Abstract
Motivation:Several strategies have been developed to predict the fold of a target protein sequence, most of which are based on aligning the target sequence to other sequences of known structure. Previously, we demonstrated that the consideration of protein–protein interactions significantly increases the accuracy of fold assignment compared with PSI-BLAST sequence comparisons. A drawback of our method was the low number of proteins to which a fold could be assigned. Here, we present an improved version of the method that addresses this limitation. We also compare our method to other state-of-the-art fold assignment methodologies.
Results: Our approach (ModLink+) has been tested on 3716 proteins with domain folds classified in the Structural Classification Of Proteins (SCOP) as well as known interacting partners in the Database of Interacting Proteins (DIP). For this test set, the ratio of success [positive predictive value (PPV)] on fold assignment increases from 75% for PSI-BLAST, 83% for HHSearch and 81% for PRC to >90% for ModLink+at the e-value cutoff of 10−3. Under this e-value, ModLink+can assign a fold to 30–45% of the proteins in the test set, while our previous method could cover <25%. When applied to 6384 proteins with unknown fold in the yeast proteome, ModLink+combined with PSI-BLAST assigns a fold for domains in 3738 proteins, while PSI-BLAST alone covers only 2122 proteins, HHSearch 2969 and PRC 2826 proteins, using a threshold e-value that would represent a PPV >82% for each method in the test set.
Availability: The ModLink+server is freely accessible in the World Wide Web at http://sbi.imim.es/modlink/.
Contact: boliva@imim.es.
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Large-scale sequencing methods have provided a large amount of protein sequence data. However, even for well-characterized organisms, structure and function remain unknown for a significant fraction of their proteomes (Sharan et al., 2007). Traditionally, both protein function and structure have been inferred by transferring annotation from characterized proteins of similar sequence (Sanchez et al., 2000). Nevertheless, the structure of a protein generally provides more information about its function than its sequence alone (Hegyi and Gerstein, 1999; Orengo et al., 1999; Thornton et al., 1999). Moreover, two proteins can have similar structures even when their sequence similarity is low (Rost, 1997), and it is therefore important to determine or predict the structures of as many proteins as possible. Structure-based characterization of proteins has been facilitated by databases of (i) protein domain classification such as Structural Classification Of Proteins (SCOP; Andreeva et al., 2008), PFAM (Finn et al., 2008) and CATH (Greene et al., 2007); (ii) protein–protein interactions such as Database of Interacting Proteins (DIP; Salwinski et al., 2004) and IntAct (Kerrien et al., 2007) [reviewed in Shoemaker and Panchenko (2007)]; and (iii) protein structures such as PDB (Berman et al., 2000).
Most methods for structure characterization rely on either sequence comparisons or sequence to structure alignments using statistical potentials encoding features extracted from databases of protein structures. The first group uses position-specific scoring matrices (Marti-Renom et al., 2004; Mittelman et al., 2003) or hidden Markov models (HMMs) (Eddy, 1998; Madera, 2008; Soding, 2005) to construct a multiple sequence alignment of close homologs of the query that is later used to scan a database of sequences. The second group uses threading (Jones, 1997), fold recognition (Kelley et al., 2000) or secondary structure predictions (Rost, 1995). The structure of single domain proteins can be occasionally predicted with relative success (Kryshtafovych et al., 2005) by ab initio methods such as Rosetta predictions (Das and Baker, 2008) and by contact maps (Bastolla et al., 2005; Punta and Rost, 2005).
The structure of a protein can be frequently divided into one or more domains, which may interact with domains from other proteins. Moreover, homologous proteins tend to interact through similar domains. Thus, it may be possible to predict the fold of a protein from its interacting partners of known structure (Kiel et al., 2008). We developed a method, named ModLink (Espadaler et al., 2005a), which used both sequence similarity and protein–protein interactions to assign a SCOP fold and a family classification to uncharacterized proteins. An extension of ModLink was used for functional annotation of enzymes (Espadaler et al., 2008). The rationale behind ModLink is that two proteins are more likely to be homologous if they also have similar interacting partners. Nevertheless, the method could only be applied if the accuracy and number of interactions available for a query protein were high. To overcome this limitation, ModLink increased the available interactions by extrapolation: two proteins were linked by extrapolation if any members from their SCOP families interacted with each other. By design, ModLink was not able to deal successfully with proteins having a large number of different interacting partners, usually referred to as ‘hubs’. As a result, ModLink did not use such hub proteins for extrapolating links. Therefore, the performance of ModLink would improve if the method could distinguish between hub proteins whose interacting partners have similar sequences and those that do not.
Here, we describe a new version of ModLink, called ModLink+. ModLink+includes an improved procedure for extrapolating links that iteratively varies the number of interactions required to consider a protein as a hub. This new algorithm, that comprises a ‘self-adaptive’ definition of hub proteins, has increased applicability without affecting its accuracy. ModLink+is accessible via a World Wide Web server (http://sbi.imim.es/modlink/).
2 METHODS
2.1 Datasets
2.1.1 Protein data
SCOP (version, 1.71; December 2006) (Andreeva et al., 2008) was used to assign structural information. TrEMBL (release, 34.3; December 2006) (The UniProt Consortium, 2009) was employed to construct ‘position-specific scoring matrix’ (PSSM) profiles for searching similar sequences with PSI-BLAST (Altschul et al., 1997) against the SCOP database. Sequences from fly, human, worm and yeast proteomes were extracted from UniProt (release, 10.0; April 2007) (The UniProt Consortium, 2009).
2.1.2 Interaction data
A total of 55 271 protein–protein interactions from the DIP database (release 20080708; July 2008) (Salwinski et al., 2004) were used to evaluate the method. Yeast interaction data were extracted using the PIANA software (version 1.2) (Aragues et al., 2006), which contained protein–protein interactions from the following databases: BIND (April 2007) (Alfarano et al., 2005); BioGRID (version 2.0.26; May 2007) (Breitkreutz et al., 2008); DIP (release 20070219; February 2007); HPRD (release 6; January 2007) (Mishra et al., 2006); IntAct (release 2007-04-20; April 2007) (Kerrien et al., 2007); MINT (release 2007-04-05; April 2007) (Chatr-aryamontri et al., 2007); and MIPS (March 2007) (Pagel et al., 2005).
2.1.3 DIP–SCOP set
Fold, superfamily and family domain codes from the SCOP database were assigned to proteins in the DIP database using BLAST (Altschul et al., 1997). A SCOP code was assigned to a DIP sequence if it aligned to a SCOP representative with an e-value <10−8 and covered a minimum of 75% of the domain sequence. The resulting group of sequences was named DIP–SCOP.
2.1.4 Test set
ModLink+was tested using a non-redundant set of 3716 proteins from the DIP–SCOP set. Redundancy was removed at the 25% sequence identity cutoff using BLAST. The test set was used to compare ModLink+against PSI-BLAST (Altschul et al., 1997), HHSearch (Soding, 2005) and PRC (Madera, 2008). All methods were used with default parameters and protocols as previously described (Agarwal et al., 2008; Espadaler et al., 2005a; Madera, 2008).
2.2 Algorithm
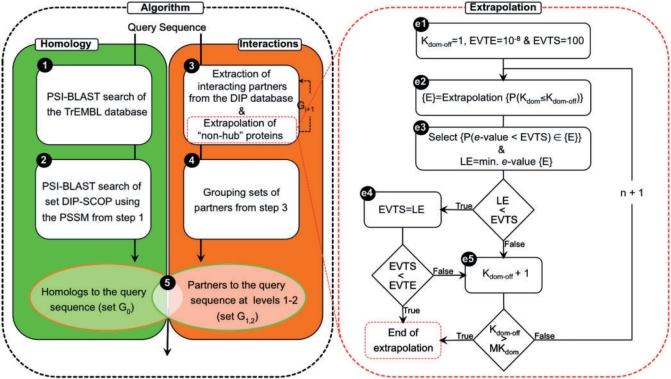
The assignment of a fold, superfamily and family to a query sequence in ModLink+is a five-step procedure, similar to ModLink (Fig. 1):
A PSSM profile of the query is obtained with PSI-BLAST by searching the TrEMBL database with a maximum of five iterations.
Putative query homologs are detected in the DIP–SCOP group by using PSI-BLAST and the PSSM profile from Step 1, assigning an e-value for the comparison and grouping them in set G0.
Interacting partners of the query (partners of the query at Level 1) and proteins that interact with them (partners of the query at Level 2) are extracted from the list of protein–protein interactions (containing known and extrapolated links).
Partners of the query at Levels 1 and 2 are grouped (set G1,2).
Members of G1,2 are ranked according to the e-value calculated in Step 2.
The algorithm can also be applied by substituting sequence similarities detected by PSI-BLAST with those obtained from HHSearch or PRC. The HMM of the query sequence is constructed with HMMER (Eddy, 1998) for HHSearch and PRC. For HHSearch, additionally, the secondary structure of the query sequence, as predicted by PSIPRED (Jones, 1999), is added to the HMM. In Step 1, the HMM substitutes the PSSM profile and in Step 2 HHSearch and PRC are used instead of PSI-BLAST for detecting putative homologs. The resulting protocols are named Modlink+combined with PSI-BLAST, HHSearch or PRC, respectively.
Fig. 1.
ModLink + flowchart. Brackets are used to indicate a set of proteins. {E} is the set of proteins obtained from the extrapolation of links for ‘non-hub’ proteins. In Step e3, extrapolated proteins (from {E}) with an e-value lower than EVTS with respect to the query are selected: {P(e-value < EVTS)}. This set is returned to Step 3 at the end of the extrapolation.
2.3 Extrapolation of links
One of the main limitations of the original ModLink algorithm was the scarcity of protein–protein interaction data, which clearly limited its applicability and was only solved by the use of predicted interactions (i.e. extrapolated links in Step 3). In ModLink, two proteins were linked by extrapolation if any members from their SCOP families interacted with each other. However, this produced false relationships caused by the extrapolation of proteins that interacted with many proteins of different families, which were considered hub proteins. Therefore, to avoid the negative impact of hubs, the extrapolation was only performed on proteins interacting with proteins classified in less than 10 different SCOP families (defined as domain degree cutoff, see below). This filtering implied that some query proteins could not benefit from the use of extrapolation, while for other query proteins the extrapolation was unnecessary and it increased the number of false fold assignments. In ModLink+, we have addressed this problem by converting the original one-step extrapolation to an iterative process that selects the best cutoff (i.e. domain degree cutoff) for a given query.
The iteration does not alter the core algorithm, but requires a few additional definitions (Fig. 1). First, two proteins are linked by extrapolation if any pair of proteins that share any SCOP domain code with them interact with each other. Thus, there are three versions of the algorithm that differ in the type of extrapolation at the level of fold, superfamily or family SCOP codes. Moreover, a link is defined between protein X and SCOP domain D if protein X interacts with a protein that contains at least one SCOP domain D. The domain degree of a protein (Kdom) is the number of different SCOP domains interacting with the protein. The domain degree cutoff (Kdom−off) is obtained as the minimum Kdom that results in a hub protein. In addition, two thresholds on the PSI-BLAST (HHSearch or PRC) e-value are established: the first one named EVTE (e-value threshold for ending the extrapolation), which controls the end of the extrapolation process, and the second one named EVTS (e-value threshold for selecting links), which ensures the accuracy of the selected extrapolated links in each iteration. Thus, the extrapolation procedure consists of five steps (Fig. 1):
(el)Kdom−off is set to 1, EVTE is set to 10−8 (where 10−8 is the maximum e-value for assigning a SCOP code to a protein in DIP with BLAST) and EVTS is set to 100 (where 100 is the maximum e-value allowed to find similarities in Step 2).
(e2)The extrapolation is performed for ‘non-hub’ proteins.
(e3)Extrapolated links that include partners aligned with the query in Step 2, with an e-value lower than EVTS, are selected to be used in Step 3. Then, the lowest e-value (LE) of the alignment between the query and its predicted partners at Levels 1 and 2 is selected: if LE is smaller than EVTS, Step e4 follows, otherwise Step e5 follows.
(e4)EVTS is swapped with LE. If EVTS is smaller than EVTE, the extrapolation ends, otherwise Step e5 follows.
(e5)Kdom−off is increased by 1. If Kdom−off is higher than the maximum domain degree among proteins interacting with the query (MKdom), the extrapolation ends, otherwise a new iteration starts with Step e2.
2.4 Statistical analysis
The number of predicted matches was defined as the number of sequences belonging to the DIP–SCOP group that aligned with a query sequence in the test set with an e-value smaller or equal to a given threshold. Among these predictions, the number of true positives was defined as the number of sequences sharing the same SCOP code with the query sequence. Moreover, the positive predictive value (PPV) was defined as the percentage of true positives over the total number of predictions. The applicability (coverage) of the method was defined as the percentage of queries that the method could assign at least one predicted hit over the total number of queries in the test set.
2.5 Server
2.5.1 Input
The following inputs are required: (i) query sequence of a protein; (ii) identifiers for interaction databases; (iii) type of extrapolation (based on fold, superfamily or family SCOP domain codes or none); and (iv) EVTE (Section 2.2). Additionally, the user can submit sequences of other proteins that interact with the query protein (if known) and a threshold on the PSI-BLAST (HHSearch or PRC) e-value (EVTH) to use homologs of the submitted sequences in the case the database does not contain interactions of the query.
2.5.2 Output
The server outputs the sequences predicted to have a SCOP domain similar to that in the query. It also provides their alignments and e-values according to PSI-BLAST. It prints the predicted SCOP domain codes for the folds, superfamilies and families of the query domains. Finally, it lists the partners of the query at Levels 1 and 2 that share a SCOP domain code with the query. In addition, the server also shows the databases describing the interactions of the proteins at these levels.
3 RESULTS AND DISCUSSION
3.1 ModLink+accuracy
ModLink+combined with PSI-BLAST (or HHSearch or PRC) was compared against the original ModLink, PSI-BLAST, HHSearch and PRC using a test set of non-redundant protein sequences.
3.1.1 PPV of fold assignment
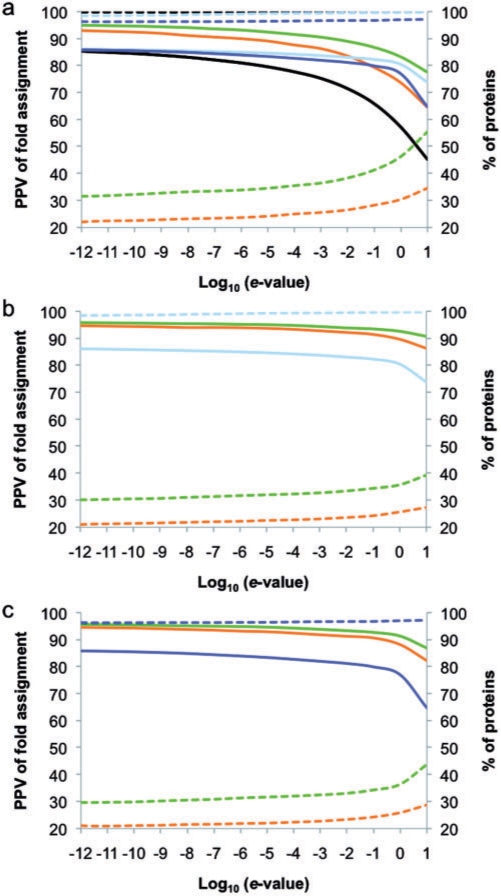
Similar to the work of Espadaler et al. (2005a), fold assignment based on sequence similarity was improved by using interaction data. For example, using sequence similarity with PSI-BLAST e-value <10−3 and extrapolation by SCOP families, ModLink+achieved a maximum PPV of 90% (Fig. 2a), while PSI-BLAST achieved only 75%, HHSearch 83% and PRC 81%. When the extrapolation was based on SCOP folds or superfamilies, the PPV of fold assignment decreased in less than 2 percentatge points (Supplementary Material). Moreover, ModLink+combined with HHSearch and PRC, using e-values <10−3 and extrapolation by SCOP families, achieved a PPV of 94% for HHSearch (Fig. 2b) and of 93% for PRC (Fig. 2c).
Fig. 2.
PPV (continuous line) and applicability (dashed line) of fold assignment is plotted as a function of the threshold on the sequence similarity e-values: (a) PSI-BLAST (black), HHSearch (cyan), PRC (blue), Modlink (orange) and Modlink+(green) combined with PSI-BLAST; (b) HHSearch (cyan), Modlink (orange) and Modlink+(green) combined with HHSearch; (c) PRC (blue), Modlink (orange) and Modlink+(green) combined with PRC. Extrapolation for Modlink+and Modlink in all plots was based in SCOP family codes.
The improvement of PPV in fold assignment with respect to PSI-BLAST, HHSearch and PRC justifies the use of less stringent e-value cutoffs in ModLink+to predict with the same confidence as any of these three individual methods, but with larger applicability (coverage). Consequently, ModLink+can be applied to proteins for which the assignment of fold with other methods fails. Moreover, higher PPVs are obtained in ModLink+compared with ModLink (Fig. 2), suggesting that the modifications introduced in ModLink+do not affect the accuracy of the original method, while increasing its coverage (Section 3.1.2).
It is known that proteins in the same SCOP group do not necessarily share the same interactions (Aloy and Russell, 2002; Keskin and Nussinov, 2007). For example, on the one hand, multidomain proteins have more than one SCOP fold code. On the other hand, databases of protein–protein interactions do not inform of the interacting domains. Therefore, the extrapolation of multidomain proteins assigns interactions between untested domains (false interactions). Consequently, ModLink+would benefit from the use of domain–domain interactions (Boxem et al., 2008; Davis and Sali, 2005; Finn et al., 2005; Jefferson et al., 2007; Ogmen et al., 2005; Stein et al., 2005; Winter et al., 2006) and methods that detect the binding regions of proteins (Aragues et al., 2007; Espadaler et al., 2005b; Guo et al., 2008; Kim et al., 2006).
3.1.2 Applicability of fold assignment
ModLink+ combined with PSI-BLAST has higher applicability than ModLink (Fig. 2). For example, using sequence similarity with PSI-BLAST e-value <10−3 and extrapolation by SCOP families, ModLink+ achieved an applicability of 35% (Fig. 2a), while ModLink achieved only 25%. When the extrapolation was based on SCOP superfamilies or folds, the applicability of the method increased to 41% and 45%, respectively (Supplementary Material). Moreover, the applicability of ModLink+ combined with HHSearch (33%) or PRC (33%) was higher than the applicability of the original ModLink method (Fig. 2b, c) but not higher than Modlink+ combined with PSI-BLAST. On the one hand, the dependence on protein–protein interaction data makes ModLink+less applicable than sequence/profile comparison methods such as PSI-BLAST, HHSearch or PRC. On the other hand, the applicability improvement with respect to the original ModLink revealed that the use of a ‘self-adaptive’ definition of hubs increases the number of query sequences for which a fold could be assigned.
3.2 Assignment of SCOP domains in yeast proteome
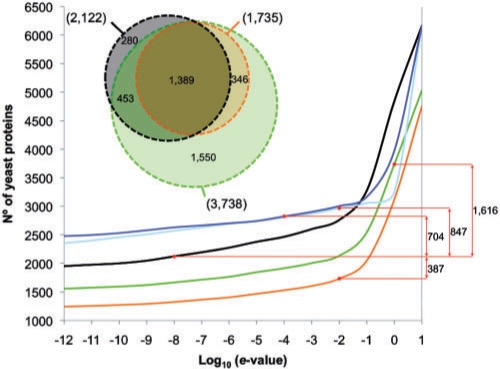
To test the applicability of ModLink+in a realistic scenario, putative folds were assigned to yeast proteins using all the protein–protein interaction data available. A total of 7463 different yeast sequences were taken from UniProt. Fold assignment was achieved for domains in 1079 proteins by following the same procedure that had been used to create the DIP–SCOP group (Section 2). Among the remaining proteins (6384), a fold could be assigned to 2122 proteins by PSI-BLAST (Fig. 3) as described in Steps 1 and 2 of ModLink+, when the e-value of their alignment was <10−8 (representing a PPV of 82% for the test set). At this e-value threshold, ModLink+ combined with PSI-BLAST could assign a fold to 1778 proteins and ModLink to 1325 proteins (Fig. 3) when extrapolating links based on SCOP families. Moreover, using 10−8 e-value cutoff, we could assign a fold to 2573 proteins using HHSearch and to 2633 using PRC (Fig. 3). However, for a PPV of 82% we could raise the cutoff e-value to 1 for ModLink+ combined with PSI-BLAST and 10−2 for ModLink, increasing the coverage to 3738 and 1735 proteins, respectively (Fig. 3). Furthermore, HHSearch achieves 82% PPV with a cutoff e-value of 10−2 and it assigns a fold to 2969 proteins, while PRC achieves a 82% PPV when applying a 10−4 e-value cutoff and it assigns a fold to 2826 proteins (Fig. 3). Besides, the combination of ModLink+ combined with HHSearch or PRC achieves PPV >90% for the cutoff e-value of 1 although decreasing the coverage to 2509 and 2952 proteins, respectively. Finally, our predictions could be classified as a function of the percentage of the sequence of the target protein being covered by SCOP domains into four groups: (i) 75–100% of coverage, (ii) 50–75% of coverage, (iii) 25–50% of coverage and (iv) 0–25% of coverage. Using PSI-BLAST, 1002 proteins were classified in Group 1, 524 in Group 2, 370 in Group 3 and 226 in Group 4. The use of protein–protein interactions in Modlink+ allowed new predictions and improved the percentage of sequence with assigned structure of some target proteins. We classified 1141 target sequences in Group 1 (allotting a putative structure for 75–100% of their sequence). Out of the 139 new proteins, 76 were new predictions with ModLink+ and 63 corresponded to targets for which a putative structure had already been predicted with PSI-BLAST for some percentage of their sequence (Groups 2, 3 and 4). Also Group 2 increased up to 797 proteins (237 were new predictions and 87 were enriched from other groups, while 473 remained unmodified from the previous classification of the PSI-BLAST predictions). Group 3 was enlarged up to 1002 proteins (652 new predictions, 53 from the enrichment of Group 4 and 292 unmodified). Finally, for 1083 target proteins, it was possible to predict the structure for <25% of their sequence (Group 4). Modlink+ assigned a putative fold to 931 new targets, while the rest of targets were predicted by PSI-BLAST (152 proteins).
Fig. 3.
Total number of proteins with assigned fold versus the threshold on the sequence similarity e-value for ModLink+ (green), ModLink (orange), PSI-BLAST (black), HHSearch (cyan) and PRC (blue). Extrapolation was based on SCOP family codes for PSI-BLAST. Red dots indicate the number of proteins with an assigned fold when applying a threshold on the e-value at which the PPV is >82% (for the test set). The difference between each method of the total number of target proteins with at least one assigned fold is shown in the right margin. The distribution of target proteins with domains with an assigned fold obtained with PSI-BLAST (black), Modlink (orange) and Modlink+ (green) is represented in a Venn diagram in the upper left corner.
In summary, with PPV of 82%, a total of 1842 target proteins had a fold assigned by both ModLink+ and PSI-BLAST. Among these, Modlink+ increased the percentage of sequence with assigned structure for 203 target proteins. Moreover, 1896 predictions were provided only by ModLink+ and 280 only by PSI-BLAST. Thus, the difference of using ModLink+ instead of PSI-BLAST yields to the prediction of fold for 1616 more target proteins (Fig. 3). We also compared the coverage of Modlink+ with HHSearch and PRC. ModLink+ combined with PSI-BLAST could be applied to 769 more targets than HHSearch and 912 more than PRC. The amount of targets for which a fold is predicted by one or more methods is shown in the Supplementary Material. We could predict a fold for 1235 proteins using ModLink+ combined with PSI-BLAST and e-values <1 (Table S1 in Supplementary Material). Among them, we could predict the fold for 365 proteins using ModLink+ combined with HHSearch and/or PRC under the same e-value cutoff (increasing the PPV confidence to >90%).
4 CONCLUSIONS
We have shown that the use of a ‘self-adaptive’ definition of hub proteins increases the number of protein sequences for which ModLink+ can assign a SCOP fold, while maintaining the accuracy of our previous version of ModLink. Therefore, ModLink+ can use cutoff e-values of little significance on the assignment of fold. In addition, the web server of ModLink+ allows the use of putative interacting partners of the query: when the databases of protein–protein interactions do not contain interactions for the query, ModLink+ can assign to the query the interactions of its homologs. Finally, we have improved the server by including the possibility of using sequence similarities obtained by means of profile–profile comparisons with HHSearch and PRC.
Our results show that ModLink+ is applicable to a significant number of sequences for which the assignment of fold with other methods fails. Moreover, we have shown that ModLink+ can enlarge the sequence coverage with structure upon the predictions of PSI-BLAST, also improving the coverage of HHSearch and PRC with the same accuracy. For example, the assignment of SCOP fold codes to the Saccharomyces cerevisiae proteome at the confidence level of 82% of PPV using ModLink+ was possible for 1896 sequences that could not be matched by PSI-BLAST. Therefore, using a PPV of 82% and assuming that the ratio on the knowledge of proteome and interactome of most well-studied organisms is similar to that of S.cerevisiae, we would be able to increase the number of targets with putative fold for 5917 proteins of Caenorhabditis elegans, 7394 of Drosophila melanogaster and 17 602 of Homo sapiens. Additionally, by combining PSI-BLAST and ModLink+, we would be able to assign a structure to almost the whole sequence (Group 1) to 3561 proteins of C.elegans, 4450 of D.melanogaster and 10 593 of H.sapiens. If the knowledge on protein–protein interactions was complete, our results in yeast suggest that ModLink+ combined with PSI-BLAST could be applied to more than 2 000 000 sequences in the UniProt database (The UniProt Consortium, 2009). In summary, we have shown that ModLink+ combined with PSI-BLAST, HHSearch or PRC surpasses state-of-the-art methods of remote homology detection in coverage and accuracy. In other words, we improved PSI-BLAST, HHSearch and PRC by using protein–protein interactions even at the expense of reducing their original coverage (applicability).
The coverage of ModLink+increased due to the extrapolation of links between proteins sharing common domains, as defined by domain codes of SCOP, assuming that these linked proteins have common interaction partners. Nevertheless, ModLink+ cannot recognize the regions in contact in the interactions that can cause exceptions in the rationale behind the method, for example, when extrapolating links in multidomain proteins. This limitation could be avoided by using more reliable and extensive protein– protein interaction data with knowledge of the binding interfaces or with information on the protein domains involved in the interaction. Besides, we have to note that similar structures do not necessarily interact in the same way and differences in very few residues can lead to different preferred associations (Keskin et al., 2008; Keskin et al., 2004; Tsai et al., 1996). Hence, ModLink+ will benefit in the future from methods capable of detecting the interacting domains or binding regions whenever these can be extracted from the network (Aragues et al., 2007) or from experimental data (Wang et al., 2007).
ACKNOWLEDGEMENTS
The authors would like to thank all members of the SBI lab, especially David Alarcon, Jaume Bonet, Javier García and Joan Planas, for helpful comments on the server construction and SQL data management.
Funding: EU funding (IST-027703 to O.F.); Spanish Ministerio de Educación y Ciencia (MEC, PROFIT PSE0100000-2007-1 grant to R.A.); Research program of the Barcelona Supercomputing Center (BSC) (to access the facilities of Mare Nostrum); Spanish Ministerio de Educación y Ciencia grant (BIO2007/66670 to M.A.M.-R.); National Institutes of Health U54 GM074945, U54 RR022220 and R01 GM54762, Sandler Family Supporting Foundation, Mike Homer, Ron Conway, IBM, Intel, Netapp and Hewlett Packard (to A.S.); Spanish Ministerio de Educación y Ciencia (MEC, BIO02008-0205 and PROFIT PSE0100000-2007-1, FIT-350300-2007-67 and FIT-350300-2006-42 to B.O.).
Conflict of Interest: none declared.
REFERENCES
- Agarwal V, et al. PDBalert: automatic, recurrent remote homology tracking and protein structure prediction. BMC Struct. Biol. 2008;8:51. doi: 10.1186/1472-6807-8-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfarano C, et al. The Biomolecular Interaction Network Database and related tools 2005 update. Nucleic Acids Res. 2005;33:D418–D424. doi: 10.1093/nar/gki051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aloy P, Russell RB. The third dimension for protein interactions and complexes. Trends Biochem. Sci. 2002;27:633–638. doi: 10.1016/s0968-0004(02)02204-1. [DOI] [PubMed] [Google Scholar]
- Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreeva A, et al. Data growth and its impact on the SCOP database: new developments. Nucleic Acids Res. 2008;36:D419–D425. doi: 10.1093/nar/gkm993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragues R, et al. PIANA: protein interactions and network analysis. Bioinformatics. 2006;22:1015–1017. doi: 10.1093/bioinformatics/btl072. [DOI] [PubMed] [Google Scholar]
- Aragues R, et al. Characterization of protein hubs by inferring interacting motifs from protein interactions. PLoS Comput. Biol. 2007;3:1761–1771. doi: 10.1371/journal.pcbi.0030178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastolla U, et al. Principal eigenvector of contact matrices and hydrophobicity profiles in proteins. Proteins. 2005;58:22–30. doi: 10.1002/prot.20240. [DOI] [PubMed] [Google Scholar]
- Berman HM, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boxem M, et al. A protein domain-based interactome network for C. elegans early embryogenesis. Cell. 2008;134:534–545. doi: 10.1016/j.cell.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitkreutz BJ, et al. The BioGRID Interaction Database: 2008 update. Nucleic Acids Res. 2008;36:D637–D640. doi: 10.1093/nar/gkm1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatr-aryamontri A, et al. MINT: the Molecular INTeraction database. Nucleic Acids Res. 2007;35:D572–D574. doi: 10.1093/nar/gkl950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R, Baker D. Macromolecular modeling with Rosetta. Annu. Rev. Biochem. 2008;77:363–382. doi: 10.1146/annurev.biochem.77.062906.171838. [DOI] [PubMed] [Google Scholar]
- Davis FP, Sali A. PIBASE: a comprehensive database of structurally defined protein interfaces. Bioinformatics. 2005;21:1901–1907. doi: 10.1093/bioinformatics/bti277. [DOI] [PubMed] [Google Scholar]
- Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- Espadaler J, et al. Detecting remotely related proteins by their interactions and sequence similarity. Proc. Natl Acad. Sci. USA. 2005a;102:7151–7156. doi: 10.1073/pnas.0500831102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espadaler J, et al. Prediction of enzyme function by combining sequence similarity and protein interactions. BMC Bioinformatics. 2008;9:249. doi: 10.1186/1471-2105-9-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espadaler J, et al. Prediction of protein-protein interactions using distant conservation of sequence patterns and structure relationships. Bioinformatics. 2005b;21:3360–3368. doi: 10.1093/bioinformatics/bti522. [DOI] [PubMed] [Google Scholar]
- Finn RD, et al. iPfam: visualization of protein-protein interactions in PDB at domain and amino acid resolutions. Bioinformatics. 2005;21:410–412. doi: 10.1093/bioinformatics/bti011. [DOI] [PubMed] [Google Scholar]
- Finn RD, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene LH, et al. The CATH domain structure database: new protocols and classification levels give a more comprehensive resource for exploring evolution. Nucleic Acids Res. 2007;35:D291–D297. doi: 10.1093/nar/gkl959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, et al. Genome-wide inference of protein interaction sites: lessons from the yeast high-quality negative protein-protein interaction dataset. Nucleic Acids Res. 2008;36:2002–2011. doi: 10.1093/nar/gkn016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegyi H, Gerstein M. The relationship between protein structure and function: a comprehensive survey with application to the yeast genome. J. Mol. Biol. 1999;288:147–164. doi: 10.1006/jmbi.1999.2661. [DOI] [PubMed] [Google Scholar]
- Jefferson ER, et al. SNAPPI-DB: a database and API of Structures, iNterfaces and Alignments for protein-protein interactions. Nucleic Acids Res. 2007;35:D580–D589. doi: 10.1093/nar/gkl836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT. Progress in protein structure prediction. Curr. Opin. Struct. Biol. 1997;7:377–387. doi: 10.1016/s0959-440x(97)80055-3. [DOI] [PubMed] [Google Scholar]
- Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- Kelley LA, et al. Enhanced genome annotation using structural profiles in the program 3D-PSSM. J. Mol. Biol. 2000;299:499–520. doi: 10.1006/jmbi.2000.3741. [DOI] [PubMed] [Google Scholar]
- Kerrien S, et al. IntAct–open source resource for molecular interaction data. Nucleic Acids Res. 2007;35:D561–D565. doi: 10.1093/nar/gkl958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keskin O, et al. Principles of protein-protein interactions: what are the preferred ways for proteins to interact? Chem. Rev. 2008;108:1225–1244. doi: 10.1021/cr040409x. [DOI] [PubMed] [Google Scholar]
- Keskin O, Nussinov R. Similar binding sites and different partners: implications to shared proteins in cellular pathways. Structure. 2007;15:341–354. doi: 10.1016/j.str.2007.01.007. [DOI] [PubMed] [Google Scholar]
- Keskin O, et al. A new, structurally nonredundant, diverse data set of protein-protein interfaces and its implications. Protein Sci. 2004;13:1043–1055. doi: 10.1110/ps.03484604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiel C, et al. Analyzing protein interaction networks using structural information. Annu. Rev. Biochem. 2008;77:415–441. doi: 10.1146/annurev.biochem.77.062706.133317. [DOI] [PubMed] [Google Scholar]
- Kim PM, et al. Relating three-dimensional structures to protein networks provides evolutionary insights. Science. 2006;314:1938–1941. doi: 10.1126/science.1136174. [DOI] [PubMed] [Google Scholar]
- Kryshtafovych A, et al. Progress over the first decade of CASP experiments. Proteins. 2005;61(Suppl. 7):225–236. doi: 10.1002/prot.20740. [DOI] [PubMed] [Google Scholar]
- Madera M. Profile Comparer: a program for scoring and aligning profile hidden Markov models. Bioinformatics. 2008;24:2630–2631. doi: 10.1093/bioinformatics/btn504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marti-Renom MA, et al. Alignment of protein sequences by their profiles. Protein Sci. 2004;13:1071–1087. doi: 10.1110/ps.03379804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra GR, et al. Human protein reference database–2006 update. Nucleic Acids Res. 2006;34:D411–D414. doi: 10.1093/nar/gkj141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittelman D, et al. Probabilistic scoring measures for profile-profile comparison yield more accurate short seed alignments. Bioinformatics. 2003;19:1531–1539. doi: 10.1093/bioinformatics/btg185. [DOI] [PubMed] [Google Scholar]
- Ogmen U, et al. PRISM: protein interactions by structural matching. Nucleic Acids Res. 2005;33:W331–W336. doi: 10.1093/nar/gki585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orengo CA, et al. From protein structure to function. Curr. Opin. Struct. Biol. 1999;9:374–382. doi: 10.1016/S0959-440X(99)80051-7. [DOI] [PubMed] [Google Scholar]
- Pagel P, et al. The MIPS mammalian protein-protein interaction database. Bioinformatics. 2005;21:832–834. doi: 10.1093/bioinformatics/bti115. [DOI] [PubMed] [Google Scholar]
- Punta M, Rost B. PROFcon: novel prediction of long-range contacts. Bioinformatics. 2005;21:2960–2968. doi: 10.1093/bioinformatics/bti454. [DOI] [PubMed] [Google Scholar]
- Rost B. TOPITS: threading one-dimensional predictions into three-dimensional structures. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1995;3:314–321. [PubMed] [Google Scholar]
- Rost B. Protein structures sustain evolutionary drift. Fold Des. 1997;2:S19–S24. doi: 10.1016/s1359-0278(97)00059-x. [DOI] [PubMed] [Google Scholar]
- Salwinski L, et al. The Database of Interacting Proteins: 2004 update. Nucleic Acids Res. 2004;32:D449–D451. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez R, et al. Protein structure modeling for structural genomics. Nat. Struct. Biol. 2000;7:986–990. doi: 10.1038/80776. [DOI] [PubMed] [Google Scholar]
- Sharan R, et al. Network-based prediction of protein function. Mol. Syst. Biol. 2007;3:88. doi: 10.1038/msb4100129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker BA, Panchenko AR. Deciphering protein-protein interactions. Part I. Experimental techniques and databases. PLoS Comput. Biol. 2007;3:e42. doi: 10.1371/journal.pcbi.0030042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soding J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005;21:951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- Stein A, et al. 3did: interacting protein domains of known three-dimensional structure. Nucleic Acids Res. 2005;33:D413–D417. doi: 10.1093/nar/gki037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The UniProt Consortium (2009) The Universal Protein resource (UniProt) 2009. Nucleic Acids Res., 37, D169–D174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton JM, et al. Protein folds, functions and evolution. J. Mol. Biol. 1999;293:333–342. doi: 10.1006/jmbi.1999.3054. [DOI] [PubMed] [Google Scholar]
- Tsai CJ, et al. A dataset of protein-protein interfaces generated with a sequence-order-independent comparison technique. J. Mol. Biol. 1996;260:604–620. doi: 10.1006/jmbi.1996.0424. [DOI] [PubMed] [Google Scholar]
- Wang H, et al. InSite: a computational method for identifying protein-protein interaction binding sites on a proteome-wide scale. Genome Biol. 2007;8:R192. doi: 10.1186/gb-2007-8-9-r192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter C, et al. SCOPPI: a structural classification of protein-protein interfaces. Nucleic Acids Res. 2006;34:D310–D314. doi: 10.1093/nar/gkj099. [DOI] [PMC free article] [PubMed] [Google Scholar]