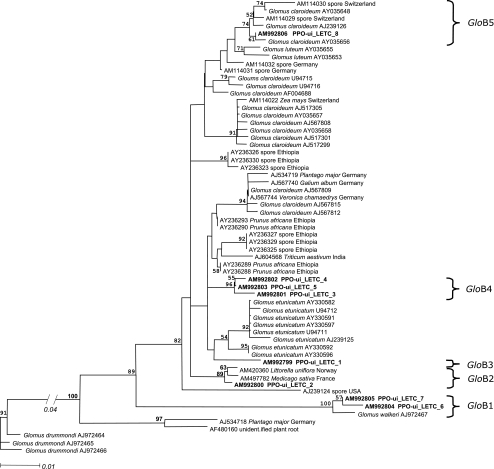
Among the total plant samples, 56% in 2004 and 93% in 2005 yielded AMF amplification products and subsequent restriction profiles. The GIGA5.8R combined with NS5 did not yield any amplification product. After phylogenetic analysis, 65 rDNA-obtained sequences were clustered in 14 phylotypes, namely, six Glomus-A, five Glomus-B, two Archaeospora, and one Paraglomus phylotypes (Figs. 2 and 3). Phylogenetic analysis of the ITS2-5.8S sequences of Glomus-A species was in agreement with the phylogenetic tree obtained for the sequences from the 18S region (Figure S1). An exception was the distinction between phylotypes GloA5 and GloA6, which were clearly distinct regarding the ITS2-5.8S region, whereas the 18S region clusters did not hold high bootstrap values.
Fig. 2.
Neighbor-joining phylogenetic analysis of arbuscular mycorrhizal fungal 5.8S-ITS2 rDNA sequences obtained from onion root samples in the Netherlands for Glomus-A (as defined by Schwarzott et al. 2001) phylotypes, rooted to Paraglomus brasilianum sequence AJO12112. The scale represents substitutions per site. Sequences obtained in the present study are shown in boldface. Sequences from databases isolated from roots were labeled with the accession number, host plant, and country of origin, whereas those isolated from spores, with fungal species and accession number. Bootstrap values from 1,000 replications larger than 50% are indicated above branches. Brackets to the right indicate phylotypes defined for the sequences obtained in this study
Fig. 3.
Neighbour-joining phylogenetic analysis of arbuscular mycorrhizal fungal 5.8S-ITS2 rDNA sequences obtained from onion root samples in the Netherlands for Glomus-B (as defined by Schwarzott et al. 2001) phylotypes, rooted to the G. drummondii sequence AJ972466. The scale represents substitutions per site. Sequences obtained in the present study are shown in boldface. Sequences from databases isolated from roots were labeled with the accession number, host plant, and country of origin, whereas those isolated from spores, with fungal species and accession number. Bootstrap values from 1,000 replications larger than 50% are indicated above branches. Brackets to the right indicate phylotypes defined for the sequences obtained in this study
Glomus-A species were predominant in onion cultivation in the Netherlands, particularly phylotype GloA6, which was present in 19 of the 20 onion fields in 2004 and all surveyed fields in 2005 (Table 3). Overall, this phylotype associated with the G. mosseae–coronatum complex was present in 70% of the individual sampled plants. Phylotype GloA5 associated with the G. caledonium–geosporum complex was the second most abundant one and was found in 31 of the 39 fields.
The other phylotypes were present in a much lower frequency, three of them being exclusively found in conventional fields and seven only found in organic fields (Table 3). Some phylotypes were only found in 2004, such as GloA1, GloA2, and GloA3, whereas others were found only in 2005, such as Arch1 (Table S1). Noticeably, onion fields of two organic farms harbored phylotypes Par1 (Paraglomus) and Arch2 (Archaeospora) in 2004, and the next year, sampling again on different fields of the same farms resulted in the finding of the same phylotypes.
The number of AMF phylotypes per field ranged from one to six. The highest diversity in phylotypes was found in two organic and two conventional fields. Organic and conventional management systems did not differ in the number of phylotypes per field (REML analysis, p = 0.48), nor did the regions (p = 0.99). In the same way, the Shannon–Weaver index calculated on the basis of the relative abundance of phylotypes per field (Table 4) did not significantly differ for the management systems studied and also not for the four management systems–region combinations (REML analysis).
Table 4.
Means and their standard errors (±SE) of the number of phylotypes per field, and the Shannon–Weaver indices based on the relative abundance of phylotypes per field, for the cultivation systems and regions in 2004 and 2005
| Diversity indices | Cultivation system | Survey 2004 | Survey 2005 | |
|---|---|---|---|---|
| Zeeland | Flevoland | Flevoland | ||
| Nr of phylotypes | Conventional | 2.4 ± 0.9 | 1.4 ± 0.5 | 2.8 ± 1.0 |
| Organic | 2.6 ± 0.5 | 2.6 ± 2.1 | 2.7 ± 0.7 | |
| Shannon index | Conventional | 0.70 ± 0.31 | 0.25 ± 0.35 | 0.89 ± 0.30 |
| Organic | 0.66 ± 0.22 | 0.66 ± 0.69 | 0.88 ± 0.20 | |
The number of years a field was under organic management was neither correlated with the number of phylotypes per field, nor with the Shannon–Weaver index. Onion yields in organic or conventional farming were also not correlated significantly with AMF diversity indices (data not shown).
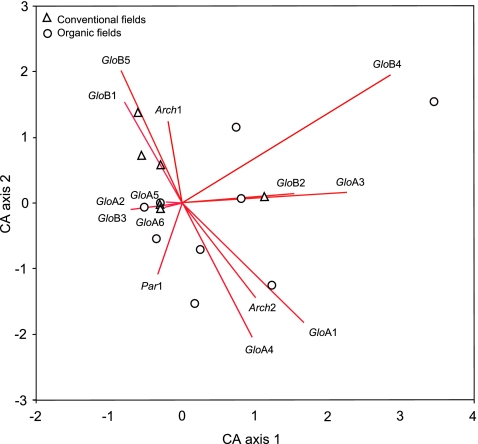
The contribution of phylotypes and sampled fields to the total variation in diversity was analyzed via correspondence analysis (CA; data matrix shown as Table S2). Figure 4 presents the biplot for the first and second CA axes, which explained 31.4% of the total variance (first axis: 16.8%, second axis 14.6%, eigenvalues 0.54 and 0.47, respectively; total inertia 3.24). Phylotypes GloA6 (G. mosseae–coronatum complex) and GloA5 (G. caledonium–geosporum complex) took a central position in the CA space, as these phylotypes did not contribute much to the variation between sampled sites. Other less frequent phylotypes, which took more extreme positions in the CA space, contributed more to the variation between fields. The larger proportion of sampled sites, 13 conventional and eight organic ones, took a central position in the chart, close to the most abundant phylotypes (GloA5 and GloA6), and irrespective of cultivation system and sampled region (Fig. 4). As a result, only a few other organic and conventional sites had larger AMF diversity, differing in community composition among them (Fig. 4).
Fig. 4.
Biplot for the first and second axis in the correspondence analysis for the number of AMF phylotypes in conventional onion fields (open triangles) and organic fields (open circles) in the Netherlands in 2004 and 2005. Some field positions overlap (same AMF species composition). AMF phylotypes are presented as vectors
Discussion
Molecular diversity
Only a few studies have addressed the question if and to what extent AMF species richness and composition differ between organic and conventional farming systems (Oehl et al. 2003, 2004; Hijri et al. 2006). To contribute to this field of knowledge, the present research was carried out involving a larger number of sites compared to previous studies and different environmental conditions like a temperate sea climate.
AMF diversity was estimated via the number of different phylotypes per field, as no reliable molecular method has been developed to determine all different AMF morphospecies yet. This estimator can be seen as a lower limit for diversity estimation, as each phylotype may contain different AMF morphospecies. Phylotypes were defined as monophyletic groups in a conservative way. Since the taxonomic position of rDNA sequences from root samples is unknown, and rDNA sequence variation within a given morphospecies or even within a single spore exists (Jansa et al. 2002b), AMF diversity may be overestimated if smaller monophyletic groups are assumed as phylotypes. Furthermore, we used the Shannon–Weaver index calculated on the basis of the number of phylotypes per field.
A total of 14 AMF phylotypes were detected in onion roots, with one to six phylotypes per field. The total number of phylotypes and number of phylotypes per field were well in line with those obtained by Hijri et al. (2006) who studied AMF diversity in arable soils using the same primer set for AMF identification and a similar sampling effort. The most abundant phylotypes GloA6 (associated with G. mosseae–coronatum species complex) and GloA5 (G. caledonium–geosporum species complex) could be, respectively, phylotypes GLOM-A3 and GLOM-A4 in Hijri et al. (2006) and Appoloni et al. (2008). Phylotype GloA3 was associated with G. intraradices and corresponds with GLOM-A1 in Hijri et al. (2006) and Sýkorová et al. (2007). Phylotype GloA2 comprised the sequence AJ872051, which is representative of GLOM-A2 (Hijri et al. 2006).
Phylotype GloA4 clustered with a sequence from naturally occurring Botrychium virginianum L. (Kovacs et al. 2007) and a Glomus constrictum sequence (Hijri et al. 2006). Phylotype Arch2 corresponds with phylotype ARCH-1 defined by Hijri et al. (2006) and ARCH-4 of Sýkorová et al. (2007), as these share the same Archaeospora trappei sequence and a sequence from a species of Podocarpaceae. Our Paraglomus sequences clustered in phylotype Par1 that corresponds to phylotype PARA-1 in Appoloni et al. (2008), as both clustered with the same sequence from Zea mays, except for our sequence PPO-ui_ARCH_7 which showed similarity to AJ564068 from Dactylis glomerata (Wirsel 2004).
Predominance of some Glomus-A phylotypes in AMF communities was in agreement with previous reports for agricultural lands. For example, analyzing SSU rDNA fragments, Helgason et al. (1998) found predominance of G. mosseae or closely related species, whereas Daniell et al. (2001) found predominantly G. caledonium–G. geosporum sequences. The dominance of Glomus-A species was also reported in studies based on the morphology of spores, being G. mosseae as the most observed AMF species (Oehl et al. 2003; Cheng and Baumgartner 2004; Sjöberg et al. 2004; Wang et al. 2008). In other studies, however, G. intraradices was the dominant AMF species (Hijri et al. 2006; Mathimaran et al. 2005). The dominance of a few Glomus-A species is probably a consequence of the strong selection pressure imposed by agricultural practices leading to the predominance of fast root-colonizing species (Oehl et al. 2004) and species able to tolerate, among others, the repeated disruption of external hyphal networks, periods without mycorrhizal host plants, and the application of fertilizers and fungicides (Gosling et al. 2006).
In contrast, other AMF phylotypes were found at very low frequencies, which make it difficult to establish associations between farming systems with specific phylotypes or community composition. While some Glomus-B, Paraglomus, and Archaeospora phylotypes were occasionally found in some Dutch farm fields, neither Acaulospora nor Scutellospora rDNA sequences were detected at all. In previous studies, the presence of Glomus-B, Archaeospora, Acaulospora, Scutellospora, and Paraglomus species in arable lands was low in comparison to grasslands, or they were absent (Oehl et al. 2004; Hijri et al. 2006).
We found that AMF diversity in organically grown onions did not differ from conventional ones, with a considerable overlap between farming systems in the canonical analysis. This result is in contrast with Oehl et al. (2003) who reported that AMF diversity in organic fields (13–18 AMF species) took an intermediate position between conventional fields (eight to ten) and grasslands (20–25). However, Oehl and co-workers analyzed the morphology and number of AMF spores in soil samples and trap cultures, and therefore, they analyzed a broader part of the AMF species richness than the present research. Hijri et al. (2006) used an approach similar to our study, relying on the same primer set for AMF identification. Among four management systems established on the same loess soil (the DOK experiment), higher AMF biodiversity in organically managed plots than in conventional ones was observed, though not significantly. This trend is not in agreement with our results. Regional characteristics and specificities in AMF community composition may explain this difference. Furthermore, these authors compared a few fields at one location, whereas our study had a much broader setup, as we compared in total 20 organic and 19 conventionally managed fields belonging to commercial farms on clay and loess–clay soils.
Among the studied farmlands in the Netherlands, only a few had larger AMF diversity indices. This is in line with Hijri et al. (2006) who found, after studying two fields apart from the DOK experiment, that AMF diversity varied also within management systems depending upon the specific agricultural history. It was noteworthy, in the present study, that AMF diversity was larger in fields belonging to two farms sampled both years, in which onions were grown each year on different pieces of land within the farm. These observations suggest that farming system as such did not influence AMF diversity, but rather specific management practices or environmental conditions may contribute to the maintenance of more diverse AMF community in some farmlands. For instance, the continuous cultivation of mycorrhizal host crops in the rotations, or the use of green cover crops instead of fallow periods might favor the presence of more AMF species in the long term (Gosling et al. 2006; Hijri et al. 2006).
Onion yields were not correlated with AMF diversity indices. Nevertheless, the practical role of AMF diversity in agricultural systems should be further studied. First indications coming up from experimental setups in which mixed AMF species were applied as inocula point to the complexity arising from multiple fungi–host interactions. For instance, Van der Heijden et al. (2006) reported variable benefit in biomass increase from mixed inocula, whereas Jansa et al. (2008) reported only improved phosphorus acquisition in comparison to the same AMF species acting separately.
Colonization in onion roots
Onion roots in Dutch agricultural fields had high AMF colonization levels in comparison to levels reported for naturally occurring AMF in other crops and environments (Ryan et al. 2000; Mäder et al. 2000). Although obtained at a single sampling date within a single growing season, our results revealed the abundance of AMF in agricultural soils in the Netherlands, including reclaimed lands.
As conventional farming in Zeeland had significantly lower AMF colonization levels compared to the other three management systems–region combinations (Table 2), the adoption of organic farming seemed to increase AMF colonization levels quickly. Indeed, the organic fields sampled in Zeeland were only 1 to 12 years taken out of conventional management. In contrast, mean colonization levels in Flevoland were high regardless of the cultivation system and not correlated with the number of years under organic management.
Furthermore, we found that colonization parameters were neither correlated with the concentration of readily available phosphorus in the soil, even when a trend toward a higher P content for conventional fields was observed (Table 1). Several authors reported larger AMF colonization for organically managed fields compared to conventionally managed fields. Among others, Ryan et al. (1994, 2000) found higher colonization levels in organically grown wheat and pastures than conventionally managed ones, whereas Mäder et al. (2000, 2002) reported higher colonization in organic winter wheat, vetch-rye, and grass-clover when comparing organic and conventional soil management systems. Similarly, Oehl et al. (2003) found higher colonization levels in plants grown on organic soils compared to conventional ones. In these studies, differences in colonization levels were explained mainly by the lower P concentration in organically managed soils, which was not clearly the case for the onion fields in the present research.
A striking finding of our survey was the significant correlation between colonization parameters of naturally occurring AMF and onion yields in conventionally managed onion farmlands, under moderate to high concentrations of readily available phosphorus (Table 1, Fig. 1). Organically managed fields followed the same trend as conventional ones, but the correlation coefficient was considerably lower and not significant for AC. The slope of both linear regression lines was 0.38 indicating that onions benefited similarly from AMF in both management systems. Alternatively, it could be speculated that AMF colonization and onion yields benefit simultaneously from yet unidentified environmental conditions. Anyhow, in order to practically exploit the fact that 10% increase in AMF colonization may represent an average yield increase of 3.8 tons·ha−1, it would be necessary to better understand farm management practices or environmental factors leading to higher vitality of indigenous AMF.
Concluding remarks
This study did not demonstrate differences in biodiversity of AMF colonizing onion roots grown under organic and conventional cultivation, and this result raises at least two important questions. Firstly, is the lack of differences in AMF community composition between cultivation systems due to the fact that nutrient levels (especially phosphorus) are moderate to high in organic fields? If this is the case, it is uncertain that a further continuation under organic cultivation (with a concomitant decrease in soil nutrient levels) would increase mycorrhizal fungal diversity over a longer temporal scale.
Secondly, the positive correlation between mycorrhizal colonization and onion yield established for conventional fields suggests a mycorrhizal benefit even at high nutrient availability. It is therefore tempting to speculate that onion, with its depauperate root system, depends on mycorrhiza even at high nutrient levels, thereby preventing the selection for reduced functioning of the symbiosis under agricultural intensification postulated by Johnson (1993) and Kiers et al. (2002). Breeding onions with improved rooting system while retaining a diversity of mycorrhizal benefits would be an urgent further research step. Otherwise, onion cultivation under nutrient-poor conditions might result in yields that are too low to be economically attractive. In this context, the research of De Melo (2003) on the genetic basis of traits describing the Allium rooting system can be seen as a promising first step.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Number of fields which harbor specific AMF phylotype per surveyed year (DOC 48.5 KB)
Presence or absence of AMF phylotypes in each onion field (DOC 46.5 KB)
Phylogenetic analysis of 18S rDNA sequences of Glomus-A (DOC 313 KB)
Acknowledgements
We thank Ir. R. van den Broek for information on onion growers provided for the survey, P. Hendrickx, B. Lavrijssen, and T. Olijnsma for technical assistance during the molecular and microscopic analyses, and Prof. Dr. R. Hoekstra for critically reading the manuscript. This work was funded by the Dutch Ministry of Agriculture, Nature and Food quality as part of Programme 388-II Plant Breeding for Organic Farming. G.A. Galván thanks the Alßan Programme of the European Union (Fellowship E03D02847UR), PDT-CONICYT Uruguay (Fellowship S/C/BE/20/09), and the NFP (the Netherlands) for their financial support. Laboratory studies by I. Parádi were done via a Huygens Fellowship awarded by Nuffic (the Netherlands).
Open Access This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
Authors Guillermo A. Galván and István Parádi contributed equally to this research and share first co-authorship.
Electronic supplementary material
The online version of this article (doi:10.1007/s00572-009-0237-2) contains supplementary material, which is available to authorized users.
References
- Altschul SF, Thomas LM, Alejandro AS, Jinghui Z, Webb M, David JL (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi:10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed]
- Appoloni S, Lekberg Y, Tercek MT, Zabinski CA, Redecker D (2008) Molecular community analysis of arbuscular mycorrhiza fungi in roots of geothermal soils in Yellowstone National Park (USA). Microb Ecol 56:649–659. doi:10.1007/s00248-008-9384-9 [DOI] [PubMed]
- Cheng X, Baumgartner K (2004) Survey of arbuscular mycorrhizal fungal communities in northern California vineyards and mycorrhizal colonization potential of grapevine nursery stock. HortScience 39:1702–1706
- Daniell TJ, Husband R, Fitter AH, Young JP (2001) Molecular diversity of arbuscular mycorrhizal fungi colonising arable crops. FEMS Microbiol Ecol 36:203–209. doi:10.1111/j.1574-6941.2001.tb00841.x [DOI] [PubMed]
- De Melo PE (2003) The root systems of onion and Allium fistulosum in the context of organic farming: a breeding approach. Ph.D. Thesis, Wageningen University, The Netherlands. 136 pp
- Douds DD, Janke RR, Peters SE (1993) VAM fungus spore populations and colonization of roots of maize and soybean under conventional and low-input sustainable agriculture. Agric Ecosyst Environ 43:325–335. doi:10.1016/0167-8809(93)90095-7 [DOI]
- Eason WR, Scullion J, Scott EP (1999) Soil parameters and plant responses associated with arbuscular mycorrhizas from contrasting grassland management regimes. Agric Ecosyst Environ 73:245–255. doi:10.1016/S0167-8809(99)00054-7 [DOI]
- Finlay RD (2004) Mycorrhizal fungi and their multifunctional roles. Mycologist 18:91–96. doi:10.1017/S0269915X04002058 [DOI]
- Gosling P, Hodge A, Goodlass G, Bending GD (2006) Arbuscular mycorrhizal fungi and organic farming. Agric Ecosyst Environ 113:17–35. doi:10.1016/j.agee.2005.09.009 [DOI]
- Hayman DS, Mosse B (1971) Plant growth responses to vesicular–arbuscular mycorrhiza. I. Growth of endogone-inoculated plants in phosphate-deficient soils. New Phytol 70:19–27. doi:10.1111/j.1469-8137.1971.tb02504.x [DOI]
- Helgason T, Daniell TJ, Husband R, Fitter AH, Young JP (1998) Ploughing up the wood-wide web? Nature 394:431. doi:10.1038/28764 [DOI] [PubMed]
- Hijri I, Sýkorová Z, Oehl F, Ineichen K, Mäder P, Wiemken A, Redecker D (2006) Communities of arbuscular mycorrhizal fungi in arable soils are not necessarily low in diversity. Mol Ecol 15:2277–2289. doi:10.1111/j.1365-294X.2006.02921.x [DOI] [PubMed]
- IFOAM (2007) The IFOAM basic standards for organic production and processing (available from www.ifoam.org, periodically updated)
- Jansa J, Mozafar A, Anken T, Ruh R, Sanders IR, Frossard E (2002a) Diversity and structure of AM communities as affected by tillage in a temperate soil. Mycorrhiza 12:225–234. doi:10.1007/s00572-002-0163-z [DOI] [PubMed]
- Jansa J, Mozafar A, Banke S, McDonald BA, Frossard E (2002b) Intra- and intersporal diversity of ITS rDNA sequences in Glomus intraradices assessed by cloning and sequencing, and by SSCP analysis. Mycol Res 106:670–681. doi:10.1017/S0953756202006032 [DOI]
- Jansa J, Smith FA, Smith SE (2008) Are there benefits of simultaneous root colonization by different arbuscular mycorrhizal fungi? New Phytol 177:779–789 [DOI] [PubMed]
- Johnson NC (1993) Can fertilization of soil select less mutualistic mycorrhizae? Ecol Appl 3:749–757. doi:10.2307/1942106 [DOI] [PubMed]
- Jongman RHG, Ter Braak CJF, Van Tongeren OFR (1995) Data analysis in community and landscape ecology. Cambridge University Press, Cambridge, UK, p 299
- Kiers ET, West SA, Denison RF (2002) Mediating mutualisms: farm management practices and evolutionary changes in symbiont co-operation. J Appl Ecol 39:745–754. doi:10.1046/j.1365-2664.2002.00755.x [DOI]
- Koide RT, Mosse B (2004) A history of research on arbuscular mycorrhiza. Mycorrhiza 14:145–163. doi:10.1007/s00572-004-0307-4 [DOI] [PubMed]
- Kovacs GM, Balazs T, Penzes Z (2007) Molecular study of arbuscular mycorrhizal fungi colonizing the sporophyte of the eusporangiate rattlesnake fern (Botrychium virginianum, Ophioglossaceae). Mycorrhiza 17:597–605. doi:10.1007/s00572-007-0137-2 [DOI] [PubMed]
- Linderman RG, Davis EA (2004) Evaluation of commercial inorganic and organic fertilizer effects on arbuscular mycorrhizae formed by Glomus intraradices. Horttechnology 14:196–202
- Mäder P, Edenhofer S, Boller T, Wiemken A, Niggli U (2000) Arbuscular mycorrhizae in a long-term field trial comparing low-input (organic, biological) and high-input (conventional) farming systems in a crop rotation. Biol Fertil Soils 31:150–156. doi:10.1007/s003740050638 [DOI]
- Mäder P, Fließbach A, Dubois D, Gunst L, Fried P, Niggli U (2002) Soil fertility and biodiversity in organic farming. Science 296:1694–1697. doi:10.1126/science.1071148 [DOI] [PubMed]
- Mathimaran N, Ruh R, Vullioud P, Frossard E, Jansa J (2005) Glomus intraradices dominates arbuscular mycorrhizal communities in a heavy textured agricultural soil. Mycorrhiza 16:61–66. doi:10.1007/s00572-005-0014-9 [DOI] [PubMed]
- McGonigle TP, Miller MH, Evans DG, Fairchild GL, Swan JA (1990) A new method which gives an objective measure of colonization of roots by vesicular-arbuscular mycorrhizal fungi. New Phytol 115:495–501. doi:10.1111/j.1469-8137.1990.tb00476.x [DOI] [PubMed]
- Merryweather J (2001) Comment: Meet the Glomales; the ecology of mycorrhiza. British Wildlife, pp 86–93
- Mosse B (1973) Plant growth responses to vesicular–arbuscular mycorrhiza. IV. In soil given additional phosphate. New Phytol 72:127–136. doi:10.1111/j.1469-8137.1973.tb02017.x [DOI]
- Mosse B, Hayman DS (1971) Plant growth responses to vesicular-arbuscular mycorrhiza. II. In unsterilized field soils. New Phytol 70:29–34. doi:10.1111/j.1469-8137.1971.tb02505.x [DOI]
- Mueller GM, Bills GF, Foster MS (2004) Biodiversity of Fungi: Inventory and Monitoring Methods. Academic, 777p
- Oehl F, Sieverding E, Ineichen K, Mäder P, Boller T, Wiemken A (2003) Impact of land use intensity on the species diversity of arbuscular mycorrhizal fungi in agroecosystems of Central Europe. Appl Environ Microbiol 69:2816–2824. doi:10.1128/AEM.69.5.2816-2824.2003 [DOI] [PMC free article] [PubMed]
- Oehl F, Sieverding E, Mäder P, Dubois D, Ineichen K, Boller T, Wiemken A (2004) Impact of long-term conventional and organic farming on the diversity of arbuscular mycorrhiza fungi. Oecologia 138:574–583. doi:10.1007/s00442-003-1458-2 [DOI] [PubMed]
- Owusu-Bennoah E, Mosse B (1979) Plant growth responses to vesicular–arbuscular mycorrhiza. XI. Field inoculation responses in barley, lucerne and onion. New Phytol 83:671–679. doi:10.1111/j.1469-8137.1979.tb02299.x [DOI]
- Redecker D (2000) Specific PCR primers to identify arbuscular mycorrhizal fungi within colonized roots. Mycorrhiza 10:73–80. doi:10.1007/s005720000061 [DOI]
- Redecker D, Hijri I, Wiemken A (2003) Molecular identification of arbuscular mycorrhizal fungi in roots: perspectives and problems. Folia Geobot 38:113–124. doi:10.1007/BF02803144 [DOI]
- Ryan MH, Chilvers GA, Dumaresq DC (1994) Colonisation of wheat by VA-mycorrhizal fungi was found to be higher on a farm managed in an organic manner than on a conventional neighbour. Plant Soil 160:33–40. doi:10.1007/BF00150343 [DOI]
- Ryan MH, Small DR, Ash JE (2000) Phosphorus controls the level of colonisation by arbuscular mycorrhizal fungi in conventional and biodynamic irrigated dairy pastures. Aust J Exp Agric 40:663–670. doi:10.1071/EA99005 [DOI]
- Schwarzott D, Walker C, Schüßler A (2001) Glomus, the largest genus of the arbuscular mycorrhizal fungi (Glomales) is nonmonophyletic. Mol Phylogenet Evol 21:190–197. doi:10.1006/mpev.2001.1007 [DOI] [PubMed]
- Scullion J, Eason WR, Scott EP (1998) The effectivity of arbuscular mycorrhizal fungi from high input conventional and organic grassland and grass-arable rotations. Plant Soil 204:243–254. doi:10.1023/A:1004319325290 [DOI]
- Sjöberg J, Persson P, Martensson A, Mattsson L, Adholeya A, Alström S (2004) Occurrence of Glomrycota spores and some arbuscular mycorrhiza fungal species in arable fields in Sweden. Acta Agric Scand B 54:202–212. doi:10.1080/09064710410030294
- Stribley DP (1990) Mycorrhizal associations and their significance. In: Rabinowitch HD, Brewster JL (eds) Onions and allied crops, Vol. II. CRC, Boca Raton, pp 85–101
- Swofford DL (2003) PAUP*: Phylogenetic analysis using parsimony (* and other methods), version 4.0b 10. Sinauer Associates, Sunderland, Massachusetts
- Sýkorová Z, Wiemken A, Redecker A (2007) Cooccurring Gentiana verna and Gentiana acaulis and their neighboring plants in two Swiss upper montane meadows harbor distinct arbuscular mycorrhizal fungal communities. Appl Environ Microbiol 73:5426–5434. doi:10.1128/AEM.00987-07 [DOI] [PMC free article] [PubMed]
- Ter Braak CJF, Smilauer P (2004) Program CANOCO Version 4.53. Plant Research International, Wageningen University and Research Centre. Box 100:6700. AC Wageningen, the Netherlands
- Van der Heijden MGA, Streitwolf-Engel R, Riedl R, Siegrist S, Neudecker A, Ineichen K, Boller T, Wiemken A, Sanders IR (2006) The mycorrhizal contribution to plant productivity, plant nutrition and soil structure in experimental grassland. New Phytol 172:739–752. doi:10.1111/j.1469-8137.2006.01862.x [DOI] [PubMed]
- Wang YY, Vestberg M, Walker C, Hurme T, Zhang X, Lindström K (2008) Diversity and infectivity of arbuscular mycorrhizal fungi in agricultural soils of the Sichuan Province of mainland China. Mycorrhiza 18:59–68. doi:10.1007/s00572-008-0161-x [DOI] [PubMed]
- Wirsel SGR (2004) Homogeneous stands of a wetland grass harbour diverse consortia of arbuscular mycorrhizal fungi. FEMS Microbiol Ecol 48:129–138. doi:10.1016/j.femsec.2004.01.006 [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Below is the link to the electronic supplementary material.
Number of fields which harbor specific AMF phylotype per surveyed year (DOC 48.5 KB)
Presence or absence of AMF phylotypes in each onion field (DOC 46.5 KB)
Phylogenetic analysis of 18S rDNA sequences of Glomus-A (DOC 313 KB)