Abstract
Motivation: Viruses employ various means to evade immune detection. One common evasion strategy is the removal of CD8+cytotoxic T-lymphocyte epitopes. We here use a combination of multiple bioinformatic tools and large amount of genomic data to compute the epitope repertoire presented by over 1300 viruses in many HLA alleles. We define the ‘Size of Immune Repertoire score’, which represents the ratio between the epitope density within a protein and the expected density. This score is used to study viral immune evasion.
Results: We show that viral proteins in general have a higher epitope density than human proteins. This difference is due to a good fit of the human MHC molecules to the typical amino-acid usage of viruses. Among different viruses, viruses infecting humans present less epitopes than non-human viruses. This selection is not at the amino-acid usage level, but through the removal of specific epitopes. Within a single virus, not all proteins express the same epitopes density. Proteins expressed early in the viral life cycle have a lower epitope density than late proteins. Such a difference is not observed in non-human viruses. The removal of early epitopes and the targeting of the cellular immune response to late viral proteins, allow the virus a time interval to propagate before its host cells are destroyed by T cells.
Contact: louzouy@math.biu.ac.il
1 INTRODUCTION
The infection of a cell by a virus can elicit a Cytotoxic T Lymphocyte (CTL) response to viral peptides presented by the Major Histocompatibility Complex (MHC) class I molecules (Ambagala et al., 2005; Gulzar and Copeland, 2004). Such a CTL response plays a critical role in the host's anti-viral immune response (McMichael et al., 1983). This role is suggested by studies indicating a drop of viral loads and the relief of the acute infection symptoms following the emergence of virus-specific CTLs (Borrow et al., 1994), as well as by data from CD8+ T cells-depleted animal models (Letvin et al., 1999; Negri et al., 2006). The CTL response is also associated with a rapid selection of viral CTL escape variants (Howley et al., 2001; Lichterfeld et al., 2005).
In order to explore the anti-viral CD8+ T cells response, one has to consider the factors affecting the CTL response, such as the kinetics of the viral protein expression, the Human Leukocyte Antigen (HLA) class I genetic background of the infected individuals and the viral gene diversifications (Lichterfeld et al., 2005). These viral genes determine the epitope repertoire presented to the immune system and consequently the immune response.
We here propose to use an immunomic methodology combining genomic data and multiple bioinformatic tools to study the anti-viral CTL response. Using this immunomic analysis, we present a novel all-virus analysis of the viral epitope repertoire and highlight the selective forces affecting viruses and their human host.
In general, CTL epitopes originate from short peptides cleaved by the proteasome (Rock et al., 2002) that can pass through the Transporter associated with Antigen Processing (TAP) and associate non-covalently with the groove of MHC-I molecules. The vast majority of these epitopes are nine-mers (although octamers and decamers and even longer epitopes can be observed). A cleaved nine-mer is presented on an MHC-I molecule only if its affinity to the MHC molecule is sufficiently high. We have recently developed and improved a set of bioinformatic tools to estimate all peptides within a virus that can be presented to the immune system (the CTL epitope repertoire). We here apply this methodology to over 1300 fully sequenced viruses and show that viruses selectively alter their epitope repertoire.
The HLA locus is the most polymorphic locus in the human genome. In the class I locus HLA-A, B and C have over 697, 1109 and 381 alleles, respectively (Robinson et al., 2003). This large polymorphism permits a rapid selection of alleles that can respond to viral threats. On the other hand, viruses can mutate rapidly. For example, the HIV mutation rate is approximately 1.e–3–1.e–4 mutations per base pair per division (Coffin, 1995), which is approximately one mutation per division for the entire viral genome. This high mutation rate coupled with a short viral life cycle [24–72 h for most viruses (Howley, et al., 2001)] allows viruses to modify their epitope repertoire within a short time.
Thus, the number of viral epitope is affected by two opposing trends: the attempts of the human immune system to recognize virally infected cells and the viral attempts to survive for a long enough time in the infected cell to bud. We here propose a direct measure of the epitope number over all fully sequenced viruses to explore the effect of these opposing forces.
2 METHODS
2.1 Genomic data
Viral and human protein sequences were used for this analysis. The human sequences were obtained from the Ensembl database (Birney et al., 2004). All human predicted protein-coding regions were used. In the current analysis, we ignored the effect of point mutations. The viral sequences were obtained from the NCBI (http://www.ncbi.nlm.nih.gov/) and LANL (Kuiken et al., 2003) databases. The proteins of some of the human viruses were divided into groups, according to the available data. Viruses were classified into human or non-human viruses, based on their main host. A virus mainly infecting non-humans that can infect humans, but is not usually transferred from one human to the other was classified as non-human.
2.2 SIR score
We have analyzed the ratio between the number of epitopes presented in viral genes and their random counterpart. The epitope number was computed using three algorithms: a homemade cleavage algorithm (Ginodi et al., 2008), a TAP-binding algorithms developed by Peters et al. (2003) and the BIMAS MHC binding (Parker et al., 1994) algorithms. We have computed epitopes for 31 common HLA alleles and weighted the results according to the allele frequency in the global human population. The algorithms' quality was systematically validated versus epitope databases and was found to induce low FP and FN error rates. A detailed description of the algorithm, their validation and the SIR score can be found in previous works (e.g. Vider-Shalit et al., 2007).
2.3 Validation
To validate our results, we checked the score of peptides present in seven different databases: IEDB (Peters et al., 2005), SYFPEITHI (Rammensee et al., 1999)—www.syfpeithi.de, MHCBN (Bhasin et al., 2003)—http://www.imtech.res.in/raghava/mhcbn/, MPID (Govindarajan et al., 2003)—surya.bic.nus.edu.sg/mpid/, MHCPEP (Brusic et al., 1998)—http://www3.oup.co.uk/nar/database/, AntiJen (Blythe et al., 2002; McSparron et al., 2003)—http://www.jenner.ac.uk/AntiJen/ and HLALigand (Sathiamurthy et al., 2003)—http://hlaligand.ouhsc.edu/LigandDB. Assuming that most peptides in the various databases are correct, we computed the threshold that would maximize the number of presented peptides from the positive databases, and minimize the number of peptides in a neutral set of 1 000 000 random peptides with the NCBI database amino-acid distribution (http://prowl.rockefeller.edu/aainfo/contents.htm). We have checked for each HLA allele the level of type-I and type-II errors and attempted to find a cutoff minimizing both. For most alleles we found cutoffs reducing both errors to less than 10% (i.e. 90% inclusion of the presented peptides and 90% exclusion of the random peptides).
2.4 Statistical analysis
The SIR score of various populations was compared. A one-way t-test with unknown and unequal variance was used to compare the SIR scores of viruses in human and non-human hosts, as well as the SIR scores of human and viral proteins. When comparing viruses, the full virus score was used. When comparing proteins, the SIR score of each protein was used. A t-test over all alleles was used to compare the average SIR score of the epitopes produced from the Hidden Markov Models (HMM) based on either human or viral amino-acid distribution properties. When comparing the early versus late proteins in a large group of viruses an ANOVA procedure was used. In the HIV analysis, we first performed an average over all sequences of each protein, and then compared the average SIR score of all proteins among different organisms. The first averaging was required since the sequence number in different HIV significantly varies.
3 RESULTS
3.1 SIR score
In order to study the human immune response to the viral epitope repertoire, we computed all predicted epitopes in each viral protein in over 1300 viruses. Predicted CTL epitopes are nine-mers fulfilling three criteria: (i) production through proteasomal cleavage. In other words, a given peptide can be potentially presented if its extreme and flanking residues enhance proteasomal cleavage and if it is not cleaved in its center (Ginodi et al., 2008). (ii) transport through the TAP machinery to the endoplasmic reticulum (ER). (iii) presentation in the context of MHC-I (Fig. 1). We have used three algorithms to predict all peptides within a protein successfully passing all these stages (Louzoun and Vider, 2004; Peters et al., 2003; Parker et al., 1994). Specifically, each viral gene was divided into all nine-mers and the appropriate flanking regions. For each nine-mer a cleavage score is computed, based on the nine-mer itself and its flanking regions. Only peptides computed to be properly cut were taken to the next stage. We then computed a TAP-binding score for all nine-mers with a positive cleavage score and choose only supra-threshold peptides. The last stage of the analysis is based on allele-specific MHC-binding scores of all TAP-binding and cleaved nine-mers. Resulting nine-mers that bind a given MHC molecule are defined as epitopes for this HLA allele. All algorithms used here were validated using a quality assurance process versus seven different databases of epitopes experimentally measured to be presented. The validation process ensured that the error levels are low enough to allow a systematic analysis of the repertoire (Ginodi, et al., 2008) (http://peptibase.cs.biu.ac.il/peptibase/validation.htm).
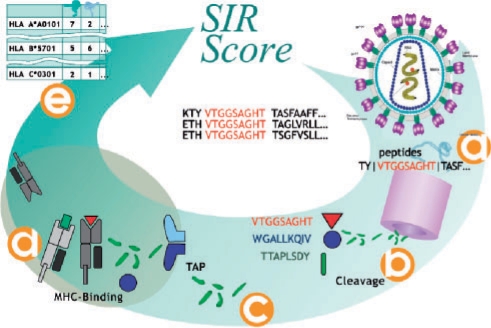
Fig. 1.
Algorithm for the SIR score computation. Each viral gene is divided into all nine-mers and the appropriate flanking regions (a). For each nine-mer a cleavage score is computed (b). We compute a TAP binding for all nine-mers with a positive cleavage score and choose only supra-threshold peptides (c). The MHC-binding score of all TAP-binding and cleaved nine-mers is computed (d). Nine-mers passing all these stages are defined as epitopes. We then compute the number of epitopes per protein per HLA allele (e).
A fraction of the epitopes is transported through a TAP-independent pathway (Yewdell et al., 1998). Since we performed a comparative analysis, we ignored this fraction, assuming that their statistics resemble those of TAP-dependent epitopes and that they should have only a minor effect on the total epitope number. We have ignored octamers and decamers for the same reasons. We estimated the number of epitopes from viral proteins on 9 HLA-A, 19 HLA-B and 3 HLA-C alleles most common in the global human population. These alleles have well-defined MHC-binding motifs. These combined HLA alleles are present in 80–90% of the human population.
3.2 Human vs viral epitope density
In order to check if the MHC allele distribution has evolved to maximize the presentation of viral epitopes, we measured the epitope density of human and viral proteins and compared them in the same allele. The epitope density of a sequence is defined as the number of predicted epitopes divided by the number of candidate nine-mers. Indeed systematically, viral proteins express more epitopes than their human counterpart over the vast majority of alleles (Fig. 2, empty bars) (P < 1.e–7).
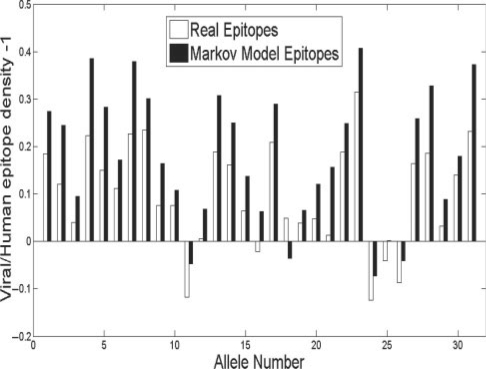
Fig. 2.
Comparison of epitope density in human and viral proteins. The white bars represent the ratio between the relative change between the viral and human epitope densities (i.e. viral epitope desnity/human epitope density –1). All positive values represent a higher viral epitope density. The black bars represent the same comparison performed on a random sequence based on human and viral amino-acid distributions.
We further checked the evolutionary selection of HLA alleles by comparing the frequency of the HLA alleles and the ratio between the density of human and viral epitopes in the same allele. The alleles with the highest ratio are the most frequent one (20% increased frequency of alleles with high ratio compared with alleles with low ratio) highlighting a possible selection at the population level of alleles presenting a large number of viral epitopes.
The most natural mechanism for such a selection is that alleles-binding residues over-represented in viral sequences are preferred. In order to test this hypothesis, we produced two random sequences with different amino-acid distributions. We trained two distinct Markov models on all human and all viral proteins, respectively and produced very long random sequences (1.e6 amino acids) based on these models. We then compared the epitope densities of these two random sequences in each HLA allele (Fig. 2). The ratio between the epitope density of the random sequence based on viral amino acids and the random sequence based on human amino acid is even larger than the ratio between the real viral and human proteins epitope densities (Fig. 2) (P < 0.02 t-test on the ratio between epitopes from the viral and human Markov models, compared with the ratio between computed epitopes from the viral and human real protein sequences). The difference between the Markov models shows that the human MHC system is evolving to recognize the viral amino-acid distribution. The smaller difference between human and viral epitope numbers shows that within the broad specificity of human MHC to preferentially bind epitopes with viral amino-acid distribution, viruses specifically mutate their epitopes to limit detection.
3.3 Human and non-human hosts
Among viruses, the attempts to mutate epitopes should only be observed in human viruses (viruses infecting a human host). We compared the epitope density in viruses living in a human host and the epitope density in their non-human counterparts. In order to ease the comparison, we used the number of epitopes in the Markov-Model-based random strand produced from all viral protein as a normalization factor. We defined a score representing the ratio between the epitope density in a given protein and the epitope density in the viral random strand and named it the ‘Size of Immune Repertoire’ (SIR) score (Almani et al., 2008; Vider-Shalit et al., 2007). This score can be interpreted as the ratio of the number of predicted CTL epitopes to the number of epitopes expected within the same number of random nine-mers with similar amino acid and amino-acid couples frequency distribution (Vider-Shalit et al., 2007). For example, assume a 308-amino acid long sequence. Such a sequence has 300 overlapping nine-mers. If a set of 300 random nine-mers with a similar amino-acid distribution is expected to have 10 HLA A*0201 epitopes and the sequence is computed to have 4 HLA A*0201 epitopes, the SIR score of the sequence for HLA A*0201 would be 0.4. The SIR score of a gene in a population is defined as the average SIR score over all HLAs, weighted by the HLA frequencies in this population. An average SIR score of < 1 represents an under-presentation of epitopes, while an average SIR score of > 1 actually represents an over-presentation of epitopes.
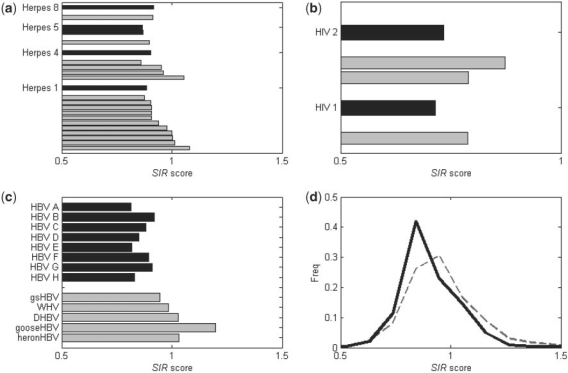
In order to compare viruses from human and non human hosts, we used three groups of viruses from different families. We compared the SIR scores of human herpes viruses and the ones of non-human herpes viruses. Five human and 18 non-human Herpes strains were tested on human MHC alleles (Vider-Shalit et al., 2007; Table 1, Fig. 3a). The average SIR score of the human herpes viruses was lower than their non-human counterparts (t-test P < 0.003). A similar result can be observed when comparing Human immunodeficiency virus (HIV) I and II, with the respective Simian immunodeficiency virus (SIV) that originated them (Vider-Shalit et al., 2008; P < 0.02). We computed the SIR score of all HIV-1, HIV-2, SIVcpz, SIVsm and SIVmac sequences in the LANL and NCBI databases (Kuiken et al., 2003). The SIVcpz, which is the ancestor of HIV-1, has a higher SIR score than average SIR score of all HIV-1 sequences. The SIR score of HIV-2 is also smaller than the SIR scores of SIVsm and SIVmac that originated it (Vider-Shalit et al., 2008) (Fig. 3b). Another virus with human strains as well as strains infecting other species is Hepatitis B virus (HBV). Viruses similar to HBV exist among others in ducks and squirrels (Table 1). As was the case for the Herpes and the HIV, the SIR score of non-human hepatitis viruses is ∼1, while the SIR score of the HBV is lower than 1 (Fig. 3c; T-test P <0.01). In the case of HBV, this comparison holds at the single protein level, for most proteins. Most HBV proteins have a lower SIR score than their non-human counterpart (X is only compared to mammalian Hepadnaviruses).
Table 1.
SIR score of human and non-human viruses used in analysis
| Virus | SIR | Virus | SIR |
|---|---|---|---|
| Human herpes virus 1 | 0.89 | HIV-1 | 0.71 |
| Human herpes virus 4 | 0.91 | HIV-2 | 0.73 |
| Human herpes virus 5-AD169 | 0.87 | SIVcpz | 0.79 |
| Human herpes virus 5-Merlin | 0.87 | SIVmac | 0.79 |
| Human herpesvirus 8 | 0.92 | SIVsm | 0.87 |
| Ostreid herpes virus 1 | 1.08 | ||
| Ictalurid herpes virus 1 | 1.01 | HBV A | 0.82 |
| Gallid herpes virus 1 | 1.00 | HBV B | 0.92 |
| Alcelaphine herpes virus 1 | 1.00 | HBV C | 0.88 |
| Meleagrid herpes virus 1 | 0.98 | HBV D | 0.85 |
| Equid herpes virus 1 | 0.94 | HBV E | 0.82 |
| Psittacid herpes virus 1 | 0.91 | HBV F | 0.90 |
| Suid herpes virus 1 | 0.91 | HBV G | 0.91 |
| Murid herpes virus 1 | 0.91 | HBV H | 0.83 |
| Bovine herpes virus 1 | 0.90 | gsHBV | 1.03 |
| Cercopithecine herpes virus 1 | 0.87 | WHV | 1.20 |
| Tupaiid herpes virus 1 | 0.86 | DHBV | 1.03 |
| Bovine herpes virus 4 | 1.06 | gooseHBV | 0.98 |
| Equid herpes virus 4 | 0.96 | heronHBV | 0.94 |
| Murid herpes virus 4 | 0.95 | ||
| Pongine herpes virus 4 | 0.86 | ||
| Bovine herpes virus 5 | 0.90 | ||
| Cercopithecine herpes virus 8 | 0.92 |
Fig. 3.
The average SIR score of human versus non-human viruses. Data are shown for three viruses: Herpes virus (HHV-1, HHV-4, HHV-5 and HHV-8), (a) HIV (HIV-1 and HIV-2) (b) and Hepatitis B (strains A-H) virus. (c) The black columns represent human strains while the gray columns represent non-human strains. In most cases, the average SIR score of the human viruses are lower than the non-human viruses. The lower right drawing represents the SIR score distributions of all full sequenced viruses from the NCBI. (d) In general, the SIR score of the non-human viruses (gray dashed line) was distributed around 1 while the human viruses (black thick line) was <1.
More generally, when comparing the SIR scores of all human-infecting viruses, it is significantly lower than the one of non-human viruses [Fig. 3d; (t-test P < 1.e–7)]. However, this last result should be taken with a grain of salt, since the SIR score of different virus families can significantly vary. Thus, the average over multiple families is affected by the number of available fully sequenced viruses in each family. The Human Papillomavirus (HPV) family (Papillomaviridae) is one of the most highly sequenced human viral families. HPV have a low SIR score, reducing the average human viruses SIR score. Thus the proper analysis is the case-by-case comparison of similar viruses, as was done in the herpes, HIV and HBV cases.
3.4 Early versus late viral proteins
The selection against viral epitopes does not end at the full virus level. Not all viral proteins are subject to the same immune pressure. An example from a different branch of the immune system would be the gp120 in HIV and Hemaglutinin in Influenza. These proteins are under the most stringent pressure from B cells, since they are directly accessible to antibodies. Similarly one could assume that different viral proteins are under more or less immune pressure from CTL. We hypothesized that early proteins should express less epitopes than late proteins. Such a difference is expected, since viruses probably attempt to delay as much as possible the destruction of infected cells to ensure the probability of budding before cellular destruction.
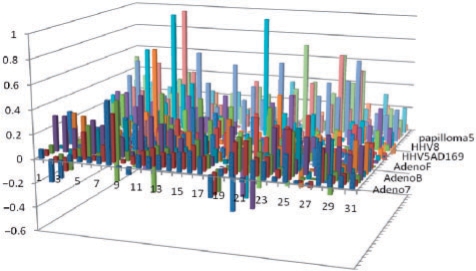
We have compared early to late proteins in 24 different viruses, mostly Adenoviruses, Herpes viruses and HPV. In most HLA alleles tested the average SIR score ratio of late to early proteins is higher than one. This result is valid for most viruses, as well as for the average of all viruses (All positive values in Fig. 4 represent allele/virus combination with a higher epitope density in late proteins than in early proteins). The difference between early and late proteins is significant, when comparing the average SIR over all alleles (t-test, P ≤ 0.0001), or when the SIR for each allele is taken into account (Anova, P < 1.e–100). Thus quite systematically, late viral proteins express more epitopes than early ones. Assuming, viruses would make all possible efforts to evade detection, one could assume that viruses would remove all epitopes through mutations. However, given the large number of epitopes resulting from the MHC polymorphism and the probable cost of mutations, viruses are probably limited in their attempts to reduce epitopes. Given these restrictions, our results show that most of the effort is targeted to early proteins that are probably the most dangerous for the virus.
Fig. 4.
SIR score of early versus late proteins. The data is shown for 24 viruses of all candidates HLA alleles (31 alleles). Each column represents the ratio between the difference of the late and early SIR score to their sum ([SIR(late) − SIR(early)]/[SIR(late) + SIR(early)]). For most HLA\virus the ratio is more than 0, indicating a significant positive difference in the number of presented epitopes between these groups.
Note that the observed effect is the average over a large number of proteins. At the single protein, many factors can affect the SIR score. For example, we have shown that proteins down-regulating MHC presentation have a high SIR score, while critical latent Herpes proteins have a very low SIR score (Vider-Shalit et al., 2007).
To summarize, the SIR score of a protein is affected at the most general level by its amino-acid distribution. At the next level, it is affected by the type of the virus and the protein expression time in the viral life cycle and finally by specific properties of the protein. To that one must add a very strong random element resulting from viral mutations that did not pass selection (such as mutations not affecting epitopes in the specific host MHC allele.).
4 DISCUSSION
The precision of CD8+ T-cell epitope presentation and processing algorithms has reached the level allowing a systematic analysis of the detailed epitope repertoire of a given organism. We have combined such algorithms and the large amount of available viral genomic data to produce for the first time a systemic analysis of viral epitope repertoires.
Viruses and the immune system play an intricate evolutionary game. Viruses attempt to evade immune detection, while the human population is constantly driven by viral epidemics to use HLA alleles presenting viral epitopes. Viruses use a large variety of immune evasion mechanisms, such as (among other): down-regulation of MHC-I expression (Hilleman, 2004), self mimicry (Alcami, 2003), down-regulation of CD1d surface expression (Yuan et al., 2006) and mutation of T-cell epitopes (Bowen and Walker, 2005; McMichael and Phillips, 1997). If the selective pressure affecting viruses and the immune system was balanced, the epitope density of viruses and of random sequences should be similar.
We have measured this epitope density and shown that MHC molecules are actually selected to present more viral epitopes than human epitopes. This selection is based on the typical amino-acid usage of viruses, since a random amino-acid sequence with a distribution resembling the viral amino-acid distribution has the highest epitope density among all cases studied here. Viruses in general attempt to avoid this bias by mutating epitopes. However, not all viral proteins have the same epitope density. We have defined a normalized score to compare the epitope density of different viruses. In a given protein/genome, libraries of all presented epitopes were devised. These libraries' size was compared to the size of their counterpart in a random sequence of similar length and amino-acid distribution. The size ratio was named the SIR score.
The SIR score of all human viruses was found to be lower than the one of non-human viruses. Note that highly acute viruses, such as ebola and smallpox had a SIR score higher than 1. A reduction in the number of presented epitopes can have crucial effects on the immune response, since the average number of epitopes is of the order of one per protein per HLA (Louzoun et al., 2006). A reduction in this number can simply prevent the immune detection of a given protein. A single epitope could theoretically induce an immune response. Thus theoretically, as long as the SIR score is not 0, an immune response is possible. However, the immune response is a stochastic process. The viral effort to reduce the epitope number accompanied by other immune evasion methods are probably meant to reduce the probability that an infected cell should be destroyed to a level allowing some of the infected cell to produce virions. Even if most cells are indeed destroyed, a few surviving infected cells can be enough to ensure a successful infection.
Even within a given virus, not all proteins have the same SIR score. We compared early to late proteins in 24 viruses and found that proteins expressed early in the viral life cycle had a lower SIR score than late proteins. An early presentation of epitopes may give the immune system enough time to kill the infected cell before budding, while a late destruction may not prevent budding and the infection of new cells. The difference between early and late proteins disappears when looking at the non-human counterparts of the viruses checked. The different SIR score seems to directly result from the negative selection of epitopes in the appropriate host. We have compared in a few viruses the effect of the protein concentration on the SIR score, but never found a consistent relation. The lack of such a correlation may stem from the small number of epitopes required to induce an immune response.
The SIR score has beyond its explanatory power, important applications. The detection of outliers in the distribution allows us to detect proteins that the virus tries to ‘hide’ from the immune system. Their epitopes are optimal targets for immunotherapy or simply for vaccination. One can assume that if viruses attempt to hide a protein from the immune system, the detection of this protein would maximize the immune impact. Thus, the SIR score can track the important proteins for immunotherapy even if their function is not known. It can be used for any virus. The full list of epitopes for other viruses for any HLA alleles can be obtained from the PEPTIBASE server at peptibase.cs.biu.ac.il.
Funding: National Institutes of Health (1 R01 AI61062-01 to Y.L., K.M., R.L. and T.V.).
Conflict of Interest: none declared.
REFERENCES
- Alcami A. Viral mimicry of cytokines, chemokines and their receptors. Nat Rev Immunol. 2003;3:36–50. doi: 10.1038/nri980. [DOI] [PubMed] [Google Scholar]
- Almani M, et al. Human self protein CD8+ T cell epitopes are both positively and negatively selected. Eur. J. Immunol. 2008;39:1056–1065. doi: 10.1002/eji.200838353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambagala AP, et al. Viral interference with MHC class I antigen presentation pathway: the battle continues. Vet. Immunol. Immunopathol. 2005;107:1–15. doi: 10.1016/j.vetimm.2005.04.006. [DOI] [PubMed] [Google Scholar]
- Bhasin M, et al. MHCBN: a comprehensive database of MHC binding and non-binding peptides. Bioinformatics. 2003;19:665–666. doi: 10.1093/bioinformatics/btg055. [DOI] [PubMed] [Google Scholar]
- Birney E, et al. An overview of ensembl, Ensembl 2004. Genome Res. 2004;14:925–928. doi: 10.1101/gr.1860604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blythe MJ, et al. JenPep: a database of quantitative functional peptide data for immunology. Bioinformatics. 2002;18:434–439. doi: 10.1093/bioinformatics/18.3.434. [DOI] [PubMed] [Google Scholar]
- Borrow P, et al. Virus-specific CD8+cytotoxic T-lymphocyte activity associated with control of viremia in primary human immunodeficiency virus type 1 infection. J. Virol. 1994;68:6103–6110. doi: 10.1128/jvi.68.9.6103-6110.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen DG, Walker CM. Mutational escape from CD8+ T cell immunity: HCV evolution, from chimpanzees to man. J. Exp. Med. 2005;201:1709–1714. doi: 10.1084/jem.20050808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brusic V, et al. MHCPEP, a database of MHC-binding peptides: update 1997. Nucleic Acids Res. 1998;26:368–371. doi: 10.1093/nar/26.1.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffin JM. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science. 1995;267:483–489. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- Ginodi I, et al. Precise score for the prediction of peptides cleaved by the proteasome. Bioinformatics. 2008;24:477–483. doi: 10.1093/bioinformatics/btm616. [DOI] [PubMed] [Google Scholar]
- Govindarajan KR, et al. MPID: MHC-Peptide Interaction Database for sequence-structure-function information on peptides binding to MHC molecules. Bioinformatics. 2003;19:309–310. doi: 10.1093/bioinformatics/19.2.309. [DOI] [PubMed] [Google Scholar]
- Gulzar N, Copeland KF. CD8+ T-cells: function and response to HIV infection. Curr HIV Res. 2004;2:23–37. doi: 10.2174/1570162043485077. [DOI] [PubMed] [Google Scholar]
- Hilleman MR. Strategies and mechanisms for host and pathogen survival in acute and persistent viral infections. Proc. Natl Acad. Sci. USA. 2004;101:14560–14566. doi: 10.1073/pnas.0404758101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howley PM, et al. Fields Virology. Philadelphia, USA: Lippincott Williams & Wilkins; 2001. [Google Scholar]
- Kuiken C, et al. HIV sequence databases. AIDS Rev. 2003;5:52–61. [PMC free article] [PubMed] [Google Scholar]
- Letvin NL, et al. Cytotoxic T lymphocytes specific for the simian immunodeficiency virus. Immunol Rev. 1999;170:127–134. doi: 10.1111/j.1600-065x.1999.tb01334.x. [DOI] [PubMed] [Google Scholar]
- Lichterfeld M, et al. Immunodominance of HIV-1-specific CD8(+) T-cell responses in acute HIV-1 infection: at the crossroads of viral and host genetics. Trends Immunol. 2005;26:166–171. doi: 10.1016/j.it.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Louzoun Y, Vider T. Score for Proteasomal Peptide Production Probability. Immunology. 2004;1:45–50. [Google Scholar]
- Louzoun Y, et al. T-cell epitope repertoire as predicted from human and viral genomes. Mol. Immunol. 2006;43:559–569. doi: 10.1016/j.molimm.2005.04.017. [DOI] [PubMed] [Google Scholar]
- McMichael AJ, et al. Cytotoxic T-cell immunity to influenza. N. Engl. J. Med. 1983;309:13–17. doi: 10.1056/NEJM198307073090103. [DOI] [PubMed] [Google Scholar]
- McMichael AJ, Phillips RE. Escape of human immunodeficiency virus from immune control. Annu. Rev. Immunol. 1997;15:271–296. doi: 10.1146/annurev.immunol.15.1.271. [DOI] [PubMed] [Google Scholar]
- McSparron H, et al. JenPep: a novel computational information resource for immunobiology and vaccinology. J. Chem. Inf. Comput Sci. 2003;43:1276–1287. doi: 10.1021/ci030461e. [DOI] [PubMed] [Google Scholar]
- Negri DR, et al. Identification of a cytotoxic T-lymphocyte (CTL) epitope recognized by Gag-specific CTLs in cynomolgus monkeys infected with simian/human immunodeficiency virus. J. Gen. Virol. 2006;87:3385–3392. doi: 10.1099/vir.0.81934-0. [DOI] [PubMed] [Google Scholar]
- Parker KC, et al. Scheme for ranking potential HLA-A2 binding peptides based on independent binding of individual peptide side-chains. J. Immunol. 1994;152:163–175. [PubMed] [Google Scholar]
- Peters B, et al. Identifying MHC class I epitopes by predicting the TAP transport efficiency of epitope precursors. J. Immunol. 2003;171:1741–1749. doi: 10.4049/jimmunol.171.4.1741. [DOI] [PubMed] [Google Scholar]
- Peters B, et al. The immune epitope database and analysis resource: from vision to blueprint. PLoS Biol. 2005;3:e91. doi: 10.1371/journal.pbio.0030091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rammensee H, et al. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics. 1999;50:213–219. doi: 10.1007/s002510050595. [DOI] [PubMed] [Google Scholar]
- Robinson J, et al. IMGT/HLA and IMGT/MHC: sequence databases for the study of the major histocompatibility complex. Nucleic Acids Res. 2003;31:311–314. doi: 10.1093/nar/gkg070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock KL, et al. Protein degradation and the generation of MHC class I-presented peptides. Adv. Immunol. 2002;80:1–70. doi: 10.1016/s0065-2776(02)80012-8. [DOI] [PubMed] [Google Scholar]
- Sathiamurthy M, et al. Population of the HLA ligand database. Tissue Antigens. 2003;61:12–19. doi: 10.1034/j.1399-0039.2003.610102.x. [DOI] [PubMed] [Google Scholar]
- Vider-Shalit T, et al. AIDS. in press; 2008. The HIV hide and seek game: An immunogenomic analysis of the HIV epitope repertoire. [DOI] [PubMed] [Google Scholar]
- Vider-Shalit T, et al. Phase-dependent immune evasion of herpesviruses. J. Virol. 2007;81:9536–9545. doi: 10.1128/JVI.02636-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yewdell JW, et al. TAP-independent delivery of antigenic peptides to the endoplasmic reticulum: therapeutic potential and insights into TAP-dependent antigen processing. J. Immunother. 1998;21:127–131. doi: 10.1097/00002371-199803000-00006. [DOI] [PubMed] [Google Scholar]
- Yuan W, et al. Herpes simplex virus evades natural killer T cell recognition by suppressing CD1d recycling. Nat. Immunol. 2006;7:835–842. doi: 10.1038/ni1364. [DOI] [PubMed] [Google Scholar]