Summary
Myogenic regulatory factors of the Myod family (MRFs) are transcription factors essential for mammalian skeletal myogenesis. However, the roles of each gene in myogenesis remain unclear, owing partly to genetic linkage at the Myf5/Mrf4 locus and to rapid morphogenetic movements in the amniote somite. In mice, Myf5 is essential for the earliest epaxial myogenesis, whereas Myod is required for timely differentiation of hypaxially derived muscle. A second major subdivision of the somite is between primaxial muscle of the somite proper and abaxial somite-derived migratory muscle precursors. Here, we use a combination of mutant and morphant analysis to ablate the function of each of the four conserved MRF genes in zebrafish, an organism that has retained a more ancestral bodyplan. We show that a fundamental distinction in somite myogenesis is into medial versus lateral compartments, which correspond to neither epaxial/hypaxial nor primaxial/abaxial subdivisions. In the medial compartment, Myf5 and/or Myod drive adaxial slow fibre and medial fast fibre differentiation. Myod-driven Myogenin activity alone is sufficient for lateral fast somitic and pectoral fin fibre formation from the lateral compartment, as well as for cranial myogenesis. Myogenin activity is a significant contributor to fast fibre differentiation. Mrf4 does not contribute to early myogenesis in zebrafish. We suggest that the differential use of duplicated MRF paralogues in this novel two-component myogenic system facilitated the diversification of vertebrates.
Keywords: Muscle, Zebrafish, Myosin, Slow, Fibre, Fast, Head, Fin, mrf4 (myf6), myod, myf5, Myogenin, Hedgehog, prdm1, pax3, meox1, hsp90, mef2d
INTRODUCTION
Motility is a key determinant of evolutionary success. During vertebrate evolution, somitic, appendicular and head muscle has undergone significant adaptation as fish evolved and moved onto land. One major division shared by all vertebrates is the dorsoventral segregation of the somitic myotome into epaxial and hypaxial halves with distinct innervation. A second division of somitic myogenesis that emphasises the attachments of muscles to skeletal/connective tissue elements of somitic (primaxial) or lateral plate (abaxial) origin has also been recognised (Burke and Nowicki, 2003). Human diseases and murine genetic manipulations that affect specific muscle groups suggest that such differences in muscle form are underlain by distinct populations of myogenic cells (Sambasivan and Tajbakhsh, 2007). How muscle diversified at a molecular evolutionary level is unclear, but changes in the utilisation and function of myogenic regulatory factors of the myod family (MRFs) are possible contributors. Analysis of murine MRF function has revealed that Myf5, Myod and Mrf4 (Myf6) are not, individually, essential for viability (Kassar-Duchossoy et al., 2004; Kaul et al., 2000; Rudnicki et al., 1992; Zhang et al., 1995). Yet mice lacking function of all three of these genes fail to make skeletal muscle (Kassar-Duchossoy et al., 2004; Rudnicki et al., 1993). Lack of one MRF usually delays myogenesis until another MRF is expressed, although there is debate about the cell autonomy of compensation (Braun and Arnold, 1996; Braun et al., 1994; Gensch et al., 2008; Haldar et al., 2008; Kassar-Duchossoy et al., 2004; Tajbakhsh et al., 1997). Owing to the extensive growth of secondary muscle fibres, it has been difficult to determine whether specific differentiated muscle cells are lacking in single MRF mutant adults. An exception is the lack of tail epaxial muscle in Myf5-null alleles (Kassar-Duchossoy et al., 2004), which emphasises one overriding theme to emerge from murine MRF knockout studies of somite myogenesis: that Myf5 function is initially required for myogenin (Myog) and Myod expression and myogenesis in the epaxial somite domain (Kablar et al., 1997; Kassar-Duchossoy et al., 2004; Tajbakhsh et al., 1997). Perturbation of the Myf5/Mrf4 locus also prevents early hypaxial Myod expression, possibly owing to loss of Mrf4 function (Kassar-Duchossoy et al., 2004; Tajbakhsh et al., 1997). Myod function is required for early hypaxial myogenesis, which is probably dependent on Pax3 (Kablar et al., 1997; Tajbakhsh et al., 1997). Consistent with this, Myod is the only MRF thought to be able to drive myogenesis in the absence of any other MRF, but is not efficient (Kassar-Duchossoy et al., 2004; Rawls et al., 1995; Valdez et al., 2000). The consensus view remains that the combined activity of several MRFs promotes myogenesis (Weintraub, 1993).
Null mutation in Myog, however, is perinatal lethal owing to a failure of terminal differentiation of a large proportion of myoblasts in some muscles. Mutants have significant myogenesis at early stages of development and there is efficient terminal differentiation of Myog-null myoblasts in cell culture (Hasty et al., 1993; Nabeshima et al., 1993; Venuti et al., 1995). Early reports presumed that Mrf4, being structurally related to Myog and abundantly expressed in differentiated muscle fibres, permitted this differentiation. However, pairwise ablation of Myog and either Mrf4, Myod or Myf5/Mrf4 in mice did not worsen the Myog-null phenotype (Rawls et al., 1995; Rawls et al., 1998). The role and relationship of Myog to the other MRFs remains ambiguous.
Here, we turn to zebrafish in the hope of finding shared and divergent roles for individual MRFs in myogenesis. Each zebrafish somite generates at least four populations of muscle fibres within the first 15 hours of somitogenesis, a timing and diversity that are akin to amniote somite myogenesis (Kahane et al., 2001; Kassar-Duchossoy et al., 2004). Two Hedgehog (Hh)-dependent medial mononucleate slow muscle fibre types derive from the myf5- and myod-expressing adaxial cells lying adjacent to the notochord (Blagden et al., 1997; Coutelle et al., 2001; Weinberg et al., 1996; Wolff et al., 2003). Approximately three muscle pioneer fibres remain at the dorsoventral midline, whereas ∼20 superficial slow fibres migrate to the lateral myotome surface (Devoto et al., 1996). More lateral paraxial cells within the somite also express the MRFs myf5 and myod (Coutelle et al., 2001; Weinberg et al., 1996) and give rise to two kinds of multinucleate fast fibre: the Fgf8-dependent lateral fast fibres and the Fgf8-independent medial fast fibres, a subset of which becomes the fast Engrailed-expressing cells as a result of later Hh signalling (Groves et al., 2005; Wolff et al., 2003). Both slow and fast lineage cells also express myog and mrf4 (Hinits et al., 2007; Weinberg et al., 1996). Using predicted null mutations in myf5 and mrf4 and morpholino antisense oligonucleotides (MOs) to knockdown myod and myog function, we show that distinct muscle cell lineages use different combinations of MRFs to drive myogenesis. myf5 mutants die during larval growth, whereas mrf4 mutants are viable. Medial slow and fast fibres require either Myf5 or Myod to undergo myogenesis. Lateral myogenesis is driven by Myod alone. No Mrf4-initiated myogenesis was observed. Our data show that Myf5 and Myod are likely to have been the ancestral triggers of vertebrate myogenesis. However, neither Myf5/Myod- nor Myod/Myog-requiring cell types are restricted to epaxial or hypaxial somite domains. We speculate that the common ancestor of teleosts and amniotes had several kinds of somite skeletal muscle cell that utilised Myf5 and Myod differently.
MATERIALS AND METHODS
Zebrafish lines and maintenance
Wild-type and transgenic lines Tg(acta1:GFP)zf13 (Higashijima et al., 1997) and Tg(mylz2:GFP) (Moore et al., 2007) were maintained on King's College wild-type background, and staging and husbandry were as described (Westerfield, 1995). myf5hu2022 and mrf4hu2041 were on a Tl background and were genotyped by sequencing of PCR products amplified from fin clip or embryo genomic DNA using primers 5′-GCAACTTGCGCTTCGTCTCC-3′ and 5′-CATCGGCAGGCTGTAGTAGTTCTC-3′ for myf5, and 5′-TGAATCTGAAGCCCCGCAAC-3′ and 5′-ATTGCTCTGCTCCTGCTCATCC3′-for mrf4.
In situ mRNA hybridisation, immunohistochemistry and western analysis
In situ mRNA hybridisation and immunohistochemistry were performed as described (Hinits and Hughes, 2007). The fluorescein- or digoxigenin-tagged probes used were mef2ca and mef2d (Ticho et al., 1996), mrf4 (Hinits et al., 2007), myf5 (Groves et al., 2005), myod and myog (Weinberg et al., 1996), eng1a and eng2a (Ekker et al., 1992), mylz2, tpma and myhz1 (Xu et al., 2000), pax3 (Hammond et al., 2007), meox1 and lbx2 (Neyt et al., 2000), smyhc1 (Bryson-Richardson et al., 2005), actin (actc1, IMAGE 7284336), hsp90a (IMAGE 7259827), prdm1 (Baxendale et al., 2004), twist2 (Morin-Kensicki and Eisen, 1997) and ptc1 (Concordet et al., 1996). Antibodies were against Myog (Devoto et al., 2006), Myod (Hammond et al., 2007), Mef2 (Hinits and Hughes, 2007), MyHC (A4.1025), fast MyHC (EB165) (Blagden et al., 1997), slow MyHC [F59 or S58 (Devoto et al., 1996)] and GFP (Torrey Pines). Western analysis was performed as described (Blagden et al., 1997).
Morpholino knockdown
MOs (Gene-Tools) were injected into 1- to 2-cell stage embryos unless otherwise stated. MO sequences are: myog MO-1, 5′-GCTGGTTTAGAGTCCACCCGCTGTG-3′; myog MO-2, 5′-GGGTTGGTCTTCGAAAAGCTCCATGT-3′; myod, 5′-ATATCCGACAACTCCATCTTTTTTG-3′; and myf5, 5′-GATCTGGGATGTGGAGAATACGTCC-3′.
RESULTS
Myf5 cooperates with Myod to drive slow myogenesis
Knockdown of both Myf5 and Myod in zebrafish ablates adaxial myog, mrf4, slow myosin heavy chain 1 (smyhc1) and desmin expression and slow muscle differentiation (Hammond et al., 2007; Hinits et al., 2007; Maves et al., 2007) (Fig. 1). As myog is expressed in adaxial cells, we tested its requirement for slow myogenesis in morphants. Two non-overlapping myog MOs blocked Myog protein accumulation as detected by western analysis and ablated nuclear Myog immunoreactivity (see Fig. S1 in the supplementary material). At the 15-somite stage (15 ss), myog morphants showed no change in any of the adaxial markers examined (Fig. 1). A myod MO also reduced Myod accumulation (see Fig. S1 in the supplementary material) (Hammond et al., 2007). Injection of sufficient doses of myod MO reduced myog expression and mildly delayed, but did not prevent, smyhc1 expression and slow muscle differentiation (Fig. 1). Expression of the thin filament genes α-actin and α-tropomyosin (tpma) was not affected [Fig. 1; as for desmin (Maves et al., 2007)]. Myf5 knockdown did not affect any of the markers examined (Fig. 1). Although mrf4 is Myf5/Myod-dependent, it is not expressed until late in slow muscle differentiation and, therefore, does not trigger slow myogenesis (Hinits et al., 2007) (see below). Pairwise loss-of-function of myog with either myf5 or myod did not prevent slow fibre formation (see below). We conclude that either Myf5 or Myod alone can drive slow myogenesis, but that Myod activity might be rate limiting in the wild-type condition.
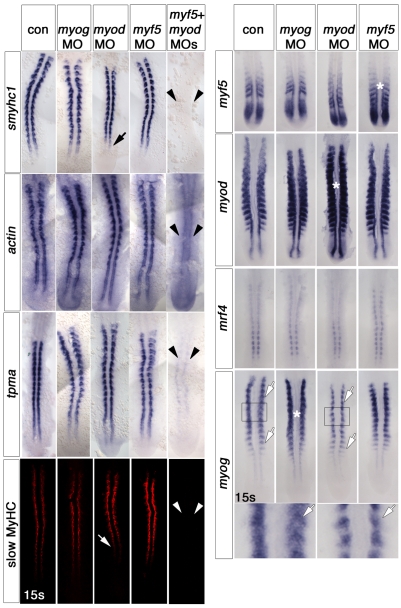
Fig. 1.
Myf5 or Myod are required for slow fibre formation. Dorsal flatmounts of zebrafish embryos injected with the indicated MOs and uninjected controls (con), analysed at the 13- to 15-somite stage by whole-mount immunohistochemistry for MyHC (red) or in situ mRNA hybridisation for the indicated mRNAs (blue). Anterior to top. (Left panels) myf5+myod MOs ablated all slow myosin heavy chain (smyhc1 and slow MyHC, arrowheads), but did not completely prevent actin or tpma expression (arrowheads). myod MO slightly delayed smyhc mRNA and MyHC accumulation (arrows), whereas other single MOs had little effect. (Right panels) Single MRF MOs had no effect on adaxial expression of the other MRFs. However, the myod MO reduced myog signal in fast muscle precursors (white arrows). The box region is shown magnified beneath. Note the increase in signal for each MRF caused by its cognate MO (asterisks). s, somites.
Myf5 is dispensable for embryonic and larval development
Injection of both myf5 and myod MOs blocked expression of late myogenesis markers and diminished, but did not entirely eliminate, expression of mRNAs encoding the earlier-expressed thin filament proteins (Fig. 1) (Hinits et al., 2007). As we have no antibody to Myf5, we were concerned that the myf5 MO might not ablate production of all Myf5 protein. The zebrafish strain myf5hu2022, in which cysteine 59 of the myf5 gene has changed to a stop codon, was obtained from the Sanger Centre TILLING screen (Stemple, 2004) (see Fig. S2 in the supplementary material). myf5hu2022 is a predicted null, truncating Myf5 upstream of the basic helix-loop-helix (bHLH) domain (Fig. 2A). Heterozygous myf5+/hu2022 incrosses yielded clutches of normal size and the survival of embryos and larvae was good throughout early development. No gross morphological defects were observed and larvae moved and fed normally (Fig. 2B). The similarity of myf5hu2022/hu2022 (hereafter called myf5hu2022) and myf5 morphants suggests that the latter represent strong loss-of-function (see below).
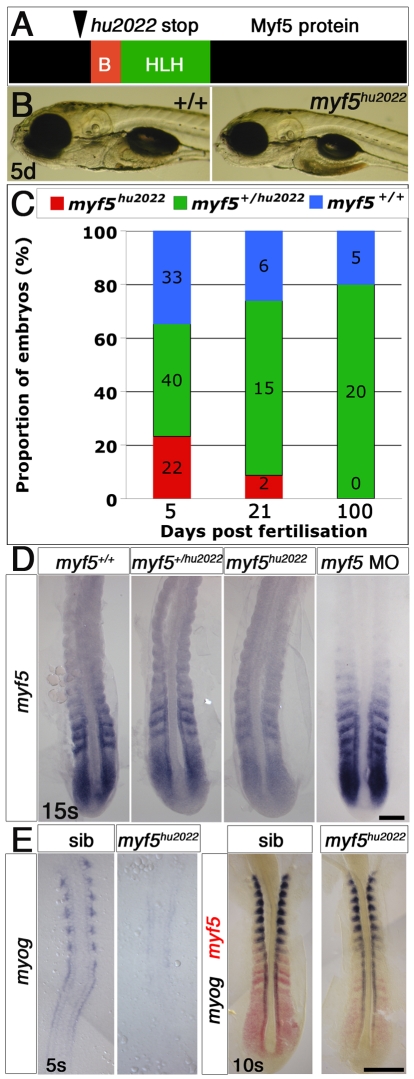
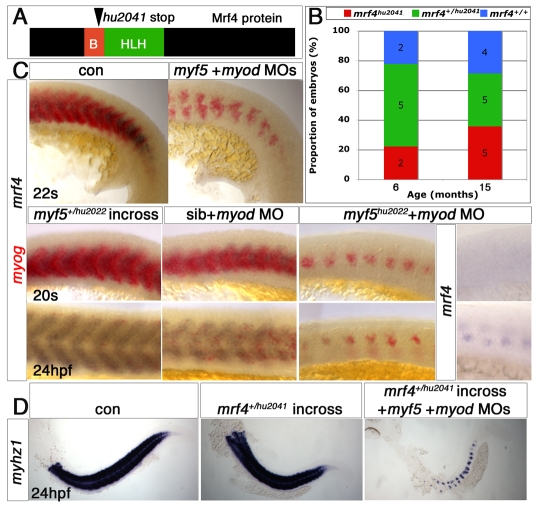
Fig. 2.
myf5hu2022 mutants appear normal as embryos but fail to mature into adults. (A) Myf5 truncation predicted from the myf5hu2022 allele. The location of the introduced stop codon is shown; B, basic domain; HLH, helix-loop-helix domain. (B) Genotyped zebrafish embryos from a myf5+/hu2022 carrier incross show no phenotype at 5 days post-fertilisation (5d) as compared with the wild type (+/+). (C) Survival at various ages of fish of each genoptype obtained from myf5+/hu2022 incrosses. The numbers of embryos are shown. (D) In situ mRNA hybridisation for myf5 revealed mildly reduced signal in putative myf5+/hu2022 (13/28) and lower levels in putative myf5hu2022/hu2022 (9/28) embryos. No such effect was observed in myf5 morphants (0/27). (E) Embryos from an myf5+/hu2022 incross lack adaxial myog mRNA early (15/49), and later have a mild delay in myog mRNA accumulation in fast muscle precursors correlated with reduced myf5 mRNA (20/74). Scale bars: 100 μm in D; 150 μm in E.
myf5hu2022 homozygotes survive to adulthood less well than their siblings; no adults have been found (Fig. 2C). Despite the lack of early muscle defects, in situ mRNA hybridisation of myf5 at 15 ss revealed a reduction in myf5 mRNA accumulation, presumably reflecting nonsense-mediated decay, which was not observed in myf5 morphants (Isken and Maquat, 2007) (Fig. 2D). We conclude that a wild-type zygotic myf5 locus is not essential for early myogenesis, but might confer a selective advantage during growth and maturation of fish into adulthood.
Murine Myf5 is essential for Myog expression and the first epaxial myogenesis (Braun et al., 1994; Kassar-Duchossoy et al., 2004; Kaul et al., 2000). Analysis of zebrafish myf5hu2022 mutants revealed a 1- to 2-hour delay in early adaxial myog expression and a ∼30-minute delay in expression in fast muscle precursors (Fig. 2E). Thus, Myf5 contributes to the rate of myogenesis.
We used myf5hu2022 to examine the role of Myf5 in muscle formation. All embryos from myf5hu2022 heterozygote crosses expressed myod, myog, mrf4, mef2ca, pax3, prdm1, ptc1, smyhc1, α-actin and tpma genes normally at 15 ss (see Fig. S2C in the supplementary material; data not shown). Where analysed, injection of myf5 MO gave the same result (Fig. 1; data not shown). When myod MO was injected into a myf5+/hu2022 incross, 25% of the resulting embryos lacked expression of slow terminal differentiation markers, confirming the previous conclusion based on myf5 MO (Fig. 3A; 59/243, for all quantitative data see Table S1 in the supplementary material). Thus, Myf5 and Myod cooperate to drive adaxial slow myogenesis in zebrafish.
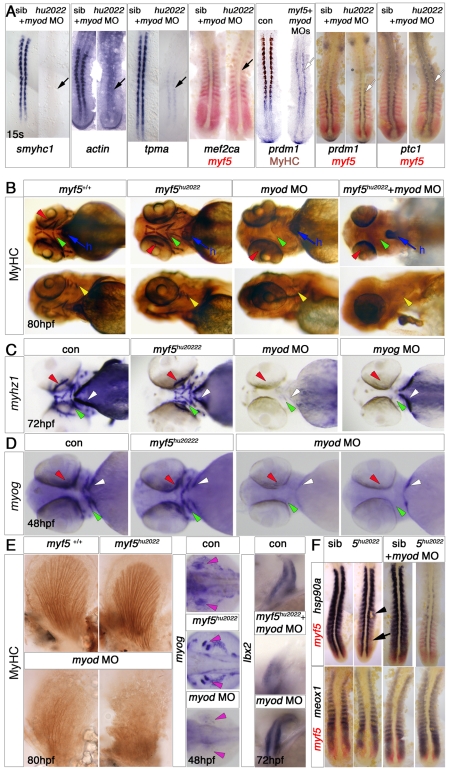
Fig. 3.
Myf5 requirement differs between somitic, head and fin muscle development. In situ mRNA hybridisation and/or MyHC immunohistochemistry of control and morphant wild-type or myf5+/hu2022 incross embryos. Dorsal flatmounts with anterior to top (A,F), or whole-mounts with anterior to left in ventral (B, upper row, C,D), lateral (B, lower row) or dorsal (E) view. (A) myf5hu2022 mutants injected with myod MO show a loss of the adaxial slow muscle differentiation markers smyhc1, actin, tpma and mef2ca and MyHC immunostaining (black arrows), but retain other adaxial markers such as prdm1 and ptc1 (white arrows). (B-D) Head muscles are present in 80 hpf myf5hu2022 mutants but are missing from embryos injected with myod (B,C) or myog MO (C). Arrowheads indicate lack of ventral (green), dorsal (yellow), extra-ocular (red) and sternohyoideus (sh, white) muscles. All embryos show normal heart (h, blue arrow). Note the presence of sternohyoideus in myog morphants. All embryos from a myf5+/hu2022 incross have normal expression of myhz1 at 72 hpf (C) and myog at 48 hpf (D) in the head. myod morphants lack myog mRNA (19/21), although sternohyoideus was still detected in some embryos (6/21). (E) In pectoral fin, myog mRNA in muscle masses (pink arrowheads) and MyHC are ablated or reduced by loss of Myod, but not by loss of Myf5. Ibx2 mRNA is unaffected by myod MO. (F) Expression of hsp90a is delayed in fast muscle precursors (arrow) of the myf5hu2022 mutant and recovers in somites (arrowhead), but is drastically reduced throughout the axis in myf5hu2022 mutants injected with myod MO, both in slow and fast precursors. meox1 expression is unchanged in fast precursors.
MRFs are not required for adaxial prdm1 expression
Which step in slow myogenesis is affected by loss of Myf5 and Myod function? Expression of most regulatory genes implicated in early myogenesis, such as myog and mef2d (presomitic adaxial markers) and mrf4 and mef2ca (adaxial slow fibre markers), is ablated in double morphants or myf5 mutant;myod morphants (Fig. 1, Fig. 3A; data not shown) (Hinits and Hughes, 2007; Hinits et al., 2007; Weinberg et al., 1996). By contrast, expression of prdm1 and ptc1 mRNAs, which encode proteins required for proper Hh-dependent slow fibre differentiation (Baxendale et al., 2004; Concordet et al., 1996), was unaffected by loss of Myf5 and Myod function, showing that the slow muscle precursor cells were still present (Fig. 3A). Thus, Myf5 and Myod drive a specific module of gene expression within slow muscle precursors.
Myod, but not Myf5, drives cranial and pectoral fin myogenesis
It has been reported, based on MO knockdown of myf5, that Myf5 is required for myogenesis of specific head muscles and brain morphogenesis (Chen and Tsai, 2002; Lin et al., 2006). We analysed cranial, fin and hypaxial myogenesis in myf5hu2022 embryos and larvae and did not detect any defects, consistent with the viability of some 3-week-old fry (Fig. 3B-D). Moreover, using the myf5 ATG MO, with a sequence identical to that reported previously (Lin et al., 2006), we only obtained head defects at high, probably toxic, MO concentrations (data not shown). As maternally expressed myf5 was not detected (Chen et al., 2001; Coutelle et al., 2001), myf5 is dispensable for non-somitic myogenesis in zebrafish embryos.
We next asked whether Myod or Myog is required for cranial myogenesis. Most myod morphants completely lacked head muscle, myhz1 and myog expression and myosin accumulation prior to 72 hours post-fertilisation (hpf), without accompanying gross morphological defects of the brain and head (Fig. 3B-D). Some myod morphants had small amounts of residual muscle, possibly arising from incomplete knockdown (see below). Injecting myod MO into myf5hu2022 mutants gave a distribution of phenotypes indistinguishable from that upon injection into siblings (Fig. 3C,D). myog is highly expressed in head muscle anlage at 48 hpf, and myog morphants showed defects in head myogenesis similar to myod morphants, with the exception that the sternohyoideus was less affected (Fig. 3D). mrf4 is not expressed in head muscle until well after 48 hpf (Hinits et al., 2007). We conclude that Myod is the primary MRF driver of myog expression and early myogenesis in cranial muscles.
Unlike most cranial muscle, fin muscle is somite derived and composed exclusively of fast fibres that express the lbx2 gene (previously known as lbx1 or lbx1h) at early stages (Neyt et al., 2000; Patterson et al., 2008; Wotton et al., 2008). Myod knockdown prevented myog mRNA and myosin accumulation but not lbx2 expression in fin buds, whereas myf5hu2022 mutants appeared normal (Fig. 3E). Therefore, Myod is the major MRF required for fast pectoral fin myogenesis.
Myf5 drives hsp90a expression in fast muscle precursors
We have shown that Myod is important for the formation of certain fast fibres in cranial, fin and somitic muscle. Yet myf5 is the first MRF expressed in early presomitic mesoderm and tailbud and persists in fast muscle precursors of the posterior somite (Chen et al., 2001; Coutelle et al., 2001; Stellabotte et al., 2007). Loss of myf5 function decreased expression of hsp90a in the anterior presomitic mesoderm, but other markers of this tissue, such as meox1 and myod, were unaffected (Fig. 3F; see Fig. S2C in the supplementary material). Anteriorly, in the nascent somites, hsp90a mRNA expression was upregulated, dependent on Myod activity (Fig. 3F). No defect in late muscle gene expression was found in myf5 mutants. Hsp90a is required for proper sarcomere assembly (Du et al., 2008; Hawkins et al., 2008). Thus, early Myf5 function may prime the somite for fast myogenesis, but is not essential for it.
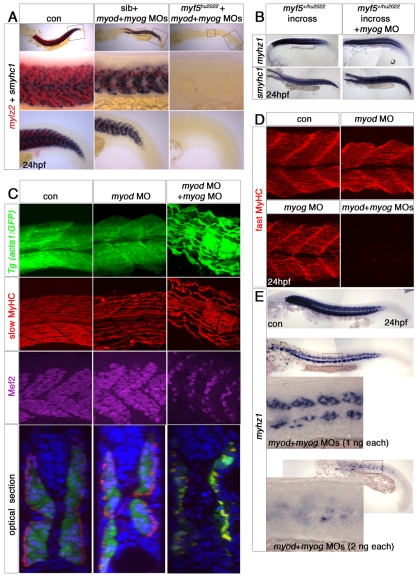
Myf5 and Myod are essential for fast myogenesis
myf5 and myod double morphants lacked myog mRNA at 10 ss and muscle at 15 ss (Hammond et al., 2007; Maves et al., 2007) (Fig. 1, Fig. 4A). By 24 hpf, however, small amounts of fast muscle formed in the medial somite (Fig. 4B,C). Knockdown of Myod in myf5hu2022 homozygotes further reduced fast muscle (Fig. 4B,D). The addition of myf5 MO did not worsen the phenotype of myod MO; myf5hu2022 mutants. Thus, the myf5 mutant revealed that myf5 morphants are hypomorphic.
Fig. 4.
Myf5 or Myod is required for medial fast myogenesis. In situ hybridisation for myog, myhz1, tpma, mylz2 or mef2d or immunohistochemistry for the indicated myosins in wild-type or myf5+/hu2022 incross zebrafish injected with the indicated MOs. Lateral views, anterior to left unless otherwise stated. (A) myog expression was delayed in embryos injected with myf5+myod MOs. Dorsolateral views, anterior to top. (B) MyHC was reduced in both epaxial and hypaxial domains in myod MO embryos, grossly reduced in double myf5+myod morphants and further diminished in myf5hu2022 mutants injected with myod MO. Note the complete absence of residual cells in some individual somites (dotted outline). (C) Residual differentiated muscle in myf5+myod morphants expressed fast but not slow MyHC at 24 hpf. Lateral view of confocal stack of somites 9-10. (D) Fast muscle markers were indistinguishable in 24 hpf myf5hu2022 homozygous mutants and their siblings (left column), reduced by myod MO in siblings (middle) and almost eliminated by myod MO in myf5hu2022 mutants (right). Inset illustrates that myf5 MO does not worsen the myod MO plus myf5hu2022 phenotype. mylz2 mRNA accumulation (red, arrowheads) and myog downregulation (blue) are delayed at ∼20 ss in fast muscle. (Bottom panels) Transverse sections of double knockdowns show residual fast muscle (arrowheads) immediately lateral to adaxial and/or twist2-expressing sclerotomal cells (arrows). (E) (Left, top) mef2d mRNA initiates normally in pbcs in the myod morphant, but is restricted to the medial region of rostral somites (yellow brackets). In myf5+myod MOs, mef2d mRNA is absent. Dorsal flatmounts, anterior to top. (Left, bottom) Lack of both myf5 and myod ablates mef2d expression. Lateral views, anterior to top. (Right) By 22 ss, transverse sections (at the level of the bar on lateral flatmounts) show reduction of mef2d mRNA (purple) in myod or myog, but not in myf5, single morphants. Mef2 position is shown relative to slow fibres (brown). Note the expression of mef2d lateral to migrating slow fibres in the control and in myf5 and myog, but not myod, morphants (arrows).
It is likely that myod morphants also retain low levels of Myod function, even though we detected little Myod protein (see Fig. S1 in the supplementary material). In the myf5hu2022 homozygote background, myod MO either completely ablated or greatly reduced fast muscle in individual somites. However, embryos entirely lacking fast muscle were never observed; a few residual fast muscle cells were present in the medial region of many somites, but lateral to undifferentiated cells that presumably represent adaxial cells or sclerotome (Fig. 4D). Residual fast muscle cells expressed myog by 20 ss, and went on to differentiate and accumulate myhz1, tpma and mylz2 mRNA at 24 hpf, but did not elongate or fuse (Fig. 4A,D). Lower doses of myod MO left more fast muscle in each somite. Therefore, the level of Myod in myf5hu2022 mutants determines the quantity of medial fast cells that can express myog and differentiate. Clearly, the medially located residual muscle required less Myf5/Myod activity than did other fast muscle.
Mrf4 is not required for viability
In the mouse, Mrf4 supports myogenesis of a subset of early muscle cells (Kassar-Duchossoy et al., 2004). Could Mrf4 activity account for the residual muscle in Myf5 plus Myod knockdown fish? TILLING yielded the mrf4hu2041 allele, a predicted null mutation in which a stop codon in the basic domain eliminates the helix-loop-helix region essential for dimerisation and myogenic function (Fig. 5A; see Fig. S3 in the supplementary material). mrf4hu2041 mutant fish were viable and fertile (Fig. 5B). Myosin accumulation in slow, fast, cranial and fin muscles occurred on time. Among a series of myogenic or muscle-specific genes examined at various developmental stages, none was altered in mrf4hu2041 homozygotes. Although we have no proof that the mrf4hu2041 allele is null, it appears that mrf4 is not an essential gene in zebrafish.
Fig. 5.
Mrf4 has no role in trunk muscle differentiation. (A) Schematic illustrating the location of the mutation encoded by mrf4hu2041. (B) Proportion of mrf4hu2041/hu2041 (red), mrf4hu2041/+ (green) or mrf4+/+ (blue) in progeny from heterozygote crosses. The number of embryos of each genotype is shown. Homozygote mutants survive well and without obvious defects. (C) Whole-mount in situ mRNA hybridisation of myf5+myod morphants or myod MO-injected myf5hu2022 mutants reveal that residual medial fast fibres express myog (red) but not mrf4 (purple) at 22 ss, but accumulate mrf4 mRNA after terminal differentiation at 24 hpf. Lateral view, anterior to left. (D) In situ mRNA hybridisation for myhz1 (purple) in embryos from a mrf4+/hu2041 incross reveals that injection of myf5+myod MOs fails to ablate all muscle irrespective of mrf4 genotype.
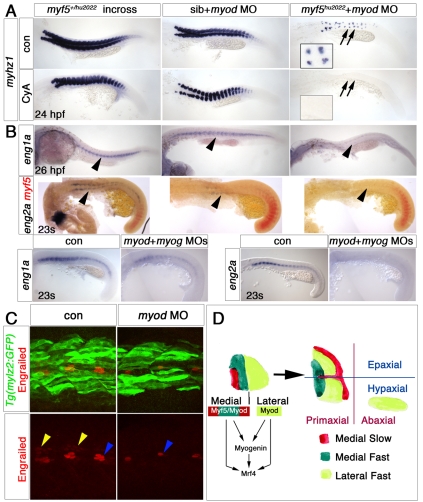
In myf5;myod morphants, which lack slow fibres, mrf4 mRNA was not detected in the residual myog-expressing cells (Fig. 5C). To eliminate the possibility that low-level Mrf4 drives residual medial fast myogenesis, we injected myf5 and myod MOs into mrf4hu2041mutants (owing to linkage of myf5 and mrf4, double mutants could not be made). No additional loss of muscle was observed above that caused by the MOs in a wild-type background (Fig. 5D). Thus, Mrf4 does not drive the residual fast, or apparently any other, myogenesis prior to 24 hpf.
Both epaxial and hypaxial somitic compartments have two modes of fast myogenesis
In mouse somites, Mrf4 drives some lateral/hypaxial myogenesis, but Myod is also involved in this region (Kassar-Duchossoy et al., 2004). In fish, Myod is the major myogenic gene in the lateral somite. Knockdown of Myod alone depletes lateral muscle in both epaxial and hypaxial somites (Fig. 4B,D) (Hammond et al., 2007). Expression of Myod was first observed in the lateral somite in posterior border cells (pbcs), but these cells only accumulated myog and mef2d mRNAs at 15 ss, just prior to becoming fast muscle (Fig. 1, Fig. 4E). Myod is required for pbc myog expression (Maves et al., 2007). However, to ablate mef2d mRNA, Myf5, which is transiently expressed in pbcs, also had to be removed (Fig. 4E). Rostrally, however, myod morphants lacked lateral mef2d mRNA, suggesting that without Myod lateral cells cannot express myog, maintain mef2d or undergo terminal differentiation (Fig. 4E).
Regulation of myog and mef2d in the medial somite is different. Both Myod and Myf5 are involved in this first fast myogenesis, because myog mRNA was decreased at 10 ss in both myf5 mutants and myod morphants (Fig. 1, Fig. 2E). Yet, neither Myod nor Myf5 is essential for medial fast myogenesis because expression of myog and mef2d was detected medially at 15 ss in single morphants and myf5hu2022 (Fig. 1, Fig. 4E). As the fast muscle differentiation markers mylz2 and myhz1 were almost absent at late somitogenesis stages from embryos lacking Myf5 and Myod (Fig. 4), the simplest interpretation of these observations is that, as in adaxial cells, Myf5 and Myod each contribute to medial fast myogenesis but, given time, either alone is sufficient. Thus, the medial cells that make the first fast muscle in the somite employ a distinct mode of myogenesis involving Myf5 that distinguishes them from lateral fast precursors. Importantly, as with the lateral Myod-dependent mode, the medial Myf5/Myod-dependent mode generates fast muscle in the medial region of both epaxial and hypaxial compartments (Fig. 4B,D).
Myod and Myog drive fast muscle differentiation
Whereas myog is expressed at low levels in slow muscle precursors, it is abundant just before terminal differentiation in both medial and lateral fast precursors, both in wild-type and manipulated embryos (Figs 1 and 4). For example, myog mRNA accumulated prior to mylz2 mRNA in residual medial fast fibres of myod MO-injected myf5hu2022 mutant embryos (Fig. 4D). Upon injecting both myog and myod MOs into myf5hu2022 mutant embryos, all muscle was ablated (Fig. 6A). We conclude that Myog accumulation is essential for residual medial fast fibre differentiation.
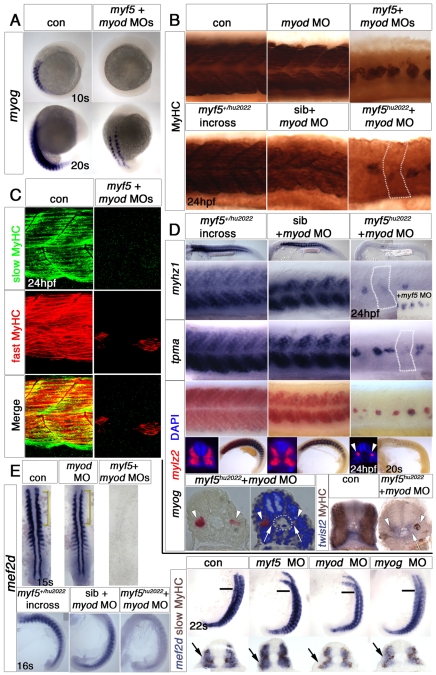
Fig. 6.
Loss of Myod and Myog ablates somitic fast muscle. In situ mRNA hybridisation (A,B,E) or immunofluorescent confocal stacks (C,D) of 24 hpf wild-type (con), Tg(acta1:GFP) or myf5+/hu2022 incrosses injected with the indicated MOs. Lateral views, anterior to left. (A) Magnified regions (boxed) showing that lack of Myf5, Myod and Myog results in loss of all muscle. Knockdown of Myod and Myog results in loss of most fast, but not slow, muscle. Note that mylz2 (red) is weakly expressed in immature slow muscle. (B) Loss of Myog and Myf5 has little effect, if any, on fast myhz1 or slow smyhc1 expression. (C) Injection of myod+myog MOs into Tg(acta1:GFP) fish results in loss of fast muscle, as shown by loss of Mef2 staining and GFP medial to the slow fibre layer accompanied by somitic apoptosis as revealed with DAPI (blue in the optical transverse sections). Elongated mononucleate slow fibres differentiate and express GFP, slow MyHC and Mef2. myod morphants confirm the lateral reduction of fast muscle mass. (D) myod+myog MOs ablate fast MyHC in somites, whereas single MOs have a lesser effect. (E) Titration of myod+myog MOs progressively diminishes residual myhz1-expressing cells. The lower panels are high-magnification images of mid-trunk somites (boxed).
Could activation of myog expression in the medial somite of myf5 mutant;myod morphant embryos occur independent of residual Myod activity? We think not, for three reasons. First, although myog mRNA appears in medial fast fibres prior to their differentiation, Myog knockdown alone has no detectable effect on fast myogenesis (Fig. 1, Fig. 6D). Second, knockdown of Myf5 and Myod prevents all early myog expression, including that in the medial somite region that is destined to make the first fast muscle. Third, the occurrence of individual somites entirely lacking muscle in myf5 mutant;myod morphant embryos argues against a specific myog-dependent population in the medial region of each somite. We propose that residual Myod activity drives myog expression, but proof will await a null myod mutant.
We next tested whether Myod or Myf5 alone can drive fast myogenesis. Injection of myog MO into myf5hu2022 mutant embryos did not detectably reduce muscle (Fig. 6B). By contrast, when myog and myod MOs were co-injected into α-actin:GFP transgenic embryos, essentially all fast muscle was ablated (Fig. 6C,D). Slow muscle fibres were still present in normal numbers and accumulated GFP, slow Myosin heavy chain (MyHC) and Mef2. However, the somewhat U-shaped somites had shorter, disorganised slow fibres and less Engrailed, a marker of muscle pioneer cells (Fig. 6C, Fig. 7C). Both myhz1 transcript and fast MyHC were essentially absent from myog;myod morphants (Fig. 6D). Therefore, Myod drives fast muscle differentiation in the absence of Myog. Moreover, Myf5 drives significant fast muscle differentiation in the absence of Myod through activation of myog expression.
Fig. 7.
Residual fast fibres after Myf5 plus Myod knockdown are hedgehog dependent. In situ mRNA hybridisation (A,B) and immunofluorescent confocal stacks (C) of 23 ss, 24-26 hpf wild-type, Tg(mylz2:GFP) or myf5+/hu2022 incross embryos. Lateral views, anterior to left. (A) Cyclopamine (CyA) treatment of embryos from a myf5+/hu2022 incross injected with myod MO ablates residual myhz1 expression (arrows, magnified in insets). Fast muscle still forms in the absence of Myod and Hh signals. Note the altered somite shape, paralleling the absence of slow fibres. (B) Expression of eng1a and eng2a (arrowheads) is missing from myf5hu2022 mutants injected with myod MO and is reduced in myod+myog double morphants. Eng1a is not affected by myf5 mutation (revealed by weak myf5 mRNA expression) or myod MO alone, although eng2a is drastically reduced in the myod morphant. (C) Tg(mylz2:GFP) embryos injected with myod MO show reduction of both the weak Engrailed protein in MFF nuclei (yellow arrowheads) and the strong Engrailed expression of muscle pioneers (blue arrowheads). (D) Schematic illustrating how the cells generated in medial and lateral modes of myogenesis in the early zebrafish somite (left) each contribute to several later compartments (right). Note that dermomyotomal contributions are not defined because cell lineage is largely unknown.
Hedgehog signalling can drive medial fast myogenesis
A unique population of medial fast fibres, defined by weak Engrailed expression and previously designated MFFs, are dependent on midline Hh signalling at relatively late stages (Wolff et al., 2003). We therefore examined the Hh dependence of our residual medial fast cells. The Hh-signal-blocking drug cyclopamine completely ablated residual myhz1 expression in myf5hu2022 mutants injected with myod MO (Fig. 7A). Thus, Hh can promote medial fast cell differentiation, perhaps by enhancing residual Myod activity.
Hh is not required for most medial fast myogenesis. Treatment of myod morphants with cyclopamine produced little if any change in myhz1 expression (Fig. 7A). Thus, Myf5 and Myog can trigger fast muscle differentiation without Hh.
We could not detect Engrailed in residual fast fibres in myf5hu2022 mutants injected with myod MO or in myf5;myod double morphants (Fig. 7B; data not shown). Thus, the residual cells are not MFFs, as originally defined. However, the continued presence of unmigrated adaxial cells expressing ptc1 in myf5;myod morphants might reduce Hh signal strength in the region of the residual fast fibres, consistent with the original proposal that MFF Engrailed expression is triggered by enhanced Hh signalling once slow fibres migrate (Wolff et al., 2003). Interestingly, myod MO alone prevented eng2a expression and reduced Engrailed protein in the somite, without obviously affecting eng1a mRNA (Fig. 7B). Moreover, myod;myog double morphants lacked eng2a and had little eng1a mRNA (Fig. 7B).
DISCUSSION
By providing for a first comparison of a vertebrate species with the mouse, our data give insight into the evolution of MRF function. The single MRF gene in Drosophila and C. elegans is not required for muscle differentiation (Balagopalan et al., 2001; Chen et al., 1994). In deuterostomes, MRFs have greater importance: the single Ciona MRF drives myogenesis (Meedel et al., 2007). The duplicated MRFs in mice and zebrafish are essential for skeletal myogenesis, but can partially compensate for one another.
There are two major modes of myogenesis in the early zebrafish (Fig. 7D). In the `medial' mode, Myf5 and Myod act together to drive slow myogenesis and the first fast myogenesis in the medial somite. In the `lateral' mode, Myod is uniquely required for lateral muscle precursor myogenesis, including that in the pectoral fin. We also show that myog expression is dependent on Myf5 and Myod and contributes to both medial and lateral modes of fast muscle differentiation. Zebrafish cranial muscles develop by the Myod-dependent mode, in contrast to murine head muscle. The data reveal strong similarities, but also significant differences, in the myogenic programmes regulating specific fibre populations across vertebrate phylogeny.
Role of zebrafish MRFs
Predicted null mutations in myf5 and mrf4 demonstrate that these genes are not essential for early myogenesis. Loss of Myf5 delayed myog and hsp90a expression in slow and fast muscle precursors, but this soon recovered due to Myod action. In mice, Mrf4, not Myod, appears to permit recovery of epaxial muscle in Myf5loxP/loxP mutants, after a longer delay (Kassar-Duchossoy et al., 2004). In fish, unlike mice, ablation of both myf5 and myod led to loss of essentially all muscle formation. Interpretation of murine Myf5 knockouts has been plagued by cis effects on the linked Mrf4 gene (Kassar-Duchossoy et al., 2004). Because our myf5-null allele is a single base change and can be phenocopied by MO knockdown, we can rule out cis effects on mrf4, which is likewise linked to myf5 in zebrafish. Perhaps unsurprisingly, mutants are more effective than morphants. In myf5hu2022 mutants injected with myod MO, medial Hh signalling permits low levels of residual Myod to activate myog and thereby drive some myogenesis. Mrf4 does not account for residual muscle. We conclude that either Myf5 or Myod is required for myog expression and muscle differentiation throughout the early zebrafish embryo.
As with mice lacking Mrf4, fish mrf4 mutants are viable and fertile, in contrast to a recent MO analysis (Wang et al., 2008). Our mutation is predicted to disrupt both mrf4 transcripts upstream of the HLH domain required for myogenesis (Hinits et al., 2007; Wang et al., 2008). Murine Mrf4 initiates myogenesis in certain hypaxial cells in embryonic somites (Kassar-Duchossoy et al., 2004). Fish mrf4 expression is restricted to differentiated muscle fibres (Hinits et al., 2007). Even though ectopic Mrf4 expression is potently myogenic (see Fig. S3B in the supplementary material), we have no evidence for an early myogenic role of Mrf4 in zebrafish.
Zebrafish lacking myf5 function fail to thrive during larval phases. Although it has been reported that Myf5 is required for the formation of most head muscles (Lin et al., 2006), we observe no such defects in myf5 mutants. Head, fin and trunk muscles appear to form normally in myf5 mutants, as in our myf5 morphants. We suspect, therefore, that the defects in brain and head musculature previously reported after Myf5 knockdown (Chen and Tsai, 2002; Lin et al., 2006) reflect MO toxicity. To date, the cause of death in myf5hu2022 mutants is unknown. Because the myf5hu2022 allele was found by TILLING in mutagenised individuals, mutation of a linked gene could account for the phenotype. The mrf4 coding sequence is, however, wild-type in myf5hu2022 mutants (data not shown), and mrf4 is expressed normally.
MRFs and slow myogenesis
MRFs are an essential part of the slow muscle programme. Knockdown of Myf5 and Myod ablates early slow myogenesis (Hammond et al., 2007; Maves et al., 2007) (Fig. 1). The myf5 mutation confirms this result. Loss-of-function of either myf5 or myod alone delays but ultimately does not prevent slow (and medial fast) myogenesis. Myog is not essential, supporting the original view that the combined activity level of various MRFs drives muscle formation (Weintraub, 1993).
Several genes important for slow myogenesis require little or no MRF. Like myf5 and myod themselves, prdm1 and ptc1 are Hh-dependent genes involved in adaxial slow myogenesis (Baxendale et al., 2004; Concordet et al., 1996; Coutelle et al., 2001; Koudijs et al., 2008; Weinberg et al., 1996). In MRF knockdown embryos, adaxial prdm1 expression is normal at 15 ss, apparently recovering from an earlier defect (Liew et al., 2008). As Prdm1 is not required for MRF expression (Baxendale et al., 2004) (our unpublished observations), Hh signalling may bifurcate, promoting MRF expression to trigger muscle formation and prdm1 to drive slow character. The reduction of Engrailed in myod morphants (Fig. 7) suggests that MRF activity might also contribute to aspects of differentiated slow fibre character.
Differential MRF activity in medial and lateral fast myogenesis
In wild-type fish, terminal differentiation of fast fibres is delayed until well after somite border formation and follows myog expression in fast precursors (Blagden et al., 1997; Devoto et al., 1996; Weinberg et al., 1996; Xu et al., 2000). Maves et al. (Maves et al., 2007) showed that Myod promotes timely myog and myhz1 expression in fast muscle precursors in vivo, which we confirm. However, knockdown of Myod alone does not prevent fast muscle differentiation (Hammond et al., 2007). To ablate fast muscle it is necessary to knockdown both Myod and Myf5, which prevents myog expression. Indeed, knockdown of Myod and Myog ablates fast muscle differentiation, but leaves slow fibres intact. As myf5 mRNA accumulates only transiently as somites form, it seems that myog is a target of both Myf5 and Myod that amplifies MRF activity during fast myogenesis, as first speculated for specific muscle lineages by Weintraub (Weintraub, 1993).
Lateral somite myogenesis is distinct from medial based on (1) a specific requirement for Myod to drive myog expression and trigger differentiation (Hammond et al., 2007; Maves et al., 2007), (2) the differential requirement for Fgf signalling (Groves et al., 2005; Hamade et al., 2006) and (3) the finding that myf5 and myod sequentially activate mef2d expression in lateral cells just prior to their differentiation. Mef2d is not essential for fast myogenesis (Hinits and Hughes, 2007). We hypothesise that, together with other Myod targets, Mef2d works to promote lateral myogenesis in fish, as suggested from studies in culture (Penn et al., 2004).
Expression of both myog and mef2d persists in medial somite cells after Myod knockdown, but is lost if Myf5 is also removed. Medial fast fibre precursors require the lowest levels of Myf5 and Myod. The Hh dependence of residual myogenesis in myf5hu2022;myod morphants suggests that Hh promotes Myod-driven myog expression. Hh signalling is required in several aspects of fast fibre differentiation (Henry and Amacher, 2004; Wolff et al., 2003). Perhaps some medial fast fibres depend on two phases of Hh action for their differentiation, analogous to the two phases of Hh action in slow myogenesis (Wolff et al., 2003).
Myod drives fin and head myogenesis
Myod has an important role in abaxial pectoral fin and head muscle formation in zebrafish. Mice lacking Myod are viable and fertile without reported head muscle defects, but do show a delay in limb myogenesis, similar to, but less severe than, that in fish (Kablar et al., 1998; Kablar et al., 1997; Rudnicki et al., 1992). In both species, both Myf5 and Myod are expressed in fin and head muscle precursors. However, in fish myf5 becomes downregulated in head muscle anlage prior to the expression of muscle structural genes (Lin et al., 2006). Myod is essential at these locations for myog expression, which itself is required for normal head myogenesis. Interestingly, Myog knockdown failed to phenocopy Myod knockdown in primaxial muscle, despite affecting mef2d expression, perhaps indicating greater Myod activity in the somite.
Pectoral fin muscle derives from the rostral somites (Neyt et al., 2000). As with lateral fast primaxial muscle, Myod is required for early fin muscle, which is entirely fast (Patterson et al., 2008). Thus, fin muscle fibres may have an origin, molecular mechanism of myogenesis and fate similar to those of the lateral somitic fibres lacking in myod morphants (Hammond et al., 2007). As compared with mammals, teleosts retain a bodyplan more similar to that of primitive teleostomi (bony fish and tetrapods), so we suggest that lateral Myod-driven myogenesis was an ancestral trait of teleostomi.
Modes of myogenesis provide insight into somite evolution
Medial and lateral somite myogenesis are fundamentally distinct (Fig. 7D). Our findings do not reveal differences in MRF function that correspond either to the traditional epaxial/hypaxial division of the somite, or to the more recent primaxial/abaxial proposal (Burke and Nowicki, 2003). Zebrafish have retained an ancestral mode of somite myogenesis in which midline-derived Hh signalling induces slow muscle fibres that contribute to both dorsal/epaxial and ventral/hypaxial regions, which differ in innervation (Devoto et al., 2006; Eisen, 1991; Lacalli, 2002). Similarly, Hh-dependent MFFs (Wolff et al., 2003) and additional `medial mode' fast fibres are present in both epaxial and hypaxial regions. These medial fibre populations are primaxial and form through either Myf5 or Myod activity. However, Myod-dependent `lateral mode' fibres [which appear Fgf8-dependent (Hammond et al., 2007)] also form primaxially, again in both epaxial and hypaxial portions. We speculate that duplication of the ancestral myf5/myod gene in an invertebrate might have facilitated the evolution of novel lateral myogenesis under distinct myod regulation.
Muscle in the pectoral fin, hypaxial yolk region and possibly sternohyoid, arises from migratory muscle precursors that originate in the lateral somite (Haines et al., 2004; Neyt et al., 2000). The majority of such migratory precursors, reminiscent of the abaxial division of amniote somites (Burke and Nowicki, 2003), are Myod-dependent. Moreover, the intrinsic head muscles share Myod dependence with these abaxial populations. As the abaxial division arises from the lateral somite, Myod dependence appears to be the common theme in zebrafish lateral somitic muscle.
Are homologues of adaxial cells and/or medial fast fibres present in amniotes? These medial mode populations are unified by Myf5/Myod and Hh dependence. Unlike in fish, mouse Myod does not contribute to early epaxial/medial myogenesis in the trunk or tail (Kassar-Duchossoy et al., 2004). However, other tetrapods, such as birds and frogs, have retained Myod expression in Hh-dependent medial cells (Borycki et al., 1998; Grimaldi et al., 2004; Hacker and Guthrie, 1998; Hopwood et al., 1991). In mice, nevertheless, epaxial myogenesis is dependent on Hh signalling from the ventral midline (Borycki et al., 1999). Thus, changes in MRF utilisation were selected during evolution. Perhaps mammals have lost adaxial cells, leaving Myf5- and Hh-dependent homologues of medial fast fibres to generate the early myotome.
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/cgi/content/full/136/3/403/DC1
Supplementary Material
We thank many colleagues for reagents, and particularly Robert Knight for head muscle identification and Kuoyu Li for Fig. 2E. The TILLED mutants were generously provided by E. Busch-Nentwich and D. Stemple through EU ZF-MODELS funding. S.M.H. is a member of MRC Scientific Staff with Programme Grant and EU MYORES support. Deposited in PMC for release after 6 months.
References
- Balagopalan, L., Keller, C. A. and Abmayr, S. M. (2001). Loss-of-function mutations reveal that the Drosophila nautilus gene is not essential for embryonic myogenesis or viability. Dev. Biol. 231, 374-382. [DOI] [PubMed] [Google Scholar]
- Baxendale, S., Davison, C., Muxworthy, C., Wolff, C., Ingham, P. W. and Roy, S. (2004). The B-cell maturation factor Blimp-1 specifies vertebrate slow-twitch muscle fiber identity in response to Hedgehog signaling. Nat. Genet. 36, 88-93. [DOI] [PubMed] [Google Scholar]
- Blagden, C. S., Currie, P. D., Ingham, P. W. and Hughes, S. M. (1997). Notochord induction of zebrafish slow muscle mediated by Sonic Hedgehog. Genes Dev. 11, 2163-2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borycki, A.-G., Mendham, L. and Emerson, C. P., Jr (1998). Control of somite patterning by Sonic hedgehog and its downstream signal response genes. Development 125, 777-790. [DOI] [PubMed] [Google Scholar]
- Borycki, A.-G., Brunk, B., Tajbakhsh, S., Buckingham, M., Chiang, C. and Emerson, C. P., Jr (1999). Sonic hedgehog controls epaxial muscle determination through Myf5 activation. Development 126, 4053-4063. [DOI] [PubMed] [Google Scholar]
- Braun, T. and Arnold, H.-H. (1996). Myf-5 and myoD genes are activated in distinct mesenchymal stem cells and determine different skeletal muscle cell lineages. EMBO J. 15, 310-318. [PMC free article] [PubMed] [Google Scholar]
- Braun, T., Bober, E., Rudnicki, M. A., Jaenisch, R. and Arnold, H.-H. (1994). MyoD expression marks the onset of skeletal myogenesis in Myf-5 mutant mice. Development 120, 3083-3092. [DOI] [PubMed] [Google Scholar]
- Bryson-Richardson, R. J., Daggett, D. F., Cortes, F., Neyt, C., Keenan, D. G. and Currie, P. D. (2005). Myosin heavy chain expression in zebrafish and slow muscle composition. Dev. Dyn. 233, 1018-1022. [DOI] [PubMed] [Google Scholar]
- Burke, A. C. and Nowicki, J. L. (2003). A new view of patterning domains in the vertebrate mesoderm. Dev. Cell 4, 159-165. [DOI] [PubMed] [Google Scholar]
- Chen, L., Krause, M., Sepanski, M. and Fire, A. (1994). The Caenorhabditis elegans MYOD homologue HLH-1 is essential for proper muscle function and complete morphogenesis. Development 120, 1631-1641. [DOI] [PubMed] [Google Scholar]
- Chen, Y. H. and Tsai, H. J. (2002). Treatment with Myf5-morpholino results in somite patterning and brain formation defects in zebrafish. Differentiation 70, 447-456. [DOI] [PubMed] [Google Scholar]
- Chen, Y. H., Lee, W. C., Liu, C. F. and Tsai, H. J. (2001). Molecular structure, dynamic expression, and promoter analysis of zebrafish (Danio rerio) myf-5 gene. Genesis 29, 22-35. [DOI] [PubMed] [Google Scholar]
- Concordet, J.-P., Lewis, K. E., Moore, J. W., Goodrich, L. V., Johnson, R. L., Scott, M. P. and Ingham, P. W. (1996). Spatial regulation of a zebrafish patched homologue reflects the roles of sonic hedgehog and protein kinase A in neural tube and somite patterning. Development 122, 2835-2846. [DOI] [PubMed] [Google Scholar]
- Coutelle, O., Blagden, C. S., Hampson, R., Halai, C., Rigby, P. W. and Hughes, S. M. (2001). Hedgehog signalling is required for maintenance of myf5 and myoD expression and timely terminal differentiation in zebrafish adaxial myogenesis. Dev. Biol. 236, 136-150. [DOI] [PubMed] [Google Scholar]
- Devoto, S. H., Melancon, E., Eisen, J. S. and Westerfield, M. (1996). Identification of separate slow and fast muscle precursor cells in vivo, prior to somite formation. Development 122, 3371-3380. [DOI] [PubMed] [Google Scholar]
- Devoto, S. H., Stoiber, W., Hammond, C. L., Steinbacher, P., Haslett, J. R., Barresi, M. J., Patterson, S. E., Adiarte, E. G. and Hughes, S. M. (2006). Generality of vertebrate developmental patterns: evidence for a dermomyotome in fish. Evol. Dev. 8, 101-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, S. J., Li, H., Bian, Y. and Zhong, Y. (2008). Heat-shock protein 90alpha1 is required for organized myofibril assembly in skeletal muscles of zebrafish embryos. Proc. Natl. Acad. Sci. USA 105, 554-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen, J. S. (1991). Determination of primary motoneuron identity in developing zebrafish embryos. Science 252, 569-571. [DOI] [PubMed] [Google Scholar]
- Ekker, M., Wegner, J., Akimenko, M. A. and Westerfield, M. (1992). Coordinate embryonic expression of three zebrafish engrailed genes. Development 116, 1001-1010. [DOI] [PubMed] [Google Scholar]
- Gensch, N., Borchardt, T., Schneider, A., Riethmacher, D. and Braun, T. (2008). Different autonomous myogenic cell populations revealed by ablation of Myf5-expressing cells during mouse embryogenesis. Development 135, 1597-1604. [DOI] [PubMed] [Google Scholar]
- Grimaldi, A., Tettamanti, G., Martin, B. L., Gaffield, W., Pownall, M. E. and Hughes, S. M. (2004). Hedgehog regulation of superficial slow muscle fibres in Xenopus and the evolution of tetrapod trunk myogenesis. Development 131, 3249-3262. [DOI] [PubMed] [Google Scholar]
- Groves, J. A., Hammond, C. L. and Hughes, S. M. (2005). Fgf8 drives myogenic progression of a novel lateral fast muscle fibre population in zebrafish. Development 132, 4211-4222. [DOI] [PubMed] [Google Scholar]
- Hacker, A. and Guthrie, S. (1998). A distinct developmental programme for the cranial paraxial mesoderm in the chick embryo. Development 125, 3461-3472. [DOI] [PubMed] [Google Scholar]
- Haines, L., Neyt, C., Gautier, P., Keenan, D. G., Bryson-Richardson, R. J., Hollway, G. E., Cole, N. J. and Currie, P. D. (2004). Met and Hgf signaling controls hypaxial muscle and lateral line development in the zebrafish. Development 131, 4857-4869. [DOI] [PubMed] [Google Scholar]
- Haldar, M., Karan, G., Tvrdik, P. and Capecchi, M. R. (2008). Two cell lineages, myf5-dependent and myf5-independent, participate in mouse skeletal myogenesis. Dev. Cell 14, 437-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamade, A., Deries, M., Begemann, G., Bally-Cuif, L., Genet, C., Sabatier, F., Bonnieu, A. and Cousin, X. (2006). Retinoic acid activates myogenesis in vivo through Fgf8 signalling. Dev. Biol. 289, 127-140. [DOI] [PubMed] [Google Scholar]
- Hammond, C. L., Hinits, Y., Osborn, D. P., Minchin, J. E., Tettamanti, G. and Hughes, S. M. (2007). Signals and myogenic regulatory factors restrict pax3 and pax7 expression to dermomyotome-like tissue in zebrafish. Dev. Biol. 302, 504-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasty, P., Bradley, A., Morris, J. H., Edmondson, D. G., Venuti, J. M., Olson, E. N. and Klein, W. H. (1993). Muscle deficiency and neonatal death in mice with targeted mutation in the myogenin gene. Nature 364, 501-506. [DOI] [PubMed] [Google Scholar]
- Hawkins, T. A., Haramis, A. P., Etard, C., Prodromou, C., Vaughan, C. K., Ashworth, R., Ray, S., Behra, M., Holder, N., Talbot, W. S. et al. (2008). The ATPase-dependent chaperoning activity of Hsp90a regulates thick filament formation and integration during skeletal muscle myofibrillogenesis. Development 135, 1147-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry, C. A. and Amacher, S. L. (2004). Zebrafish slow muscle cell migration induces a wave of fast muscle morphogenesis. Dev. Cell 7, 917-923. [DOI] [PubMed] [Google Scholar]
- Higashijima, S., Nose, A., Eguchi, G., Hotta, Y. and Okamoto, H. (1997). Mindin/F-spondin family: novel ECM proteins expressed in the zebrafish embryonic axis. Dev. Biol. 192, 211-227. [DOI] [PubMed] [Google Scholar]
- Hinits, Y. and Hughes, S. M. (2007). Mef2s are required for thick filament formation in nascent muscle fibres. Development 134, 2511-2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinits, Y., Osborn, D. P., Carvajal, J. J., Rigby, P. W. and Hughes, S. M. (2007). Mrf4 (myf6) is dynamically expressed in differentiated zebrafish skeletal muscle. Gene Expr. Patterns 7, 738-745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopwood, N. D., Pluck, A. and Gurdon, J. B. (1991). Xenopus Myf-5 marks early muscle cells and can activate muscle genes ectopically in early embryos. Development 111, 551-560. [DOI] [PubMed] [Google Scholar]
- Isken, O. and Maquat, L. E. (2007). Quality control of eukaryotic mRNA: safeguarding cells from abnormal mRNA function. Genes Dev. 21, 1833-1856. [DOI] [PubMed] [Google Scholar]
- Kablar, B., Krastel, K., Ying, C., Asakura, A., Tapscott, S. J. and Rudnicki, M. A. (1997). MyoD and Myf-5 differentially regulate the development of limb versus trunk skeletal muscle. Development 124, 4729-4738. [DOI] [PubMed] [Google Scholar]
- Kablar, B., Asakura, A., Krastel, K., Ying, C., May, L. L., Goldhamer, D. J. and Rudnicki, M. A. (1998). MyoD and Myf-5 define the specification of musculature of distinct embryonic origin. Biochem. Cell Biol. 76, 1079-1091. [PubMed] [Google Scholar]
- Kahane, N., Cinnamon, Y., Bachelet, I. and Kalcheim, C. (2001). The third wave of myotome colonization by mitotically competent progenitors: regulating the balance between differentiation and proliferation during muscle development. Development 128, 2187-2198. [DOI] [PubMed] [Google Scholar]
- Kassar-Duchossoy, L., Gayraud-Morel, B., Gomes, D., Rocancourt, D., Buckingham, M., Shinin, V. and Tajbakhsh, S. (2004). Mrf4 determines skeletal muscle identity in Myf5:Myod double-mutant mice. Nature 431, 466-471. [DOI] [PubMed] [Google Scholar]
- Kaul, A., Koster, M., Neuhaus, H. and Braun, T. (2000). Myf-5 revisited: loss of early myotome formation does not lead to a rib phenotype in homozygous Myf-5 mutant mice. Cell 102, 17-19. [DOI] [PubMed] [Google Scholar]
- Koudijs, M. J., den Broeder, M. J., Groot, E. and van Eeden, F. J. (2008). Genetic analysis of the two zebrafish patched homologues identifies novel roles for the hedgehog signaling pathway. BMC Dev. Biol. 8, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacalli, T. C. (2002). The dorsal compartment locomotory control system in amphioxus larvae. J. Morphol. 252, 227-237. [DOI] [PubMed] [Google Scholar]
- Liew, H. P., Choksi, S. P., Wong, K. N. and Roy, S. (2008). Specification of vertebrate slow-twitch muscle fiber fate by the transcriptional regulator Blimp1. Dev. Biol. 324, 226-235. [DOI] [PubMed] [Google Scholar]
- Lin, C. Y., Yung, R. F., Lee, H. C., Chen, W. T., Chen, Y. H. and Tsai, H. J. (2006). Myogenic regulatory factors Myf5 and Myod function distinctly during craniofacial myogenesis of zebrafish. Dev. Biol. 299, 594-608. [DOI] [PubMed] [Google Scholar]
- Maves, L., Waskiewicz, A. J., Paul, B., Cao, Y., Tyler, A., Moens, C. B. and Tapscott, S. J. (2007). Pbx homeodomain proteins direct Myod activity to promote fast-muscle differentiation. Development 134, 3371-3382. [DOI] [PubMed] [Google Scholar]
- Meedel, T. H., Chang, P. and Yasuo, H. (2007). Muscle development in Ciona intestinalis requires the b-HLH myogenic regulatory factor gene Ci-MRF. Dev. Biol. 302, 333-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, C. A., Parkin, C. A., Bidet, Y. and Ingham, P. W. (2007). A role for the Myoblast city homologues Dock1 and Dock5 and the adaptor proteins Crk and Crk-like in zebrafish myoblast fusion. Development 134, 3145-3153. [DOI] [PubMed] [Google Scholar]
- Morin-Kensicki, E. M. and Eisen, J. S. (1997). Sclerotome development and peripheral nervous system segmentation in embryonic zebrafish. Development 124, 159-167. [DOI] [PubMed] [Google Scholar]
- Nabeshima, Y., Hanaoka, K., Hayasaka, M., Esumi, E., Li, S., Nonaka, I. and Nabeshima, Y.-i. (1993). Myogenin gene disruption results in perinatal lethality owing to severe muscle defect. Nature 364, 532-535. [DOI] [PubMed] [Google Scholar]
- Neyt, C., Jagla, K., Thisse, C., Thisse, B., Haines, L. and Currie, P. D. (2000). Evolutionary origins of vertebrate appendicular muscle. Nature 408, 82-86. [DOI] [PubMed] [Google Scholar]
- Patterson, S. E., Mook, L. B. and Devoto, S. H. (2008). Growth in the larval zebrafish pectoral fin and trunk musculature. Dev. Dyn. 237, 307-315. [DOI] [PubMed] [Google Scholar]
- Penn, B. H., Bergstrom, D. A., Dilworth, F. J., Bengal, E. and Tapscott, S. J. (2004). A MyoD-generated feed-forward circuit temporally patterns gene expression during skeletal muscle differentiation. Genes Dev. 18, 2348-2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawls, A., Morris, J. H., Rudnicki, M., Braun, T., Arnold, H.-H., Klein, W. H. and Olson, E. N. (1995). Myogenin's functions do not overlap with those of MyoD or Myf-5 during mouse embryogenesis. Dev. Biol. 172, 37-50. [DOI] [PubMed] [Google Scholar]
- Rawls, A., Valdez, M. R., Zhang, W., Richardson, J., Klein, W. H. and Olson, E. N. (1998). Overlapping functions of the myogenic bHLH genes MRF4 and MyoD revealed in double mutant mice. Development 125, 2349-2358. [DOI] [PubMed] [Google Scholar]
- Rudnicki, M. A., Braun, T., Hinuma, S. and Jaenisch, R. (1992). Inactivation of MyoD in mice leads to up-regulation of the myogenic HLH gene Myf-5 and results in apparently normal muscle development. Cell 71, 383-390. [DOI] [PubMed] [Google Scholar]
- Rudnicki, M. A., Schnegelsberg, P. N., Stead, R. H., Braun, T., Arnold, H.-H. and Jaenisch, R. (1993). MyoD or Myf-5 is required for the formation of skeletal muscle. Cell 75, 1351-1359. [DOI] [PubMed] [Google Scholar]
- Sambasivan, R. and Tajbakhsh, S. (2007). Skeletal muscle stem cell birth and properties. Semin. Cell Dev. Biol. 18, 870-882. [DOI] [PubMed] [Google Scholar]
- Stellabotte, F., Dobbs-McAuliffe, B., Fernandez, D. A., Feng, X. and Devoto, S. H. (2007). Dynamic somite cell rearrangements lead to distinct waves of myotome growth. Development 134, 1253-1257. [DOI] [PubMed] [Google Scholar]
- Stemple, D. L. (2004). TILLING-a high-throughput harvest for functional genomics. Nat. Rev. Genet. 5, 145-150. [DOI] [PubMed] [Google Scholar]
- Tajbakhsh, S., Rocancourt, D., Cossu, G. and Buckingham, M. (1997). Redefining the genetic hierarchies controlling skeletal myogenesis: Pax-3 and Myf-5 act upstream of MyoD. Cell 89, 127-138. [DOI] [PubMed] [Google Scholar]
- Ticho, B. S., Stainier, D. Y. R., Fishman, M. C. and Breitbart, R. E. (1996). Three zebrafish MEF2 genes delineate somitic and cardiac muscle development in wild-type and mutant embryos. Mech. Dev. 59, 205-218. [DOI] [PubMed] [Google Scholar]
- Valdez, M. R., Richardson, J. A., Klein, W. H. and Olson, E. N. (2000). Failure of Myf5 to support myogenic differentiation without myogenin, MyoD, and MRF4. Dev. Biol. 219, 287-298. [DOI] [PubMed] [Google Scholar]
- Venuti, J. M., Morris, J. H., Vivian, J. L., Olson, E. N. and Klein, W. H. (1995). Myogenin is required for late but not early aspects of myogenesis during mouse development. J. Cell Biol. 128, 563-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. H., Li, C. K., Lee, G. H., Tsay, H. J., Tsai, H. J. and Chen, Y. H. (2008). Inactivation of zebrafish mrf4 leads to myofibril misalignment and motor axon growth disorganization. Dev. Dyn. 237, 1043-1050. [DOI] [PubMed] [Google Scholar]
- Weinberg, E. S., Allende, M. L., Kelly, C. S., Abdelhamid, A., Murakami, T., Andermann, P., Doerre, O. G., Grunwald, D. J. and Riggleman, B. (1996). Developmental regulation of zebrafish MyoD in wild-type, no tail and spadetail embryos. Development 122, 271-280. [DOI] [PubMed] [Google Scholar]
- Weintraub, H. (1993). The MyoD family and myogenesis: redundancy, networks, and thresholds. Cell 75, 1241-1244. [DOI] [PubMed] [Google Scholar]
- Westerfield, M. (1995). The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio rerio). Eugene, OR: University of Oregon Press.
- Wolff, C., Roy, S. and Ingham, P. W. (2003). Multiple muscle cell identities induced by distinct levels and timing of hedgehog activity in the zebrafish embryo. Curr. Biol. 13, 1169-1181. [DOI] [PubMed] [Google Scholar]
- Wotton, K. R., Weierud, F. K., Dietrich, S. and Lewis, K. E. (2008). Comparative genomics of Lbx loci reveals conservation of identical Lbx ohnologs in bony vertebrates. BMC Evol. Biol. 8, 171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Y., He, J., Wang, X., Lim, T. M. and Gong, Z. (2000). Asynchronous activation of 10 muscle-specific protein (MSP) genes during zebrafish somitogenesis. Dev. Dyn. 219, 201-215. [DOI] [PubMed] [Google Scholar]
- Zhang, W., Behringer, R. R. and Olson, E. N. (1995). Inactivation of the myogenic bHLH gene MRF4 results in up-regulation of myogenin and rib anomalies. Genes Dev. 9, 1388-1399. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.