Abstract
SMN1 and SMN2 (survival motor neuron) encode identical proteins. A critical question is why only the homozygous loss of SMN1, and not SMN2, results in spinal muscular atrophy (SMA). Analysis of transcripts from SMN1/SMN2 hybrid genes and a new SMN1 mutation showed a direct relationship between presence of disease and exon 7 skipping. We have reported previously that the exon-skipped product SMNΔ7 is partially defective for self-association and SMN self-oligomerization correlated with clinical severity. To evaluate systematically which of the five nucleotides that differ between SMN1 and SMN2 effect alternative splicing of exon 7, a series of SMN minigenes was engineered and transfected into cultured cells, and their transcripts were characterized. Of these nucleotide differences, the exon 7 C-to-T transition at codon 280, a translationally silent variance, was necessary and sufficient to dictate exon 7 alternative splicing. Thus, the failure of SMN2 to fully compensate for SMN1 and protect from SMA is due to a nucleotide exchange (C/T) that attenuates activity of an exonic enhancer. These findings demonstrate the molecular genetic basis for the nature and pathogenesis of SMA and illustrate a novel disease mechanism. Because individuals with SMA retain the SMN2 allele, therapy targeted at preventing exon 7 skipping could modify clinical outcome.
Autosomal recessive spinal muscular atrophy (SMA) is clinically categorized as type I (severe), II (intermediate), or III (mild), depending on age of onset and clinical progression. SMA occurs in approximately 1 in 10,000 live births and has a carrier frequency of 1 in 50 (1). Two SMN (survival motor neuron) genes typically are present on 5q13: SMN1 and SMN2. Homozygous absence of the telomeric copy, SMN1 (also known as SMNtel), correlates with development of SMA (2–5). Although several additional genes are located within the SMA region, a number of intragenic SMN1 mutations including frameshifts and point mutations have been identified that conclusively identified SMN1 as the SMA-determining gene (reviewed in ref. 6). These neighboring genes may function as potential candidates for SMA-modifying genes (7–10). The nearly identical centromeric copy, SMN2, appears to modify disease severity in a dose-dependent manner as SMN protein levels from this gene correlate with disease severity (11, 12) but, alone, cannot provide protection from SMA (6, 13, 14).
The SMN transcript has 9 exons and encodes a 294-aa protein. SMN localizes within discrete subcellular compartments termed Gems that colocalize with coiled bodies, which contain components of the pre-mRNA splicing machinery (15). Recent studies suggest a role for SMN in the maturation of small nuclear ribonucleoproteins through an association with SMN-interacting protein 1 (SIP-1) and Sm proteins (16). Furthermore, SMN also may serve to regenerate the pre-mRNA splicing machinery as evidenced by SMN′s ability to stimulate in vitro pre-mRNA splicing (17). SMN binds RNA and DNA, a property mediated by a peptide encoded by exon 2 (18). Exon 6, a 37-aa region containing several Y-G elements (19), is absolutely required for self-association (20). A small number of SMA patients with nonhomozygous SMN deletions possess mutations in exon 6 (6, 21). We have reported previously that these mutations decreased SMN self-association and that this property correlates with disease severity (20). Although these recently identified functions are intriguing beginnings to understanding the molecular nature of SMA, it is unclear how SMN defects result in the highly specific α-motor neuron degeneration associated with SMA.
Because SMN1 and SMN2 encode identical proteins, it is unclear why only SMN1 mutations and not SMN2 result in SMA. There are five nonpolymorphic nucleotide differences within the 3′ region of the genes that do not alter protein-coding sequences (2, 22). Their functional significance and role in the development of SMA have not been determined. Three alternatively spliced transcripts have been observed: SMNΔ5, SMNΔ7, and SMNΔ5,7, which lack exon 5, 7, or both, respectively (2, 23). SMN1 primarily produces the full-length form (FL SMN) and no detectable SMNΔ7, whereas SMNΔ7 is believed to be the primary product of SMN2. In ≈95% of SMA patients SMN1 exon 7 is homozygously deleted or gene converted to SMN2 (2, 6, 13), implying that the low levels of full-length SMN protein produced by SMN2 are insufficient to protect against disease development (2, 11). We have reported that SMNΔ7 is not functionally equivalent to FL SMN as evidenced by SMNΔ7’s significantly reduced ability to self-associate (20). Clearly, the total amount of full-length, oligomerization-competent SMN protein is a critical SMA determinant (11, 12, 24). Characterization of the mechanism governing the production of SMN vs. SMNΔ7 by SMN1 and SMN2 is a critical step in elucidating the pathogenesis of SMA.
MATERIALS AND METHODS
Patient Samples.
All patients fulfilled the diagnostic criteria for proximal spinal muscular atrophy defined by the International SMA Consortium (1992). Informed consent was obtained from all subjects. Patients 1, 2a, and 2b carrying hybrid genes have been described previously (25). The patient carrying the novel splice-site mutation has type III SMA (34). DNA isolation from whole peripheral blood, RNA isolation from lymphoblastoid cell lines, haplotype analysis with the markers C212 and Ag1-CA, molecular genetic analysis of the SMN gene, quantitative analysis of the SMN1 copies, and sequencing were performed as described (4, 25, 34).
Plasmids.
SMN minigenes were created by Expand High Fidelity (Boehringer Mannheim) PCR amplification of 250 ng of genomic DNA (Boehringer Mannheim) by using primers Ex6FX (5′-CGA TCT CGA GAT AAT TCC CCC ACC ACC TC-3′) and Ex8RS (5′-GCT ACC CGG GCA CAT ACG CCT CAC ATA CA-3′). Products were digested with XhoI and SmaI and cloned directionally into XhoI and SmaI sites within pCI and sequenced across the five nucleotide polymorphisms to identify SMN1 and SMN2 clones. Double- and single-nucleotide conversion was introduced by oligonucleotide site-directed mutagenesis by using Thermo Pol Vent polymerase (New England Biolabs): pSMN1/pSMN2ΔIn6 (5′-GCT ATA TAG AC/TA TAG ATA GC-3′; 5′-GCT ATC TAT G/ATC TAT ATA GC-3′); pSMN1/pSMN2ΔEx7 (5′-GAT TTT GTC TG/AA AAC CCT GTA AG-3′; 5′-CTT ACA GGG TTT C/TAG ACA AAA TC-3′); pSMN1/pSMN2ΔIn7+100 (5′-CTT TCA ACT TTT/C TAA CAT CTG-3′; 5′-CAG ATG TTA A/GAA AGT TGA AAG-3′); pSMN1ΔIn7+214 (5′-CTT CCA CAT/C AAC CAA CCA G-3′; 5′-CTG GTT GGT TA/GT GTG GAA G-3′). Identical primer sets were used to generate the nucleotide substitutions except for appropriate SMN1- or SMN2-derived nucleotides (as indicated in bold).
Reverse Transcription–PCR (RT-PCR).
Total cellular RNA was isolated over a CsCl gradient 48 h posttransfection with 2 μg of pSMN1, pSMN2, and derivatives by Lipofectamine (Life Technologies, Gaithersburg, MD). One microgram of total RNA was used to generate first-strand cDNA by using oligo(dT) and Super Script II reverse transcriptase (Life Technologies). Plasmid-derived cDNAs were amplified by using the primer set pCIFwd (5′-GCT AAC GCA GTC AGT GCT TC-3′); pCIRev (5′-GTA TCT TAT CAT GTC TGC TCG-3′). Fifty-microliter reactions included 20 pmol of each primer, 200 μM dNTP, and 1 unit of Thermo Pol Vent(-exo) polymerase (NEB, Beverly, MA). Cycling conditions were an initial denaturation step (94°C/2 min), 30 cycles (94°C/30 sec; 56°C/1.5 min; 72°C/1 min), and a final extension step (72°C/10 min). Reaction products were resolved by electrophoresis through a 1.9% agarose gel and visualized by ethidium bromide staining. Products were cloned and sequenced. RNA was isolated from fibroblasts or Epstein–Barr virus-transformed lymphocytes by using the TRIzol kit (Life Technologies). First-strand synthesis was initiated with oligo(dT) primer and 4 μg of RNA by using M-MLV reverse transcriptase (Life Technologies). SMN cDNAs were amplified with the primers Ex5Fwd (5′-CTA TCA TGC TGG CTG CCT-3′) and Ex8Rev (5′-CTA CAA CAC CCT TCT CAC AG-3′). Fifty-microliter reactions contained the same components as above except with 2 units of Taq polymerase, and the annealing time was for 30 sec. Reaction products were digested with DdeI and resolved by electrophoresis into a 2.0% agarose gel. SMN2-derived transcripts contain a unique DdeI restriction element introduced because of a nucleotide polymorphism not present in SMN1 and are differentiated from SMN1-derived transcripts because of the faster migration of the SMN2 products.
RESULTS
Alternative Splicing of SMN Exon 7 by SMN-Hybrid Genes.
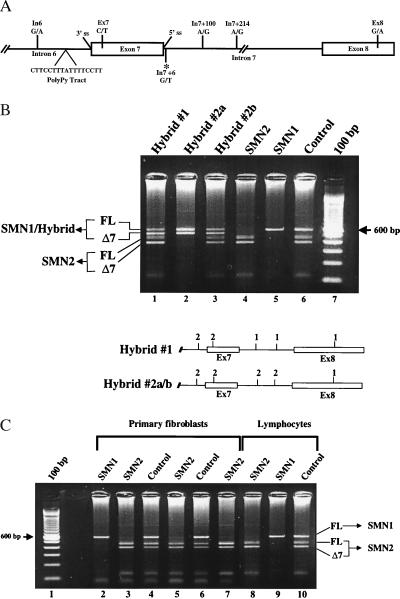
To evaluate the nucleotide differences between SMN1 and SMN2 and their effects on SMN RNA exon 7 alternative processing, lymphoblastoid cell lines from SMA patients carrying SMN hybrid genes were analyzed (25). The five nucleotide differences between SMN1 and SMN2 known to exist are at the following positions: intron 6(-45); exon 7(+6); intron 7(+100); intron 7(+214); exon 8(+245) (In6; Ex7; In7+100; In7+214; Ex8, respectively) (Fig. 1A). RT-PCR was performed with an SMN exon 5 forward primer and an SMN exon 8 reverse primer to generate cDNAs that subsequently were digested with DdeI. One of the nucleotide polymorphisms between SMN1 and SMN2 in exon 8 results in a DdeI restriction site unique to SMN2, thereby allowing SMN1- and SMN2-derived transcripts to be distinguished by gel electrophoresis (Fig. 1B, lanes 4–6). SMN hybrid gene 1, which contains SMN2-derived nucleotides at the In6 and Ex7 positions and SMN1 nucleotides at the remaining positions, produced an abundant level of SMNΔ7 and decreased FL SMN compared with the typical SMN1 transcript pattern (Fig. 1B, compare lanes 1, 5, and 6). SMN hybrid gene 2, which contains SMN2-derived nucleotides at all positions except Ex8, was isolated from two unrelated SMA patients (hybrid genes 2a and 2b) and also resulted in high levels of the exon 7 skipped transcript (Fig. 1B, compare lanes 2–4 and 6). SMN2-derived transcripts were detectable at levels equivalent to control individuals in hybrid genes 1 and 2b, but were absent in the patient carrying hybrid gene 2a because of a deletion of SMN2. These results demonstrate that the transcript profiles in cells from SMA individuals with SMN1/SMN2 hybrid genes have high-level SMNΔ7 expression and reduced FL SMN, thereby establishing a direct correlation between the development of SMA and alternative processing of SMN exon 7. Analyses of the previously reported SMA patient SMN-hybrid genes revealed that only the exon 7+6 nucleotide “T” was SMN2-derived in all instances, suggesting that this position influences exon skipping and the subsequent development of SMA.
Figure 1.
Endogenous SMN hybrid gene transcript analysis. (A) Expanded view of the 3′ region of SMN genes. Intron (lines) and exon (boxes) boundaries are indicated. Positions of the five nucleotide differences between SMN1 and SMN2 within intron 6 (In6, G/A), exon 7 (Ex7, C/T), intron 7 (In7+100, A/G; In7+214, A/G), and exon 8 (Ex8, G/A) are shown (SMN1/SMN2 sequence). The new intron 7 mutation (*In7 +6), and splice elements are shown (3′ ss, 3′ splice site; 5′ ss, 5′ splice site; PolyPy Tract, polypyrimidine tract). (B) RT-PCR amplification of SMN transcripts from Epstein–Barr virus transformed from SMA and control individuals by using oligonucleotides located in SMN exons 5 and 8. Products were digested with DdeI and resolved in a 2% agarose gel. The SMN2 nucleotide within exon 8 creates a DdeI site, resulting in faster-migrating species for SMN2 transcripts. The positions of full-length (FL) and exon 7-skipped (Δ7) transcripts are indicated. Graphic representations of the 3′ end of the hybrid genes and the origin of the five nucleotide polymorphisms are indicated (1, SMN1; 2, SMN2; In, intron; Ex, exon): Hybrid #1 (In6/SMN2, Ex7/SMN2, In7+100/SMN1, In7+214/SMN1, Ex8/SMN1); Hybrid #2a/b (In6/SMN2, Ex7/SMN2, In7+100/SMN2, In7+214/SMN2, Ex8/SMN1). (C) RT-PCR amplification of SMN transcripts from primary fibroblasts and EBV-transformed lymphocytes from SMA and control individuals (see B).
Because SMA typically is associated with the specific degeneration of α-motor neurons, several cell types were examined to determine whether a tissue-specific factor was required for exon 7 skipping. The cellular context did not affect alternative splicing patterns of SMN1 and SMN2, because SMN transcripts from fibroblasts and lymphoblastoid cell lines from SMA and control individuals were similar (Fig. 1C). Sixteen additional tissues were examined and found to produce similar exon 7 splicing patterns for the two genes (data not shown). Although we cannot assess RNA transcripts from SMA motor neurons, alternative splicing of exon 7 does not appear to be cell type-restricted.
Transient Expression of Synthetic SMN Minigenes.
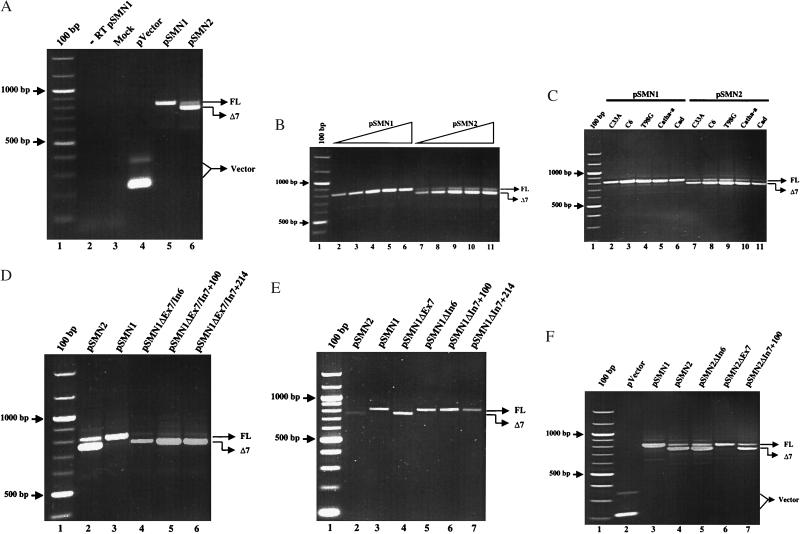
To evaluate systematically the role of the nucleotide differences between SMN1 and SMN2 on SMN exon 7 alternative processing, a plasmid-based SMN system was developed. Genomic SMN1 and SMN2 DNA from and including exons 6–8 was cloned into the mammalian expression vector pCI, downstream of the constitutively expressing cytomegalovirus promoter. This system allowed synthetic hybrid genes to be engineered and transcripts derived from them to be analyzed specifically because the oligonucleotides used in the amplification step of the RT-PCR annealed only to transcript sequences within the plasmid backbone. The plasmid-based SMN1 (pSMN1) and SMN2 (pSMN2) constructs recapitulated the endogenous SMN1 and SMN2 genes’ exon 7 splicing patterns; pSMN1 expressed an abundant FL SMN transcript with no detectable SMNΔ7, and pSMN2 produced a low level of FL SMN and abundant SMNΔ7 (Fig. 2A, lanes 5 and 6). As expected, endogenous-derived SMN transcripts were not observed (Fig. 2A, lanes 2 and 3). To determine whether the splicing patterns of SMN1 and SMN2 were consistent over a broad range of plasmid concentrations, 100 ng to 10 μg was transiently transfected. Even at the highest concentration, processing of the plasmid-derived SMN1 and SMN2 transcripts were consistent (Fig. 2B). RNA processing within a variety of cell types also was examined. In all instances, pSMN1 and pSMN2 splicing patterns were similar to the transcript production of the respective endogenous SMN gene (Fig. 2C). These results demonstrate that the plasmid and endogenous genes are subject to equivalent processing mechanisms and that further analysis by using the plasmid-based system represents a useful tool to characterize SMN exon 7 alternative splicing.
Figure 2.
Analysis of plasmid-based SMN transcripts. (A) RT-PCR amplification of total RNA isolated from neuroblastoma C6 cells 48 h posttransfection with plasmid-based SMN1 (pSMN1), SMN2 (pSMN2), vector alone (pVector), or mock-transfected vector by using Lipofectamine (Life Technologies). FL SMN and SMNΔ7 transcripts are indicated and have been sequenced to ensure fidelity of splicing events. Two vector transcript species are a result of incomplete excision of the plasmid-based intron. No reverse transcriptase (-RT pSMN1) served as a control. (B) pSMN1 or pSMN2 (100 ng, 500 ng, 1 μg, 5 μg, and 10 μg) was transiently transfected into C6 cells. RT-PCR was performed (see A), and FL SMN and SMNΔ7 transcripts are indicated. (C) Transient expression of pSMN1 or pSMN2 in C33A (cervical carcinoma), C6 (neuroblastoma), T98G (glioblastoma), and Cath.a and CAD (murine neuroblastoma) cell lines and RT-PCR analysis of plasmid SMN transcripts (see A). (D) Synthetic SMN1 hybrid constructs. Two SMN2 nucleotides were introduced into a pSMN1 construct (In, intron; Ex, exon): pSMN1ΔIn6/Ex7; -ΔEx7/In7+100; -ΔEx7/In7+214. SMN2-derived nucleotides follow “Δ.” (E) Single SMN2 nucleotides introduced into pSMN1: pSMN1ΔEx7; -ΔIn6; -ΔIn7+100; -ΔIn7+214. SMN2-derived nucleotides follow “Δ.” (F) Single SMN1 nucleotides introduced into pSMN2: pSMN2ΔIn6; -ΔEx7; -ΔIn7+100. SMN2-derived nucleotides follow “Δ.” A 100-bp ladder is indicated.
Identification of a Single Nucleotide Difference Between pSMN1 and pSMN2 Regulating Exon 7 Alternative Processing.
Next, we generated a series of synthetic hybrid SMN genes and systematically characterized their expression pattern after transfection. Using pSMN1 as a template, two SMN2-specific nucleotide changes were introduced into an otherwise SMN1 construct (refer to Fig. 1A). Converting the In6/Ex7, Ex7/In7+100, and Ex7/In7+214 positions to SMN2-derived nucleotides dramatically altered the transcript pattern, resulting in a high level of SMNΔ7 and low level of FL SMN, similar to the pattern observed for pSMN2 (Fig. 2D). Importantly, each synthetic hybrid construct possessed the SMN2-derived nucleotide at the exon 7 position. Furthermore, the introduction of a single SMN2-derived nucleotide into an otherwise SMN1 background at the exon 7 position conferred the pSMN2 alternative splicing pattern: substantial reduction in FL SMN and a dramatic increase in SMNΔ7 production (Fig. 2E, lanes 3 and 4). In contrast, single-nucleotide SMN2-to-SMN1 conversions at the In6, In7+100, and In7+214 positions individually had no detectable effect on exon 7 processing (Fig. 2E). In the reciprocal experiments, introduction of the SMN1 nucleotide from exon 7 into an otherwise pSMN2 construct resulted in high levels of FL SMN and undetectable SMNΔ7, similar to the transcript pattern from pSMN1 (Fig. 2F, lanes 4 and 6). The replacement of either SMN1-derived nucleotides flanking exon 7, In6 and In7+100, did not detectably alter exon 7 processing (Fig. 2F). Taken together these results demonstrate that a single nucleotide in exon 7 that differs between SMN1 and SMN2 dictates the alternative splicing pattern for SMN genes and provides a molecular basis for understanding why all previously reported SMA hybrid genes contain an SMN2-derived nucleotide at the exon 7 position (6).
Identification and Characterization of a Novel SMA Allele and Its Effects on Exon 7 Processing.
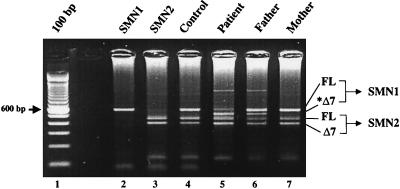
Although the nucleotide difference in exon 7 between SMN1 and SMN2 genes has a critical role in exon 7 skipping, clearly additional regulatory elements exist in and around exon 7. To this end, we screened heterozygously deleted SMA patients with no coding alterations in their remaining SMN1 allele for intronic mutations. A novel SMN mutation was identified that contains a single nucleotide exchange within intron 7, c.922+6 T/G (34), which is predicted to disrupt the consensus exon 7 splice donor motif (26). Quantitative SMN1 analysis demonstrated that the father has only one SMN1 gene, whereas the mother carries two SMN1 copies. The mother carries the new SMN1 mutation in addition to a normal SMN1 gene, as evidenced by the detection of an SMN-derived SMNΔ7 species (Fig. 3, lane 6). Analyzing SMN transcripts in lymphoblastoid cell lines from the SMA patient carrying only the new SMA allele revealed that FL SMN is reduced dramatically and SMNΔ7 is highly expressed (Fig. 3, lane 5). The identification of an intronic SMN mutation that results in the abundant production of SMNΔ7 and low levels of FL SMN further underscores the critical role that SMN exon 7 skipping has in the development of SMA.
Figure 3.
SMNΔ7 production from a novel SMN1 spice-site mutation. RT-PCR analysis (see Fig. 1B) of endogenous SMN transcripts from SMA and control individuals’ EBV-transformed lymphocytes. The father is carrier-ΔSMN1, and the mother is carrierΔc.922+6 T/G. Each has an additional, single SMN1 copy. The patient carries an SMN1 deletion and an SMN1 copy with the splice-site mutation. The SMN1-derived, exon-skipped transcript is shown in lanes 5 and 6 (*Δ7).
DISCUSSION
Transcripts derived from SMA patients’ leukocytes and fibroblasts, synthetic SMN minigenes, and a newly identified SMN splice-site mutation clearly demonstrate that aberrant splicing of SMN exon 7 determines SMA development. This conclusion answers the question of why SMN1, as opposed to SMN2, is the disease gene. This C-to-T transition in exon 7 disrupts a putative “exon-splicing enhancer” (ESE), a cis element that promotes inclusion of specific exons (27). Because of the genetic instability of the SMA region, gene-conversion events are believed to account for a high percentage of mutated SMA alleles. In light of these results, gene conversions that encode the SMN2-derived nucleotide at the exon 7+6 position would behave as an SMA allele, and hybrid genes that contain the SMN1-derived nucleotide at this position would be protective against the development of SMA. To date, all reported SMA-causing hybrid genes contain the SMN2-derived nucleotide at the exon 7 position (2, 25, 28). SMN1 is the disease gene because it produces FL SMN protein. The SMN2 allele is the disease-modifying gene because of a single nucleotide difference in exon 7 that results in alternative processing of its mRNA and editing out of exon 7. These results demonstrate conclusively the underlying molecular genetic mechanism for the production of a functional vs. nonfunctional SMN transcript in the majority of SMA cases.
Genetic diseases resulting from donor and acceptor splice-site or lariat branch-point mutations that cause exon skipping or involve aberrant 5′ or 3′ splice-site selection represent a common mutational mechanism (29). The situation with SMN1 and SMN2 differs notably in that the exon 7 C-to-T transition is exonic and a translationally silent mutation. This nucleotide polymorphism is present in a partially functional copy gene capable of encoding identical proteins (2, 22). It is interesting that in other systems, C-to-T transitions within ESEs also result in dramatically altered splicing patterns (30). ESEs serve as binding sites for regulatory trans-acting factors, typically, a class of splicing regulatory factors: arginine/serine-rich (SR) proteins. SR proteins are recruited by CA- and purine-rich enhancer cis elements. SR proteins facilitate splicing through a series of protein–protein and protein–RNA bridges and are required for the recruitment of essential splicing factors U1 small nuclear ribonucleoproteins and U2AF to the 5′ splice site and the polypyrimidine (polyPy) tract, respectively. ESEs have been identified in exons that are flanked by one or more suboptimal cis-splice signals, such as polyPy tracts, and donor or acceptor sites. Positively acting ESEs can compensate for these suboptimal splice elements, thereby allowing constitutive inclusion of nonconsensus exons. Consistent with this, the polyPy tract, which optimally consists of a long, uninterrupted succession of “U” nucleotides (with “C” less preferred), upstream of exon 7 (SMN1 and SMN2) is interrupted by several “C” and nonconsensus “A” residues. Identification of the factor(s) mediating exon 7 processing would provide mechanistic insight into SMA and potential targets for therapy.
Neuronal-specific splicing factors that regulate the inclusion/exclusion of specific exons have been identified (31). SMN exon 7 processing does not appear to be mediated by a neuronal-specific splicing factor because SMNΔ7/FL ratios are similar in a variety of widely divergent cell lines. However, it remains possible that a specific factor is quantitatively, spatially, or temporally restricted in developing motor neurons that could account for the specific defect present in SMA.
The identification of a new SMN1 mutation with a single nucleotide substitution at the intron 7+6 (922+6 G/T) position emphasizes the critical role of exon 7 alternative splicing. This mutation disrupts the 5′ splice site (donor site) in intron 7 that is recognized by U1 small nuclear ribonucleoproteins early in the formation of the spliceosomal complex. The principle unit of definition by the vertebrate pre-mRNA splicing machinery is believed to be the exon. Based on the exon-definition model, exon 7, as other exons, must be “defined” primarily by relatively short cis signals, including the flanking 5′ and 3′ splice sites, the branch point, and the polypyrimidine tract. The intron 7+6 mutation disrupts the 5′ splice site; therefore, exon 7 is poorly recognized by the splicing machinery and subsequently is “skipped,” allowing the exon 6 donor to splice with the exon 8 acceptor. Although the mutation exists within the context of a SMN1 gene, it is, at best, functionally equivalent to SMN2 because only very low levels of the full-length transcript were detectable. The mutation was isolated from an SMA type III patient, consistent with the notion that SMNΔ7 produced from SMN2 or SMN2 equivalents function in a dose-dependent manner to decrease the severity of the disease. Because this patient also carries at least two copies of SMN2, low levels of FL and abundant SMNΔ7 are produced from SMN2. However, it cannot be determined conclusively whether the low levels of FL produced or whether the SMNΔ7 affords the partial protection from severe SMA. Recently, a correlation has been established between the levels of SMNΔ7 transcript and disease severity, suggesting a pathogenic contribution for SMN2 (32). Intronic mutations such as this and other mutations that alter the SMNΔ7/FL ratio indicate the need to fully characterize the SMN genes in compound heterozygotes.
These results may prove a useful tool in the development of an SMA animal model. Because there is a single copy of SMN analogous to SMN1 in the mouse and its targeted disruption results in early embryonic lethality (33), construction of transgenic mice with SMN2 or SMN genes with similar splicing patterns would be predicted to result in viable mice with the SMA phenotype. Furthermore, because all 5q-linked SMA patients possess at least a single copy of SMN2, increasing FL SMN expression from SMN2 might represent a potential therapy (6, 13, 24). Based on these findings, future efforts in gene therapy specifically could target the single nucleotide conversion at the exon 7+6 position, thereby functionally converting SMN2 to SMN1.
Acknowledgments
We thank S. Young for technical assistance, Arthur Burghes for helpful discussions, David Lazinski for C6 cells, Susan Berget for T98G cells, and Dona Chikaraishi for Catha.a and Cad cells. E.H. was supported by a fellowship from the Deutscher Akademischer Austauschdienst. C.L.L. was supported by a fellowship from the Families of SMA. Funding for these studies was provided by the Deutsche Forschungsgemeinschaft to B.W. and the Muscular Dystrophy Association to E.J.A.
ABBREVIATIONS
- SMN
survival motor neuron
- SMA
spinal muscular atrophy
- ESE
exon-splicing enhancer
- FL
full length
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Pearn J. Lancet. 1980;1:919–922. doi: 10.1016/s0140-6736(80)90847-8. [DOI] [PubMed] [Google Scholar]
- 2.Lefebvre S, Burglin L, Reboullet S, Clermont O, Burlet P, Viollet L, Benichou B, Cruaud C, Millasseau P, Zeviani M, et al. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 3.Rodrigues N R, Owen N, Talbot K, Ignatius J, Dubowitz V, Davies K E. Hum Mol Genet. 1995;4:631–634. doi: 10.1093/hmg/4.4.631. [DOI] [PubMed] [Google Scholar]
- 4.Hahnen E, Forkert R, Marke C, Rudnik-Schoneborn S, Schonling J, Zerres K, Wirth B. Hum Mol Genet. 1995;4:1927–1933. doi: 10.1093/hmg/4.10.1927. [DOI] [PubMed] [Google Scholar]
- 5.Cobben J, van der Steege G, Grootscholten P, de Visser M, Scheffer H, Buys C. Am J Hum Genet. 1995;57:805–808. [PMC free article] [PubMed] [Google Scholar]
- 6.Burghes A. Am J Hum Genet. 1997;61:9–15. doi: 10.1086/513913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roy N, Mahadevan M S, McLean M, Shutler G, Yaraghi Z, Farahani R, Baird S, Besner-Johnston A, Lefebvre C, Kang X. Cell. 1995;80:167–178. doi: 10.1016/0092-8674(95)90461-1. [DOI] [PubMed] [Google Scholar]
- 8.Carter T, Bonnemann C, Wang C, Obici S, Parano E, De Fatima Bonaldo M, Ross B, Penchaszadeh G, Mackenzie A, Soares M, et al. Hum Mol Genet. 1997;6:229–236. doi: 10.1093/hmg/6.2.229. [DOI] [PubMed] [Google Scholar]
- 9.Burglen L, Seroz T, Miniou P, Lefebvre S, Burlet P, Munnich A, Pequignot E, Egly J M, Melki J. Am J Hum Genet. 1997;60:72–79. [PMC free article] [PubMed] [Google Scholar]
- 10.Scharf J M, Endrizzi M G, Wetter A, Huang S, Thompson T G, Zerres K, Dietrich W F, Wirth B, Kunkel L M. Nat Genet. 1998;20:83–86. doi: 10.1038/1753. [DOI] [PubMed] [Google Scholar]
- 11.Coovert D, Le T, McAndrew P, Strasswimmer J, Crawford T, Mendell J, Coulson S, Androphy E J, Prior T, Burghes A H M. Hum Mol Genet. 1997;6:1205–1214. doi: 10.1093/hmg/6.8.1205. [DOI] [PubMed] [Google Scholar]
- 12.Lefebvre S, Burlet P, Liu Q, Bertrandy S, Clermont O, Munnich A, Dreyfuss G, Melki J. Nat Genet. 1997;16:265–269. doi: 10.1038/ng0797-265. [DOI] [PubMed] [Google Scholar]
- 13.Melki J. Curr Opin Neurol. 1997;10:381–385. doi: 10.1097/00019052-199710000-00005. [DOI] [PubMed] [Google Scholar]
- 14.McAndrew P E, Parsons D W, Simard L R, Rochette C, Ray P N, Mendell J R, Prior T W, Burghes A H. Am J Hum Genet. 1997;60:1411–1422. doi: 10.1086/515465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Q, Dreyfuss G. EMBO J. 1996;15:3555–3565. [PMC free article] [PubMed] [Google Scholar]
- 16.Fischer U, Liu Q, Dreyfuss G. Cell. 1997;90:1023–1029. doi: 10.1016/s0092-8674(00)80368-2. [DOI] [PubMed] [Google Scholar]
- 17.Pellizzoni L, Kataoka N, Charroux B, Dreyfuss G. Cell. 1998;95:615–624. doi: 10.1016/s0092-8674(00)81632-3. [DOI] [PubMed] [Google Scholar]
- 18.Lorson C L, Androphy E J. Hum Mol Genet. 1998;7:1269–1275. doi: 10.1093/hmg/7.8.1269. [DOI] [PubMed] [Google Scholar]
- 19.Talbot K, Ponting C P, Theodosiou A M, Rodrigues N R, Surtees R, Mountford R, Davies K E. Hum Mol Genet. 1997;6:497–500. doi: 10.1093/hmg/6.3.497. [DOI] [PubMed] [Google Scholar]
- 20.Lorson C L, Strasswimmer J, Yao J-M, Baleja J D, Hahnen E, Wirth B, Thanh L, Burghes A H M, Androphy E J. Nat Genet. 1998;19:63–66. doi: 10.1038/ng0598-63. [DOI] [PubMed] [Google Scholar]
- 21.Lefebvre S, Bürglen L, Frézal J, Munnich A, Melki J. Hum Mol Genet. 1998;7:1531–1536. doi: 10.1093/hmg/7.10.1531. [DOI] [PubMed] [Google Scholar]
- 22.Burglen L, Lefebvre S, Clermont O, Burlet P, Viollet L, Cruaud C, Munnich A, Melki J. Genomics. 1996;32:479–482. doi: 10.1006/geno.1996.0147. [DOI] [PubMed] [Google Scholar]
- 23.Gennarelli M, Lucarelli M, Capon F, Pizzuti A, Merlini L, Angelini C, Novelli G, Dallapiccola B. Biochem Biophys Res Commun. 1995;213:342–348. doi: 10.1006/bbrc.1995.2135. [DOI] [PubMed] [Google Scholar]
- 24.Campbell L, Potter A, Ignatius J, Dubowitz V, Davies K. Am J Hum Genet. 1997;61:40–50. doi: 10.1086/513886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hahnen E, Schonling J, Rudnik-Schoneborn S, Zerres K, Wirth B. Am J Hum Genet. 1996;59:1057–1065. [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang M Q. Hum Mol Genet. 1998;7:919–932. doi: 10.1093/hmg/7.5.919. [DOI] [PubMed] [Google Scholar]
- 27.Berget S M. J Biol Chem. 1995;270:2411–2414. doi: 10.1074/jbc.270.6.2411. [DOI] [PubMed] [Google Scholar]
- 28.Bussaglia E, Clermont O, Tizzano E, Lefebvre S, Burglen L, Cruaud C, Urtizberea J A, Colomer J, Munnich A, Baiget M, et al. Nat Genet. 1995;11:335–337. doi: 10.1038/ng1195-335. [DOI] [PubMed] [Google Scholar]
- 29.Bai G, Lipton S A. Neuron. 1998;20:363–366. doi: 10.1016/s0896-6273(00)80979-4. [DOI] [PubMed] [Google Scholar]
- 30.Liu W, Qian C, Francke U. Nat Genet. 1997;16:328–329. doi: 10.1038/ng0897-328. [DOI] [PubMed] [Google Scholar]
- 31.Grabowski P J. Cell. 1998;92:709–712. doi: 10.1016/s0092-8674(00)81399-9. [DOI] [PubMed] [Google Scholar]
- 32.Gavrilov D K, Shi X Y, Das K, Gilliam T C, Wang C H. Nat Genet. 1998;20:230–231. doi: 10.1038/3030. [DOI] [PubMed] [Google Scholar]
- 33.Schrank B, Gotz R, Gunnersen J, Ure J, Toyka K, Smith A, Sendtner M. Proc Natl Acad Sci USA. 1997;94:9920–9925. doi: 10.1073/pnas.94.18.9920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wirth B, Herz M, Wetler A, Hahnen E, Rudnik-Schoneborn S, Wienker T, Zerres K. Am J Hum Genet. 1999;64:1340–1356. doi: 10.1086/302369. [DOI] [PMC free article] [PubMed] [Google Scholar]