Abstract
Summary:The program Fluctuation AnaLysis CalculatOR (FALCOR) is a web tool designed for use with Luria–Delbrück fluctuation analysis to calculate the frequency and rate from various mutation assays in bacteria and yeast. Three calculation methods are available through this program: (i) Ma-Sandri-Sarkar Maximum Likelihood Estimator (MSS-MLE) method, (ii) Lea-Coulson method of the median (LC) and (iii) frequency.
Availability: The FALCOR rate calculator is currently accessible at http://www.mitochondria.org/protocols/FALCOR.html. This program is written as a Java™ Applet, requiring a web browser enabled with Sun MicroSystems' Java Virtual Machine.
Contact: brandon.hall@roswellpark.org
1 BACKGROUND
Bacteria and yeast models have been engineered to facilitate the detection of many genetic events, including mutations, recombination, gross chromosomal rearrangements and repair of double-strand breaks through specific repair pathways, such as single-strand annealing. Being able to accurately determine the rate of these genetic events is fundamental to understanding the genes and pathways involved in various cellular processes.
Originally described by Luria and Delbrück (1943), fluctuation analysis has become the standard method in the field for calculating mutation rates. Briefly, a small number of cells are used to inoculate parallel cultures in a non-selective medium. The cultures are then grown to saturation to obtain equal cell densities. Cells are then plated onto selective media to obtain the number of mutants, r, and dilutions are plated onto rich medium to calculate the total number of viable cells, Nt. The number of mutants that appear in the saturated culture is a reflection of the mutation rate, as well as when the mutants arise during the growth of the culture; mutants that appear early in the growth of the culture will propagate many more mutants than those that arise later during growth. These factors cause the frequency (r/Nt) to vary greatly, even if the number of mutational events, m, is the same. As such, frequency is not a sufficiently accurate measure of mutation; mutation rate (m/Nt) should always be calculated (Rosche and Foster, 2000).
A number of statistical methods have been developed to estimate m from the observed values of r across parallel cultures. While the Lea-Coulson method of the median (LC), introduced in 1949 (Lea and Coulson, 1949), is the classic model for the estimation of mutation rates, statistical analyses have evolved to more accurately estimate m. However, the complex calculations required place these more accurate methods beyond easy reach of bench scientists. The Ma-Sandri-Sarkar Maximum Likelihood Estimator (MSS-MLE) is the best method available to date (Zheng, 2002); it is the most accurate and, unlike the LC method, is valid over all values of r and m. Furthermore, the MSS-MLE method calculates the mutation rate from the entire dataset (not just the median), providing more statistical power. A comprehensive evaluation of these methods was conducted with experimental data by Rosche and Foster (2000). To facilitate the use of these complicated methods by bench scientists, we developed a web interface to implement the three most popular methods.
Users of Fluctuation AnaLysis CalculatOR (FALCOR) should be familiar with fluctuation analysis and how to conduct experiments properly. Foster (2006) provides an excellent review of fluctuation analysis, including the underlying assumptions that shape the current mathematical models. While other programs exist for estimating mutation rates (Lang and Murray, 2008; Zheng, 2002, 2005, 2007), these require the user to implement source code into other software (i.e. Wolfram's Mathematica or The MathWorks' Matlab), whereas FALCOR is readily accessible through Java-enabled web browsers.
2 INTERFACE
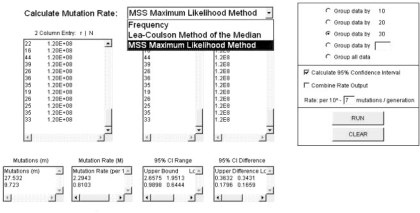
As shown in Figure 1, FALCOR is implemented as a web application for easy use. The user selects which of the three methods to execute from the pull-down menu. It is recommended that the ‘2 column entry’ box is used for data input: this is accomplished by copying data organized in Excel into the box (the first column containing values of r and the second column containing values of Nt). Alternatively, the individual columns can be copied separately into the ‘r’ and ‘N’ input boxes. Data can then be grouped according to the user's specifications, making it possible, for example, to calculate the mutation rate of multiple strains without having to run FALCOR for each strain individually. The user can also set the output's log10 value to place the rate within an appropriate range (default value is 10−7). The ‘combine rate output’ option combines the output into a single field, making it easier for the user to copy the data into a spreadsheet.
Fig. 1.
A screen shot of FALCOR implementing the MSS-MLE method. The pull-down menu has been engaged to show the various calculation methods. Sixty data points have been entered into ‘2 Column Entry’ box, representing three fluctuation analysis experiments of 10 independent cultures for two different yeast strains in a classic yeast canavanine resistance assay. The data are grouped by 30 to obtain the mutation rate for each strain, whereas grouping by 10 would produce the mutation rate per experiment for each strain. Mutation rate, as indicated, is displayed per 10−7 mutations/generation.
3 IMPLEMENTATION
The MSS-MLE method uses an initial estimate of m to generate the probability of observing r mutants on selective medium, pr. The likelihood function is the product of the pr's for each observed value of r. The value of m is then adjusted until the likelihood function reaches a maximum (Ma et al., 1992; Sarkar et al., 1992). The mutation rate, M, is then defined as m/Ñt, where Ñt represents the average of the cell counts across the cultures. The confidence intervals are calculated according to the method of Stewart (1994) who discovered that the natural log of m is normally distributed. FALCOR uses an approximation of this distribution, as described by Foster (2006). The student's t-test can be used to determine the significance between two observations; see webpage for details (Rosche and Foster, 2000).
FALCOR implements a slightly modified version of the LC method (Schmidt et al., 2006). Briefly, the value of m is calculated from r via the Lea-Coulson equation: r/m − ln(m)−1.24=0. The mutation rate, M, is then determined for each data point: m/Nt. The program then sorts the values of M and determines the median. Confidence intervals are derived from the cumulative binomial distribution of the rank values of M (Foster, 2006). The sorted values of M are displayed to facilitate significance testing via the Mann–Whitney U test (Flores-Rozas and Kolodner, 1998).
As previously discussed, frequency is highly inaccurate for measuring spontaneous mutations. However, frequencies are useful for determining the level of induced mutations, where cells are treated (e.g. with a DNA damaging agent) before the level of mutation is measured. FALCOR calculates frequency, providing statistical interpretation of the data with confidence intervals about the median, as with the LC method.
4 CONCLUSIONS
Fluctuation analysis is easily implemented and can yield highly accurate results when the data are analyzed correctly. It is our hope that this program will standardize the way that rates are calculated from fluctuation analysis by making the most powerful and accurate calculation method, the MSS-MLE method, available to everyone via the FALCOR web tool.
AKNOWLEDGEMENTS
We would like to thank Sheila Figel for her invaluable contribution to the conception of this program's name.
Funding: National Institutes of Health (R01 grant numbers 113655 and 121904 to K.K.S.).
Conflict of Interest: none declared.
REFERENCES
- Flores-Rojas H, Kolodner RD. The Saccharomyces cerevisiae MLH3 gene functions in MSH3-dependent suppression of frame shift mutations. Proc. Natl Acad. Sci. USA. 1998;95:12404–12409. doi: 10.1073/pnas.95.21.12404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster PL. Methods for determining spontaneous mutation rates. Methods Enzymol. 2006;409:195–213. doi: 10.1016/S0076-6879(05)09012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang GI, Murray AW. Estimating the per-base-pair mutation rate in the yeast Saccharomyces cerevisiae. Genetics. 2008;178:67–82. doi: 10.1534/genetics.107.071506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lea DE, Coulson CA. The distribution of the numbers of mutants in bacterial populations. J. Genet. 1949;49:264–285. doi: 10.1007/BF02986080. [DOI] [PubMed] [Google Scholar]
- Luria SE, Delbrück M. Mutations of bacteria from virus sensitivity to virus resistance. Genetics. 1943;28:491–511. doi: 10.1093/genetics/28.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma WT, et al. Analysis of the Luria-Delbrück distribution using discrete convolution powers. J. Appl. Prob. 1992;29:255–267. [Google Scholar]
- Rosche WA, Foster PL. Determining mutation rates in bacterial populations. Methods. 2000;20:4–17. doi: 10.1006/meth.1999.0901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S, et al. On fluctuation analysis: a new, simple and efficient method for computing the expected number of mutants. Genetica. 1992;85:173–179. doi: 10.1007/BF00120324. [DOI] [PubMed] [Google Scholar]
- Schmidt KH, et al. Chapter 27: analysis of gross-chromosomal rearrangements in Saccharomyces cerevisiae. Methods Enzymol. 2006;409:462–476. doi: 10.1016/S0076-6879(05)09027-0. [DOI] [PubMed] [Google Scholar]
- Stewart FM. Fluctuation tests: how reliable are the estimates of mutation rates? Genetics. 1994;137:1139–1146. doi: 10.1093/genetics/137.4.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q. Statistical and algorithmic methods for fluctuation analysis with SALVADOR as an implementation. Math. Biosci. 2002;176:237–252. doi: 10.1016/s0025-5564(02)00087-1. [DOI] [PubMed] [Google Scholar]
- Zheng Q. New algorithms for Luria-Delbruck fluctuation analysis. Math. Biosci. 2005;196:198–214. doi: 10.1016/j.mbs.2005.03.011. [DOI] [PubMed] [Google Scholar]
- Zheng Q. On Haldane's formulation of Luria and Delbruck's mutational model. Math. Biosci. 2007;209:237–252. doi: 10.1016/j.mbs.2007.03.003. [DOI] [PubMed] [Google Scholar]