Abstract
Subjective methods have been reported to adapt a general-purpose ontology for a specific application. For example, Gene Ontology (GO) Slim was created from GO to generate a highly aggregated report of the human-genome annotation. We propose statistical methods to adapt the general purpose, OBO Foundry Disease Ontology (DO) for the identification of gene-disease associations. Thus, we need a simplified definition of disease categories derived from implicated genes. On the basis of the assumption that the DO terms having similar associated genes are closely related, we group the DO terms based on the similarity of gene-to-DO mapping profiles. Two types of binary distance metrics are defined to measure the overall and subset similarity between DO terms. A compactness-scalable fuzzy clustering method is then applied to group similar DO terms. To reduce false clustering, the semantic similarities between DO terms are also used to constrain clustering results. As such, the DO terms are aggregated and the redundant DO terms are largely removed. Using these methods, we constructed a simplified vocabulary list from the DO called Disease Ontology Lite (DOLite). We demonstrated that DOLite results in more interpretable results than DO for gene-disease association tests. The resultant DOLite has been used in the Functional Disease Ontology (FunDO) Web application at http://www.projects.bioinformatics.northwestern.edu/fundo.
Contact: s-lin2@northwestern.edu
1 INTRODUCTION
A general-purpose open biomedical ontology can be thought of as a knowledge capture or knowledge representation device. According to the Open Biomedical Ontologies (OBO) Foundry principles, the ontology should contain well-defined terms with well-defined relationships (e.g. is_a, part_of) between the terms, and represent as much of the current knowledge in a given domain as possible. Disease Ontology (DO) is an OBO Foundry ontology, organized such that the path to the root is always true. DO organizes disease concepts in a directed acyclic graph (DAG) so that traversing away from the root of DO moves towards progressively more granular and specific terms. The full DO graph is very useful for organizing a wide spectrum of data, but is not necessarily optimal for specific applications such as identifying disease-gene relationships. By analogy with other biological ontologies, a trimmed-down version is very useful for building a high-level functional summary from a gene list. For instance, a simplified version of the Gene Ontology (GO) called ‘GO Slim’ provides a broad, integrative overview of molecular and cellular biology by combining and removing fine-grained terms in the GO. GO Slim has proven critical in comparing annotations across genomes (Adams et al., 2000) and interpreting biological functions of a gene list (Shah and Fedoroff, 2004).
We are especially interested in interpreting a list of genes in the context of disease, which is a critical step in translating molecular findings from microarrays, proteomics and other types of high-throughput screening methods into clinical relevance. To achieve this goal, we were part of the collaboration that has developed the general-purpose OBO Foundry DO (http://www.diseaseontology.sourceforge.net). The DO has been successfully used here at Northwestern in the NUgene project to build a detailed disease phenotype from data available in the electronic medical record. In addition, the DO has been used as a controlled vocabulary to annotate the human genome in terms of diseases (Osborne et al., 2009).
However, the DO is very complex: revision 26 of the DO contains 11 961 terms in the form of a directed acyclic graph, of which 4399 terms are internal nodes with up to 16 levels of hierarchical structures. Such a complex structure creates a special challenge for functional enrichment tests.
Functional enrichment tests were originally developed to interpret biological meanings of a gene list from microarray experiments (Dopazo, 2006). Such statistical tests are also called GO analysis, because the GO was originally used to define the functional categories. Briefly, k functional categories (diseases) are defined. For each category, a hypergeometric test is used to test the enrichment of proportions (per cent of genes belong to this category) in the identified gene list versus the genome (Falcon and Gentleman, 2007). Although more sophisticated statistical procedures are in development (Goeman and Mansmann, 2008), canonical tests assume that each category is independent and identically distributed (iid, i.e. not in a hierarchical structure). A simplified approach is to treat each term in the ontology graph as an independent category (ignoring the graph structure), which not only violates the iid assumption but also creates another problem of multiple comparisons (k = 11 961 for DO). Thus, trimming the DO into simplified categories will make the statistical test tractable as well as the results simpler for human comprehension.
A simple method of trimming DO terms is to select the DO terms at a certain level in the DAG. This method is very arbitrary and it is hard to determine a fixed level suitable for different branches in the DAG; moreover, many important DO terms might be missed. To deal with the similar problem for GO, Alterovitz et al. (2007) proposed to partition the GO database based on information content (Shannon information) of individual GO terms. But how to choose and interpret the level of information content is still a problem. Here, we prefer grouping the DO terms using semantic distance in the context of gene annotations. The semantic distances in previous studies are either information based (Jiang and Conrath, 1997; Resnik, 1999), graph-structure based (Wang et al., 2007; Wu et al., 2005) or a hybrid of both (Sheehan et al., 2008). One limitation of graph-structure-based semantic distances is that it does not consider genes that are annotated by DO terms. As the genes annotated by DO terms are direct evidence of disease classification at the molecular level, DO terms with the same or similar mapped genes are closely related and can be combined. Moreover, the combination of DO terms based on similar gene mappings will remove the redundancies from the results of an enrichment test. Therefore, we define the distance metrics of DO terms based on gene-to-DO mapping profiles. The mapping profile-based distance metric has been used to measure the similarity between two genes, but, to our knowledge, it has not been used to measure the ontological similarity. As we are interested in both the overall similarity and the subset similarity (one DO term-associated gene list is the subset of another one) between two DO terms, we separately define two types of binary distance metric. A compactness-scalable fuzzy clustering method (Du et al., 2005) is then applied to group similar DO terms based on defined distance metrics. For the clustering using subset similarity distance metric, the clustering results are further constrained by the semantic similarities between DO terms to reduce the false clustering. Following these steps, a domain expert was assigned to curate the computational results and assign new names to these DO term groups. We named these simplified controlled vocabulary lists of diseases as ‘Disease-Ontology Lite’ (DOLite). The details of each step are described in the methodology section.
2 METHODS
2.1 Overview of building the DOLite database
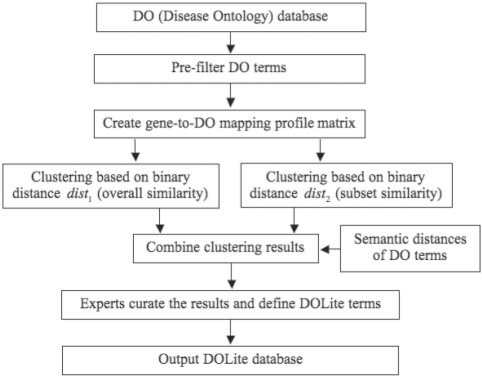
Figure 1 shows the framework of creating the DOLite database based on the DO database. Next, we describe each step in detail.
Fig. 1.
Flowchart of creating DOLite database based on DO database.
2.2 Pre-filtering the DO terms
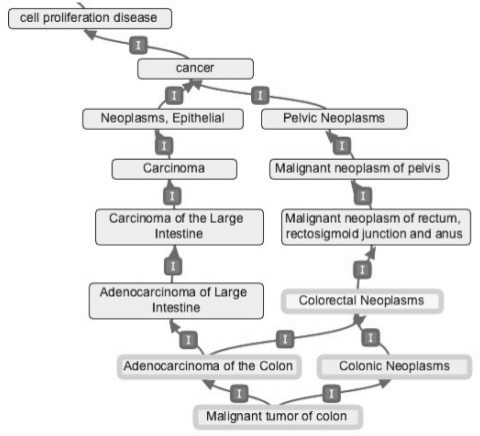
In the DO database, many of the DO terms are abstract concepts created for the purpose of the ontological reasoning; they are not used in scholarly communications and have very few genes directly associated with them. For instance, ‘cell proliferation disease’ only appears three times in 18 million MEDLINE abstracts, but it is still listed as a disease category in the DO graph (Fig. 2). A quick filtering based on the number of genes directly mapped to each DO term can easily identify these abstract DO terms.
Fig. 2.
A portion of the DO graph showing the complexity of DO.
Some other DO terms, which are not very well studied, also have very few genes associated. Because of their small sizes of gene lists, they are easy to be significant by chance in the enrichment test. To reduce the false positives and increase the computational efficiency, we pre-filter these DO terms and map their associated genes to their direct parent DO terms before further calculations.
2.3 Gene-to-DO mapping profile matrix
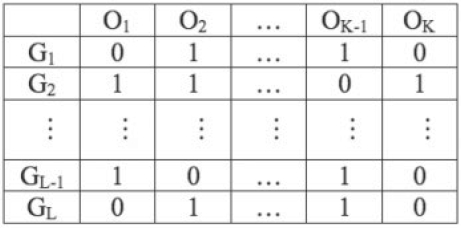
Each gene in the human genome was annotated with DO, as we reported before (Osborne et al., 2009).Figure 3 shows the mapping profile matrix M. Each element Mlk represents the mapping between gene Gl and ontology term Ok. The values of the matrix elements are as follows: for Mlk = 1, there exists evidence showing that gene Gl is involved in the disease defined by ontology term Ok and for Mlk = 0, there is a lack of evidence. Lack of evidence does not imply that there is no relationship but rather that there is no information available on a relationship. For our application, each column is a gene-to-DO mapping profile of the corresponding DO term; it is also equivalent to a DO-term-associated gene list.
Fig. 3.
An example gene-to-DO mapping matrix.
2.4 Distance metrics between gene-to-DO mapping profiles
To measure the similarity between DO terms, we first need to define the distance metrics. We chose to use the gene-to-DO mapping matrix M based on distance because we are interested in identifying associations between genes and DO terms. Kappa statistics were used to measure the similarities between genes based on their mapping profiles (rows of mapping profile matrix M in Fig. 3) (Dennis et al., 2003). As kappa statistics put equal importance on 0s and 1s, this is very problematic when applied to cluster DO terms. As shown in Figure 3, Mlk = 0 is a lack of evidence of involvement, not information that rules out an involvement between gene Gl and ontology term Ok. Therefore, our distance metrics will focus on 1s. Considering the special situation of the structured ontology, we defined two types of binary distance metrics, as shown in Equations (1) and (2). The distance values are between 0 and 1, with values closer to 0 indicating closer similarities. In Equations (1) and (2), ‘∧’ is the binary AND operator and ‘ν’ is the binary OR operator. dist1(A,B) measures the overall similarity between two binary vectors A and B. In our case, A and B represent the columns of mapping profile matrix M, and the distance is 1 minus the percentage of overlapping genes among all genes associated with DO terms A and B. dist2(A,B) measures the subset similarity between two binary vectors A and B. In our case, it is 1 minus the percentage of one DO term-associated gene list being the subset of another DO term-associated gene list. This is of interest particularly when these two ontology terms have a close semantic relationship; e.g. for parent-child relationships in DO DAG graph.
| (1) |
| (2) |
2.5 Clustering the gene-to-DO mapping profiles
With respect to the molecular basis of disease, significant redundancy and overlap also exist in DO. For example, six terms (‘carcinoma of the large intestine’, ‘adenocarcinoma of large intestine’, ‘adenocarcinoma of the colon’, ‘colorectal neoplasms’, ‘malignant tumour of colon’ and ‘colonic neoplasms’) in a small portion of the DO graph (as shown in Fig. 2) basically describe the same group of diseases centred on the adenocarcinoma of colon. Some of these terms may include other rare cancers affecting the colon, and others may also include benign colonic tumours. However, these nuances might not be differentiable by their associated genes. As such, the enrichment test will likely report all these terms redundantly, making it more difficult to interpret. In order to find the DO terms with very similar associated genes, we perform clustering based on gene-to-DO mapping matrix M. Two types of clustering based on two different binary distance metrics, as shown in Equations (1) and (2), are performed.
The first clustering is based on the binary distance metric, dist1, which measures the overall similarity of two ontology terms (i.e. their associated gene lists). As one gene can involve in multiple diseases, semantically distant DO terms may have significant biological pathway overlap and we do not want to prune semantically distant terms as these may be biologically informative. Unlike the conventional clear-cut clustering algorithms, fuzzy clustering methods allow the overlapping assignment of the DO terms to different clusters with varying degrees of membership. Therefore, fuzzy clustering is ideally suited for our situation. Another requirement for the clustering algorithm is the ability to control of cluster compactness, which means the level of similarity among cluster members can be modified by the researcher. On the basis of these considerations, we use the compactness-scalable fuzzy K-means clustering algorithm described in Du et al. (2005). This algorithm adds a scalable Gaussian window to the fuzzy membership function to control the compactness of cluster. The σ Gaussian window function controls the scale of the clustering algorithm, with smaller scales having more compact clusters. For more details, please refer to Du et al. (2005). Just as K-means clustering algorithms, the clustering result of compactness-scalable fuzzy K-means algorithm depends on the initial choice of the cluster centres. To better estimate the initial cluster centres, we first perform a hierarchical clustering using a complete linkage method. By limiting the height of the resultant hierarchical tree, we use the complete linkage method to seed the initial cluster centres for the compactness-scalable fuzzy K-means clustering algorithm.
The second clustering is based on the distance metric, dist2, which measures the subset similarity between DO terms. For the case of one DO term is a subset (or partial subset) of another, we are only interested in the case with the terms having high semantic similarity. Therefore, the clustering result will be constrained by the DO semantic distance as described in the Section 2.6). Because of the constraint on DO semantic similarity, the results of fuzzy and hierarchical clustering algorithms are very similar. Since hierarchical clustering algorithms are much more efficient, the second cluster was built by hierarchical clustering.
2.6 Semantic distances between DO terms
The estimation of semantic distance can be categorized as information-based, graph-based and hybrid methods. We select to use a graph-based Union–Intersection (UI) method, as implemented in the GOstats Bioconductor package (Falcon and Gentleman, 2007), for its simplicity, ease of interpretation and consistency with other distance metric defined in Equations (1) and (2). The semantic distance of UI method is 1 minus the percentage of overlapping nodes between two induced graphs among the total number of nodes in two graphs. The resulting distance is in the range of 0–1 with values close to 0 having better similarities. The graph corresponding to each DO term is composed of multiple paths from the DO term to the root of the DO terms (disease). An implication of this method is that two deep sister nodes (i.e. nodes that are distant from the root) will have higher levels of similarity than two shallow sister nodes. Given the design of DO, this is consistent and intuitive with the semantic similarity, or lack thereof, for shallow nodes. This also means that two neighbouring, deep nodes, one of which has two or more distinct paths to the root will be quite distinct from nodes that only have one path to the top, or have differing paths to the top. This again leverages the biological knowledge embedded in DO.
2.7 Combine clustering results
We treat the clustering result based on dist1 as the major clusters. The clustering result based on dist2 is sub-clustered based on the semantic distance between DO terms. Hierarchical clustering using the single linkage method was used for sub-clustering. Sub-clusters with single elements are filtered from further processing. Other sub-clusters are then merged with major clusters whenever there are overlapping elements.
2.8 Expert curation of DOLite
After the statistical treatments as described above, a board-certified pathologist manually edited the clustered DO terms, resolved the computational artefacts and assigned a class label for each DOLite term. DOLite is a simplified version of the DO for functional enrichment tests of genes (Table 2). Instead of using medical terminologies for the label and definitions, we have attempted to use common English names as used in Wikipedia for DOLite.
Table 2.
DO versus DOLite
| DO | DOLite | |
|---|---|---|
| Design | Ontology | Controlled vocabulary |
| Purpose | General | Specifically for functional enrichment tests of genes |
| Structure | Directed acyclic graph | Immutable list |
| Details | Finer | Coarser |
| Linked to | ULMS | Wiki |
| Number of terms | 11961 | 561 |
ULMS: Unified Medical Language System.
3 RESULTS
Following the procedures shown in Figure 1, we created DOLite based on the DO. The DO terms were filtered to include only those with at least five direct mappings. A gene-to-DO mapping profile matrix was created. In this mapping matrix, each DO term also include the mappings of its offspring. Clustering was performed based on this mapping profile matrix. Because all the distance metrics we used are in the range of 0–1 and are related with the percentage of overlapping, we used the same threshold value for different steps, which include the cut-off thresholds used in clustering and the maximum allowed semantic distances of DO terms. The scale of fuzzy clustering is selected as the half of this threshold value. In our implementation, we set this distance threshold value as 0.2 (80% of similarity). As a reference to the expert curator, we also provide the clustering results based on looser threshold values 0.3 and 0.5. The expert curator decided the final mapping between DO terms and DOLite terms. Although the general-purpose DO is designed to differentiate the nuances of different disease categories, for the application of functional enrichment test of gene lists, we decide to aggregate the related terms and assign common English names for the diseases (as used in Wikipedia). An example is shown in Table 1. Table 2 shows the overview of final DOLite database and its comparison with DO database.
Table 1.
An example of mapping DO to DOLite
| DOID | DO term | DOLite term |
|---|---|---|
| DOID:680 | Tauopathies | Alzheimer's disease |
| DOID:1307 | Dementia | Alzheimer's disease |
| DOID:10652 | Alzheimer's disease | Alzheimer's disease |
3.1 DOLite annotation of the human genome
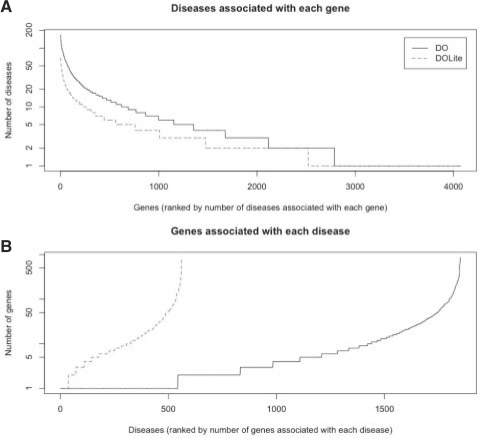
With the DO-to-DOLite mapping, we convert the previous annotation of the human genes by DO into DOLite annotations. Compared with DO, the DOLite annotation of the human genome is more compact, as evidenced by the larger number of genes assigned to individual disease category (Fig. 4B), and the smaller number of disease categories assigned to individual gene (Fig. 4A).
Fig. 4.
Comparison of DO and DOLite annotation of the human genome. (A) The number of diseases per gene is plotted for the DO and the DOLite. (B) The number of genes per disease is plotted for the DO and the DOLite.
3.2 Validation of DOLite using a benchmark microarray data set
To validate the utility of DOLite, we used a benchmark microarray data set of pancreatic cancer study (Antonov et al., 2008), which was previously utilized to test GO-based annotations (Falcon and Gentleman, 2007), for the functional enrichment test. Briefly, a list of 125 genes identified in that study was used for functional analysis. Hypergeometric test was applied to each DO (Osborne et al., 2009) and DOLite term in the database.
We show side-by-side the top 12 categories (ranked by P-values) of the DO and DOLite results in Table 3 and Figure 5. As expected, the conventional DO analysis returned highly redundant terms such as ‘cancer’, ‘carcinoma’ and ‘adenocarcinoma’, likely because a number of these genes are shared in the biological process of cancer development (Fig. 5A). However, no pancreas-specific disease entities were picked up. Instead, it returned some diseases unrelated to pancreatic adenocarcinoma, such as respiratory tract disease, soft tissue neoplasm and skin disease. These results may represent non-specific hits that contribute little to data interpretation. In contrast, the analysis based on DOLite did successfully return a number of entities closely related to the pathology of pancreas, in addition to generic cancer-related terms. These entities include diabetes mellitus, hypoglycemia and obesity and disorders frequently associated with the malfunction of pancreatic islet cells. Another entity, primary biliary cirrhosis, also causes pancreatic injury. These results support a strong pancreas conjunction of the tested gene set, validating the effectiveness of using DOLite to interpret microarray data.
Table 3.
Top 12 categories of functional enrichment tests based on DO and DOLite database
| DO | DOLite | ||||
|---|---|---|---|---|---|
| DO term | Fold-enrichment | P-value | DOLite term | Fold-enrichment | P-value |
| Cancer | 7.12 | 3.45E–36 | Cancer | 13.55 | 1.95E–25 |
| Malignant neoplasms | 6.93 | 8.74E–33 | Diabetes mellitus | 11.02 | 1.14E–09 |
| Carcinoma | 10.60 | 2.08E–32 | Breast cancer | 9.24 | 8.23E–09 |
| Respiratory tract diseases | 11.03 | 2.66E–32 | Colon cancer | 10.54 | 2.17E–07 |
| Respiratory system disease | 11.01 | 2.83E–32 | Lung cancer | 11.35 | 3.11E–06 |
| Neoplasms, epithelial | 10.22 | 9.34E–32 | Embryoma | 8.85 | 1.58E–05 |
| Adenocarcinoma | 13.30 | 1.90E–29 | Atherosclerosis | 9.73 | 3.84E–05 |
| Gastrointestinal neoplasms | 11.40 | 5.29E–28 | Stomach cancer | 11.79 | 7.06E–05 |
| Disease of skin | 9.88 | 2.36E–27 | Primary biliary cirrhosis | 32.18 | 1.12E–04 |
| Soft Tissue neoplasms | 11.28 | 4.77E–27 | Hypoglycemia | 110.84 | 1.34E–04 |
| Alimentary system disease | 8.66 | 1.05E–26 | Obesity | 9.78 | 1.70E–04 |
| Malignant neoplasm of gastrointestinal tract | 12.20 | 9.99E–26 | Pancreas cancer | 14.30 | 1.85E–04 |
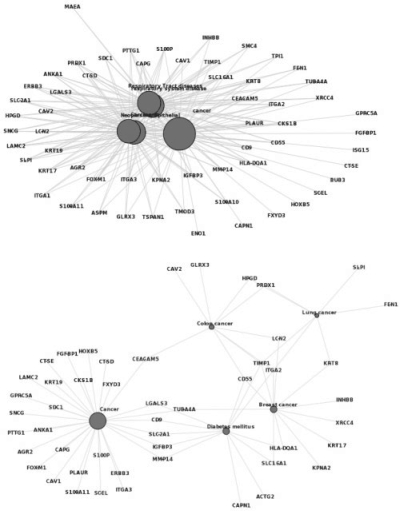
Fig. 5.
Disease-gene network analysis of the pancreatic cancer data set by (A) DO and (B) DOLite.
Results from DOLite analysis also lead to interesting discoveries that deserve further clinical investigation. We observed that breast cancer was ranked very high among the DOLite results. This could be simply because breast cancer is one of the most extensively studied cancers and therefore has more associated genes. However, the predominant type of breast cancer is adenocarcinoma, which can resemble some pancreatic adenocarcinomas on histology. So, could these two cancers share some common genetic abnormalities as well, other than those abnormalities seen in most cancers (e.g. genes involved in cell cycle control)? This may be an interesting question to explore.
4 DISCUSSION
Exploring relations between genes and diseases at the molecular level could greatly facilitate our understanding of pathogenesis, and eventually lead to better diagnosis and treatment. DO was constructed as a general-purpose ontology to define diseases. It is very useful to associate genes with diseases (e.g. a particular cancer) or disease-related processes (e.g. tumour metastasis). However, due to the complex structure of DO, the results of functional enrichment test are usually hard to interpret. In this study, we proposed statistical methods to slim the general-purpose disease ontology to a simplified controlled vocabulary list called ‘Disease-Ontology Lite’ (DOLite) specifically for the functional enrichment test of a gene list. DOLite has several advantages over the conventional DO for functional enrichment test. The complex hierarchical structure of DO is unnecessary for the desired enrichment test. DOLite contains better-defined disease entities that are easier to interpret by researchers and clinicians. The entities are also enriched with associated genes, leading to increased sensitivity of detection.
The enrichment test using the benchmark microarray data set of pancreatic adenocarcinoma suggests that DOLite returned diseases specific to the organ where original experiments were conducted on, which directly confirm the validity of the experimental results.
In terms of contributions to biological databases, we created DOLite, the first simplified version of the DO, and utilized DOLite to annotate the human genome. In terms of methodological contribution, we defined statistical methods to simplify a general-purpose ontology. In contrast, the previous construction of GO Slim from GO was largely a subjective process based on expert opinion. The major contributions in methodology include computing the ontology similarity based on gene-to-ontology mapping profiles [derived from geneRIF (Harris et al., 2004; Shah and Fedoroff, 2004)]; defining two types of binary distance metrics to separately measure the overall similarities and subset similarities and a compactness-scalable fuzzy clustering method (clustering results were verified with the constraints of semantic distance between DO terms). The methodology can be easily adapted to slim other ontologies, like GO. Currently, DOLite is utilized in the Functional Disease Ontology (http://www.projects.bioinformatics.northwestern.edu/fundo) Web application to interpret clinical relevancies of a gene list.
Previous studies have suggested that GO Slim, which is a simplified version of GO, greatly facilitates genome-wide computational analysis (Nam and Kim, 2008). Similarly, we expect that DOLite and its annotation of the human genome can be used as a foundation for the development of more computational and statistical methods to analyse the disease relevance of a gene list. In the future, we will further refine the algorithms for creating the DOLite database. For example, we can better integrate the semantic distance of DO terms into the clustering process. We also plan to add more sensitive methods to test the enrichment of each DOLite term in a gene list.
Funding: This project was supported in part by Award Number UL1RR025741 from the National Center for Research Resources. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Center for Research Resources or the National Institutes of Health.
Conflict of Interest: none declared.
REFERENCES
- Adams MD, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Alterovitz G, et al. GO PaD: the gene ontology partition database. Nucleic Acids Res. 2007;35:D322–D327. doi: 10.1093/nar/gkl799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonov AV, et al. ProfCom: a web tool for profiling the complex functionality of gene groups identified from high-throughput data. Nucleic Acids Res. 2008;36:W347–W351. doi: 10.1093/nar/gkn239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G, et al. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- Dopazo J. Functional interpretation of microarray experiments. OMICS. 2006;10:398–410. doi: 10.1089/omi.2006.10.398. [DOI] [PubMed] [Google Scholar]
- Du P, et al. Modeling gene expression networks using fuzzy logic. IEEE Trans. on SMCB (Part B) 2005;35:1351–1359. doi: 10.1109/tsmcb.2005.855590. [DOI] [PubMed] [Google Scholar]
- Falcon S, Gentleman R. Using GOstats to test gene lists for GO term association. Bioinformatics. 2007;23:257–258. doi: 10.1093/bioinformatics/btl567. [DOI] [PubMed] [Google Scholar]
- Goeman JJ, Mansmann U. Multiple testing on the directed acyclic graph of gene ontology. Bioinformatics. 2008;24:537–544. doi: 10.1093/bioinformatics/btm628. [DOI] [PubMed] [Google Scholar]
- Harris MA, et al. The gene ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32:D258–D261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang JJ, Conrath DW. Proceeding of ROCLING X. Tapei, Taiwan: Academia Sinica; 1997. Semantic similarity based on corpus statistics and lexical taxonomy. [Google Scholar]
- Nam D, Kim SY. Gene-set approach for expression pattern analysis. Brief Bioinform. 2008;9:189–197. doi: 10.1093/bib/bbn001. [DOI] [PubMed] [Google Scholar]
- Osborne JD, et al. BMC Genomics. in press; 2009. Annotating the human genome with disease ontology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnik P. Semantic similarity in a taxonomy: an information-based measure and its application to problems of ambiguity in natural language. JAIR. 1999;11:95–130. [Google Scholar]
- Shah NH, Fedoroff NV. CLENCH: a program for calculating cluster enrichment using the gene ontology. Bioinformatics. 2004;20:1196–1197. doi: 10.1093/bioinformatics/bth056. [DOI] [PubMed] [Google Scholar]
- Sheehan BE, et al. A relation-based measure of semantic similarity for gene ontology annotations. BMC Bioinformatics. 2008;9:468. doi: 10.1186/1471-2105-9-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JZ, et al. A new method to measure the semantic similarity of GO terms. Bioinformatics. 2007;23:1274–1281. doi: 10.1093/bioinformatics/btm087. [DOI] [PubMed] [Google Scholar]
- Wu H, et al. Prediction of functional modules based on comparative genome analysis and gene ontology application. Nucleic Acids Res. 2005;33:2822–2837. doi: 10.1093/nar/gki573. [DOI] [PMC free article] [PubMed] [Google Scholar]