Abstract
While effector molecules produced by activated macrophages (including nitric oxide, tumor necrosis factor α, interleukin 1, etc.) help to eliminate pathogens, high levels of these molecules can be deleterious to the host itself. Despite their importance, the mechanisms modulating macrophage effector functions are poorly understood. This work introduces two key negative regulators that control the levels and duration of macrophage cytokine production. Vacuolar-type H+-ATPase (V-ATPase) and calcineurin (Cn) constitutively act in normal macrophages to suppress expression of inflammatory cytokines in the absence of specific activation and to inhibit macrophage cytokine responses induced by bacterial lipopolysaccharide (V-ATPase), interferon γ (V-ATPase and Cn), and calcium (Ca2+) flux (Cn). Cn and V-ATPase modulate effector gene expression at the mRNA level by inhibiting transcription factor NF-κB. This negative regulation by Cn is opposite to its crucial positive role in T cells, where it activates NFAT transcription factor(s) leading to expression of interleukin 2, tumor necrosis factor α, and other cytokine genes. The negative effects of V-ATPase and Cn on NF-κB-dependent gene expression are not limited to the macrophage lineage, as similar effects have been seen with a murine fibroblast cell line and with primary astrocytes.
In the innate immune response, bacteria or bacterial products [including lipopolysaccharides (LPSs) and peptidoglycans] stimulate macrophages to produce an array of inflammatory molecules, including nitric oxide (NO), tumor necrosis factor α (TNF-α), interleukin 1 (IL-1), Macrophage inflammatory proteins (MIPs), IL-6, and IL-12, as well as known anti-inflammatory agents IL-10, interferon β (IFN-β), and prostaglandins (1). Macrophages also act as professional antigen-presenting cells (APC); they process and present antigens to T cells, thereby activating specific immune responses (2). Antigen-specific CD4+ T cells direct host effector functions by means of one of two alternative cytokine responses: Th1 (cell-mediated pro-inflammatory responses) or Th2 (antibody-mediated responses) (3, 4). The appropriate choice of Th1 or Th2 cytokine profile is crucial for the elimination of pathogens.
Macrophage cytokines induced by innate stimuli sometimes have antagonistic effects: IL-12 is known to induce and enhance Th1 responses but to suppress Th2 cytokines (5); conversely, IL-6 and IL-10 induce Th2 responses and inhibit Th1 cytokine production (6, 7). High levels of TNF-α were recently shown to suppress both Th1 and Th2 cells (8). Therefore, continuously high levels of TNF-α and IL-12 are likely to interfere with protective Th1 and Th2 responses. Prolonged high levels of TNF-α also have adverse effects on hematopoiesis, leading to aplastic anemia (9). TNF-α and IL-1 also are pathogenic in rheumatoid arthritis, multiple sclerosis, and other autoimmune diseases (10–12). Considering the importance and potential pathogenic activity of macrophage-derived cytokines, it is not surprising that the levels and duration of their production are tightly regulated. However, the mechanisms of this regulation are not well understood.
The studies described below examined signaling pathways initiated by several stimulators of macrophage cytokine responses: LPS, IFN-γ, Ca2+, and acidic intracellular pH (pHi) in the hope of identifying downstream regulatory mechanisms. LPS signals through multiple receptors, the best-characterized of which is CD14, leading to the activation of transcription factor NF-κB (13). IFN-γ is produced by natural killer (NK) cells and T cells and is a regulatory and effector molecule in both innate and specific immune responses (14–16). IFN-γ-responsive elements are present in the promoters of many genes; in macrophages IFN-γ is known to synergize with LPS for TNF-α, IL-12, and MHC class II induction (17–19). Ca2+ flux has been implicated in macrophage activation, although its role remains unclear. Calcium ionophores were shown to induce NO synthase in macrophages primed by IFN-γ or trehalose dimycolate (20), and Ca2+ was reported to be required for TNF-α induction by LPS (21, 22). However, Ca2+ flux induced by Fc-γ receptor ligation was implicated in the inhibition of IL-12 production in response to LPS (23). Conflicting reports exist as to whether LPS induces Ca2+ flux in macrophages (20, 21). pHi is controlled in macrophages by several H+ exchangers, of which vacuolar-type H+-ATPase (V-ATPase) is the major proton-extruding molecule (24). Unlike many other H+ exhangers, V-ATPase is not blocked by a low external pH, which is a likely result of the acidic bacterial inflammatory microenvironment (25). In macrophages V-ATPase is located in the plasma membrane as well as in the lysosomal membranes, and specific inhibition of V-ATPase by bafilomycin is known to lower pHi (refs. 26 and 27 and our observations).
Our results demonstrate that calcineurin (Cn) and V-ATPase suppress NF-κB activation and negatively regulate macrophage cytokine production. In contrast, in T cells Cn plays a crucial positive role: this Ca2+- and calmodulin-activated Ser/Thr phosphatase transduces the signal resulting from Ca2+ flux after T cell receptor ligation by dephosphorylating and activating NFAT transcriptional factors, leading to production of IL-2, IL-4, TNF-α, and other cytokines (28). Accordingly, specific inhibitors of Cn [cyclosporin A (CsA) and FK506] prevent T cell cytokine production and are commonly used as immunosuppressants (28).
MATERIALS AND METHODS
Reagents and Antibodies.
Bafilomycin A1, LPS (Escherichia coli O127:B8), thapsigargin, cyclosporin A, mouse recombinant IL-12, and mouse recombinant TNF-α were obtained from Sigma. Mouse recombinant IFN-γ was purchased from Boehringer Mannheim. FK506 was donated by Fujisawa Company (Melrose Park, IL). Cypermethrin was obtained from Calbiochem. Anti-IL-12 p40 mAbs C17.8 and C15.6 were provided by G. Trinchieri (Wistar Institute of Anatomy and Biology, Philadelphia, PA); anti-NF-κB p65 and p50 mAbs were purchased from Santa Cruz Biotechnology; anti-NFAT rabbit polyclonal antibodies were generously provided by G. Crabtree (Stanford University); F4/80 and Mac-1 mAb were purchased from Caltag (Burlingame, CA); anti-CD3 mAb (145.2C11 hybridoma obtained from H. McConnell, Stanford University) was purified in our laboratory. NF-κB consensus oligonucleotide was purchased from Promega.
Isolation and Culturing of Macrophages.
Seven- to 9-week-old B10.BR mice obtained from The Jackson Laboratory were injected i.p. with 2 ml of Brewster’s thioglycollate (Difco). Four to 5 days later peritoneal exudate cells were harvested with 0.34 M sucrose in phosphate-buffered saline (PBS), and after two washes in normal medium (RPMI 1640/10% FCS; GIBCO), cells were plated at 5 × 105 cells per ml. Nonadherent cells were washed off 30 min later; adherent cells were routinely identified as 85–90% pure macrophages by using Mac-1 and F4/80 cell-surface markers. The same conditions were used for isolation and culturing of resident peritoneal macrophages, except mice were not injected with thioglycollate.
T Cells, L929 Fibroblasts, and Primary Astrocytes.
T cell lines specific for mouse myelin basic protein N-terminal peptide Ac1–16 were generated and maintained as described (29). Activated T cells were obtained by culture in 24-well plates previously coated with 200 ng/ml hamster anti-mouse CD3 mAb in PBS containing 25 μg/ml goat gamma globulin. The L929 fibroblast cell line was obtained from H. McDevitt (Stanford University). Primary murine astrocytes and astrocyte/neuronal cultures (30) were generously provided by Raymond Swanson and Becky Stein (University of California at San Francisco and Veterans Affairs Hospital, San Francisco).
TNF Bioassay and IL-12 ELISA.
The TNF bioassay and double-antibody sandwich ELISA for IL-12 were carried out as described (29, 31).
Reverse Transcriptase (RT)-PCR.
Cells (1 × 106) were cultured in 1 ml of normal medium for 6–8 hr prior to harvesting. RNA was isolated by using the RNeasy kit (Qiagen, Santa Clarita, CA), and 100 μl of cDNA was synthesized from 1–3 μg of total RNA with oligo(dT) primers and Moloney murine leukemia virus RT (GIBCO). PCR was performed with 10 μl of cDNA reaction product, specific primers, and Taq DNA polymerase (GIBCO), as recommended by GIBCO.
Electrophoretic Mobility-Shift Assay (EMSA).
EMSA and supershifts with anti-p50 and anti-p65 antibodies were performed with 32P-labeled NF-κB consensus oligonucleotide (Promega), as recommended by Promega’s protocol. Nuclear extracts were prepared as described (32).
NF-κB and NFATc3 Staining.
Macrophages were plated at 5 × 105 cells per chamber in Lab-Tek chambered glass slides (Nunc) and treated with various agents. Thirty minutes later for NFATc3 and 4 hr later for NF-κB cells were washed with PBS, fixed with 2% paraformaldehyde, and stained with 0.1 μg per chamber anti-NF-κBp65 or anti-NFATc3 specific antibodies (both rabbit polyclonal IgG), or with the rabbit IgG control antibody. Cells were incubated with primary antibody overnight in PBS/1% FCS/0.1% saponin buffer. Then after two 10-min washes in the same buffer, cells were stained at room temperature for 1 hr with 1 μg per chamber FITC-labeled goat anti-rabbit IgG antibody and costained with 2.5 μg/ml propidium iodide (PI), as nuclear stain. Stained cells were analyzed on Molecular Dynamics MultiProbe confocal microscope, using dual scanning mode for FITC and PI.
Transfection.
Batches (5 × 106) of WEHI-3 myelomonocytic cells (33) were transfected by electroporation with 10 μg of plasmid DNA in 500 μl of RPMI medium 1640 in a 0.4-cm electroporation cuvette at 0.3 kV and 960 μF, by using a Bio-Rad electroporator. Transfected cells were diluted 1:20 in normal medium and cultured at 37°C under 6% CO2.
RESULTS
V-ATPase Inhibits Production of TNF-α and IL-12.
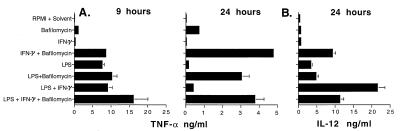
The initial series of experiments investigated how TNF-α and IL-12 production is controlled in peritoneal macrophages, either nonactivated or when activated by LPS, IFN-γ, or Ca2+ flux. The roles of V-ATPase and acidic pHi were examined by using noncytotoxic concentrations of bafilomycin. The highest levels of TNF-α were usually produced at 9 hr, whereas the highest levels of IL-12 were normally detected at 24 hr. Surprisingly, as shown in Fig. 1A, in the absence of any activating treatment, inhibition of V-ATPase with bafilomycin induced macrophages to produce an average of 1.0 ng/ml TNF-α (compare 0.05 ng/ml produced in control medium). Bafilomycin also synergized with IFN-γ to induce nearly 20 times more TNF-α than did IFN-γ alone. At 9 hr, inhibition of V-ATPase did not dramatically increase TNF-α production in response to LPS or LPS + IFN-γ. However, bafilomycin did have an effect on the duration of this response: TNF-α production normally declines to background levels by 24 hr after stimulation with LPS or LPS + IFN-γ, but when V-ATPase activity was blocked, significant levels of TNF-α were still produced at 24 hr. For IL-12, bafilomycin had little effect in the absence of specific stimuli or in the presence of LPS or LPS + IFN-γ. However, inhibition of V-ATPase dramatically augmented IL-12 production in response to IFN-γ alone (Fig. 1B).
Figure 1.
Regulation of TNF-α and IL-12 production by V-ATPase. Peritoneal macrophages (5 × 105) were cultured in 1 ml of normal medium for 2–3 hr, after which cells were treated for 9 or 24 hr with 0.1 μM bafilomycin A1, 10 μg/ml LPS, 100 units/ml IFN-γ, or combinations of reagents listed above. As bafilomycin was dissolved in DMSO, medium with an equal amount of DMSO (RPMI + Solvent) was used as control. TNF-α (A) was measured by bioassay and IL-12 (B) was determined by ELISA. Data represent means and standard deviations of duplicate cultures for each experimental condition, with TNF-α and IL-12 assays also performed in duplicate (n = 4). Similar results were obtained in four experiments with thioglycollate and in two experiments with resident peritoneal macrophages.
These results suggest that V-ATPase function inhibits TNF-α production in the absence of specific stimuli and controls the duration of the TNF-α response induced by LPS. These data also demonstrate that acidic pHi induced by V-ATPase inhibition strongly synergizes with IFN-γ signaling for TNF-α and IL-12 production, similar to the action of LPS. For IL-12, either acidic pHi or LPS synergizes with IFN-γ, but the combination of LPS + IFN-γ + bafilomycin does not result in higher levels of IL-12 than LPS + IFN-γ alone, indicating that low pHi and LPS may act through overlapping pathways.
Cn Inhibits TNF-α and IL-12 Production.
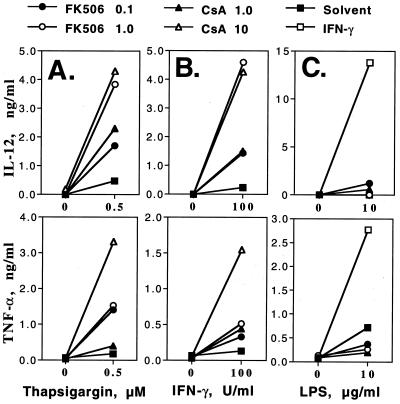
Cn is a key mediator of the inductive effects of Ca2+ flux on cytokine production by T cells (28). The role of Cn in the production of TNF-α and IL-12 induced in peritoneal macrophages by LPS, IFN-γ, or Ca2+ flux was examined by using the specific Cn inhibitors FK506 and CsA. Macrophages stimulated by thapsigargin-induced Ca2+ flux alone produced very little TNF-α or IL-12 (Fig. 2A). Surprisingly, significant induction of these cytokines occurred when Cn inhibitors were also added. Similarly, when stimulated by IFN-γ, macrophages produced significantly more TNF-α and IL-12 when Cn was inhibited (Fig. 2B). Raising the concentration of FK506 to 10 μg/ml resulted in even higher levels of TNF-α and IL-12 (not shown). In contrast, Cn inhibitors did not significantly enhance LPS-induced IL-12 production and inhibited by 2- to 3-fold LPS-induced TNF-α production (Fig. 2C). IFN-γ, as expected, augmented IL-12 and TNF-α production induced by LPS. When cells were stimulated by LPS + IFN-γ, inhibition of Cn did not change the levels of IL-12 (not shown).
Figure 2.
Regulation of TNF-α and IL-12 production by Cn. Macrophages were cultured as described in the legend of Fig. 1 and treated with 0.1 or 1 μg/ml FK506 and 1 or 10 μg/ml CsA in the presence of 0.5 μM thapsigargin (A), 100 units/ml IFN-γ (B), or 10 μg/ml LPS (C). DMSO and ethanol were used as control solvents. Data show levels of TNF-α at 9 hr and levels of IL-12 at 24 hr. TNF-α and IL-12 were assayed in duplicate, with most standard deviations within 10% of the cytokine levels. Similar results were obtained in at least three independent experiments performed, each with resident and thioglycollate peritoneal macrophages.
These data suggest that in macrophages Cn is a negative regulator of TNF-α and IL-12 production, as Cn inhibitors synergize with Ca2+ flux and IFN-γ (but not LPS) to enhance cytokine production.
Cn and V-ATPase Negatively Regulate Effector Function Gene Expression at the mRNA Level.
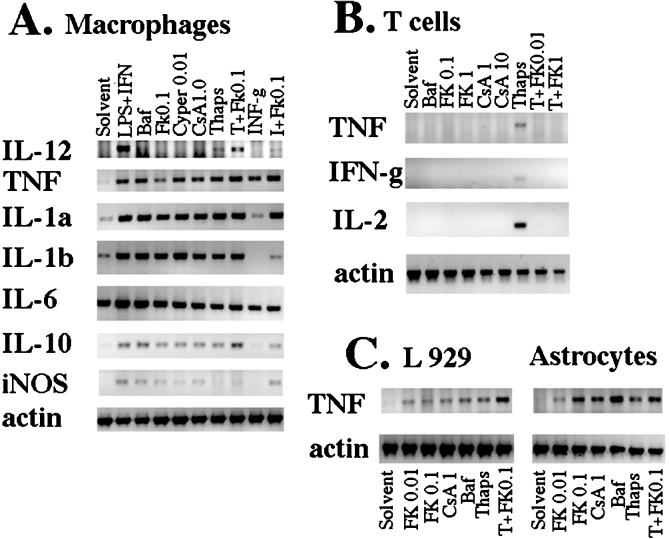
To test whether the induction of TNF-α and IL-12 protein secretion by combinations of stimuli and inhibitors of V-ATPase or Cn resulted from elevated mRNA levels, RT-PCR was performed with primers specific for these cDNAs, as well as for other macrophage effector proteins: IL-1α, IL-1β, IL-6, IL-10, and inducible NO synthase. Fig. 3A shows that mRNAs for many of these proteins were induced in macrophages simply by blocking either Cn (with the inhibitors FK506, CsA, or cypermethrin) or V-ATPase (with bafilomycin) (Fig. 3A). Inhibition of Cn resulted in levels of mRNA similar to or higher than those induced by Ca2+ flux or IFN-γ alone. In this and other experiments, IL-6 mRNA levels were often high in the absence of any stimuli.
Figure 3.
Effects of inhibition of Cn or V-ATPase on mRNA expression in macrophages, L929 fibroblasts, astrocytes, and T cells. (A) Macrophages (1 × 106) were cultured as described above and treated with 10 μg/ml LPS + 100 units/ml IFN-γ, 0.1 μM bafilomycin A1 (Baf), 0.1 μg/ml FK506, 0.01 μg/ml cypertmethrin (Cyper), 1 μg/ml CsA, 0.5 μM thapsigargin (Thaps) alone or in combinations as indicated, or with an equal volume of DMSO (Solvent). Cells were harvested 6–8 hr later, and RT-PCR was then performed with primers specific for indicated genes. One-tenth of cDNA reaction volume for effector genes and 1/500th of cDNA reaction volume for β-actin were used for the PCR amplification with specific primers. One-tenth PCR volume was run on 1% agarose gels, stained with ethidium bromide, and scanned under UV with a digital camera. The data shown are for one experiment performed with resident macrophages. Similar results were obtained in two or three independent experiments performed with thioglycollate and resident macrophages. In one of three experiment with resident cells and in experiments with thioglycollate-elicited macrophages, bafilomycin induced low levels of IL-12 (not shown). iNOS, inducible NO synthase. (B) Resting T cells (1 × 106) were plated in 1 ml of normal medium and treated as indicated. Cells were harvested for RT-PCR 6–7 hr later. (C) L929 cells (3–5 × 105) were plated in 1 ml of normal medium overnight. Astrocytes were plated at 5 × 104 cells per cm2 and were used at confluence, 14–20 days after isolation from murine forebrain. FK506, bafilomycin, and thapsigargin were added as above, and cells were harvested for RT-PCR 6–8 hr later. Three independent experiments with L929 and an additional experiment with astrocyte-neuronal cultures yielded similar results.
In contrast, in T cells no TNF-α, IFN-γ, or IL-2 mRNA expression was induced by treatment with either high or low levels of Cn inhibitors or with bafilomycin (Fig. 3B). As expected, the expression of cytokine genes (including TNF-α) induced by thapsigargin-elicited Ca2+ flux was completely blocked by FK506.
As these results indicated that Cn and V-ATPase have very different regulatory roles in macrophages and T cells, it became of interest to test the roles of V-ATPase and Cn in cytokine gene expression in other cell types. Fig. 3C shows that in L929 fibroblasts and in primary murine astrocytes, TNF-α mRNA was induced by the inhibition of V-ATPase or Cn. The levels of TNF-α induced by the thapsigargin were somewhat increased by FK506. No TNF-α secreted protein was produced by L929 cells; however, astrocytes did produce low levels of TNF-α protein in the presence of bafilomycin (not shown).
These results indicate that in macrophages V-ATPase and Cn negatively control a signaling pathway(s) involved in the induction of several effector-function genes. These pathway(s) and the roles of pHi and Cn clearly differ between T cells and the other cell types studied here: macrophages, L929 fibroblasts, and primary murine astrocytes.
Effects of V-ATPase and Cn Inhibitors on Activation of NF-κB and NFATs in Macrophages and T Cells.
The genes shown in Fig. 3 to be activated by Cn and V-ATPase inhibitors have NF-κB-responsive sites in their promoters (34). Hence, it became of interest to examine the activation status of NF-κB. This was assessed by two approaches, EMSA and fluorescence confocal microscopy.
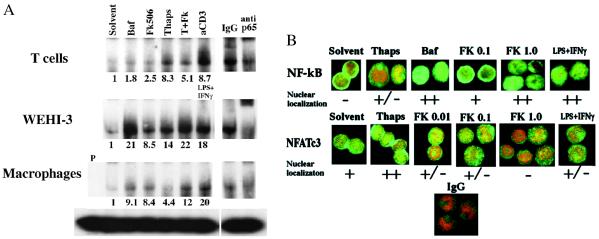
As shown in Fig. 4A, in T cells inhibition of V-ATPase and Cn resulted in about 2-fold higher levels of nuclear p65; however, total levels of this factor in the nucleus remained low, and inhibition of Cn slightly reduced p65 activation induced by thapsigargin-elicited Ca2+ flux. In normal macrophages and in the monocytic cell line WEHI-3, levels of active NF-κB p65 in nuclear extracts were increased at least 8-fold (and as much as 30-fold in some experiments) when V-ATPase or Cn was inhibited. For cells activated by Ca2+ flux induced by thapsigargin, simultaneous inhibition of Cn increased NF-κB p65 activity by at least 50%. In fact, activation of p65 in the presence of thapsigargin + FK506 was generally similar to that induced by the strong physiological stimulus LPS + IFN-γ. Inhibition of Cn in L929 cells also resulted in p65 activation (not shown).
Figure 4.
Effects of Cn or V-ATPase inhibitors on activation of NF-κB and NFATc3. (A) Cells (5 × 106 per well) were treated with control solvent (DMSO or ethanol), 0.1 μM bafilomycin (Baf), 1 μg/ml FK506, or 0.5 μM thapsigargin. As positive controls, macrophages and WEHI-3 cells were treated with 10 μg/ml LPS + 100 units/ml IFN-γ, and T cells were cultured in plates coated with anti-CD3 antibody. After 4 hr cells were harvested. EMSA was carried out with 10 μl of nuclear extract and 32P-labeled NF-κB consensus oligonucleotide. The bands containing NF-κB p65 were identified by supershift with anti-p65 antibody; isotype-matched rabbit IgG was used as a control. P indicates probe alone without nuclear extracts. Relative fold induction values indicated below the lanes are the intensities of p65 upper band in pixel density normalized by the total protein level for each lane and divided by the normalized pixel density of the p65 band of the solvent control. For T cells similar effects were observed with as little as 0.01 μg/ml FK506; for other cell types some NF-κB p65 activation was seen at 0.1 μg/ml FK506 (not shown). Two to four independent experiments with each cell type produced similar results. (B) Macrophages (3–5 × 105 per 500 μl) were plated in chambered glass slides. Cells were treated as indicated and then were stained for NF-κB p65 or for NFATc3 and costained with propidium iodide (PI). Isotype-matched IgG was used as a control. Stained cells were analyzed by confocal microscopy. PI-stained nuclei appear red, FITC staining of NF-κB p65 or NFAT is green; their colocalization in the nucleus results in combinations of red and green pixels, and when overlapping they may appear yellow. At least three independent experiments gave similar results. (×100.)
The activation of NF-κBp65 by the inhibitors of Cn and V-ATPase in macrophages was also confirmed by nuclear localization determined by fluorescence confocal microscopy. Fields shown in Fig. 4B are typical of about 70% of cells on the slide. NF-κB p65 was cytoplasmic in untreated macrophages, localized weakly in the nucleus in approximately 50% of cells treated with 0.5 μM thapsigargin, and robustly localized in the nucleus of cells treated with inhibitors of either V-ATPase or Cn or with the physiologic stimulus LPS + IFN-γ. Bafilomycin, FK506, and CsA also induced nuclear localization of NF-κB p65 in L929 cells and in primary murine astrocytes (not shown).
In T cells FK506 and CsA inhibit Cn, prevent NFAT nuclear localization, and suppress cytokine production (28). Nuclear localization was used to assess the activation status of NFAT factors in macrophages treated with inhibitors of V-ATPase and Cn. Of the NFAT transcriptional activators, NFATc1 and NFATc2 were virtually undetectable in macrophages (not shown). NFATc3 was already nuclear in about 50% of macrophages treated only with control solvent, indicating some constitutive Cn activity (Fig. 4B). Upon activation by Ca2+ flux (thapsigargin) the amount of NFATc3 in the nucleus, as well as the percentage of cells with nuclear NFATc3, increased, demonstrating increased Cn activity. As predicted from previous results (28), NFATc3 activation was inhibited by treatment with FK506. As the dose of FK506 was increased from 0.01 to 1 μg/ml, levels of nuclear NFATc3 continued to diminish, thus reflecting the range of FK506 concentration required to block Cn activity in macrophages. Treatment of cells with 0.1 μM bafilomycin or with 10 μg/ml LPS + 100 units/ml IFN-γ either did not change or somewhat decreased the levels of nuclear NFATc3.
These results show that inhibition of Cn and V-ATPase activates NF-κB in macrophages and fibroblasts, and to a lower extent in T cells. These results also suggest that in macrophages and some other cell types induction of effector gene expression is linked to NF-κB activation and appears not to depend on NFAT activation, as it does in T cells.
Constitutively Active Cn Down-Regulates TNF-α Production by the Monocytic Cell Line WEHI-3.
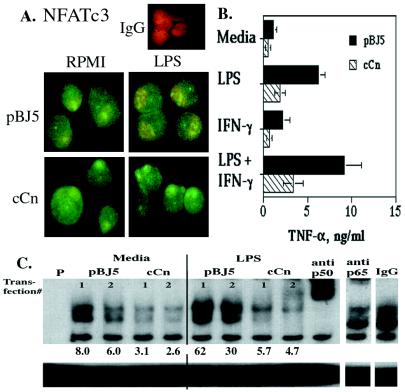
FK506 and CsA synergized with other stimuli in inducing TNF-α production by macrophages, indicating that Cn negatively controls TNF-α production. To confirm this conclusion in another experimental system, the myelomonocytic cell line WEHI-3 was transiently transfected with a vector encoding a constitutively active form of Cn (cCn), lacking its autoinhibitory domain (35, 36). The evidence for expression and activity of cCn was provided by NFATc3 staining. In contrast with cells electroporated with control plasmid, in 70–75% of cells electroporated with the cCn vector, NFATc3 was highly concentrated in the nucleus even in the presence of LPS (Fig. 5A). Cells transfected with cCn or with control vector (pBJ5) were treated with LPS, IFN-γ, LPS + IFN-γ, or medium alone, and levels of secreted TNF-α were assayed. Expression of cCn inhibited production of TNF-α (Fig. 5B) and activation of NF-κB (Fig. 5C) by cells treated with or without these stimuli. Note that untreated WEHI-3 cells and cells stimulated by IFN-γ alone produced much higher levels of TNF-α protein (0.5–1 ng/ml) than do normal mouse macrophages, resident or thioglycollate-elicited (about 0.05 ng/ml). Incomplete inhibition of TNF-α production by transfected cCn probably reflects expression of cCn in only a subset of the electroporated cells. Transfection with cCn diminished the levels of nuclear p65, shifting the NF-κB consensus oligonucleotide into the upper band (as determined by supershift with anti-p65 antibody) and also diminished levels of another nuclear rel-family protein(s) in the middle band (indicated by the supershift with anti-p50 antibody). These data confirm the role of Cn in monocyte/macrophage effector gene expression.
Figure 5.
Constitutively active Cn (cCn) down-regulates TNF-α production and NF-κB activation. WEHI-3 cells (5 × 106 ) were transfected with 10 μg of pBJ5 vector encoding cCn or with the pBJ5 control vector. After transfection, cells were plated in normal medium at 0.5–1 × 106 cells per ml for 48 hr. (A) Forty to 44 hr after electroporation, 1 μg/ml LPS was added to aliquots of WEHI-3 cells; 4–6 hr later cells were stained with anti-NFATc3 or control antibody (IgG) and analyzed by confocal microscopy, as described for Fig. 4B. (×100.) Similar results were observed in an additional experiment. (B) Seven hours after stimulation with 1 μg/ml LPS, 100 units/ml IFN-γ, LPS + IFN-γ, or control medium culture, supernatants were harvested and assayed for TNF-α. Cells transfected with cCn or pBJ5 control vector displayed similar viability by trypan blue staining. Shown are means and standard errors of results obtained in three independent transfections. (C) Four hours after stimulation with 1 μg/ml LPS or control medium, cells were harvested and EMSA was performed. Shown are results for duplicate transfections with pBJ5 and cCn. Below the lanes are pixel density for the p65 (upper band) normalized by protein level and digital input levels. Similar results were obtained in two additional experiments.
DISCUSSION
The results of these studies support the conclusions that (i) Cn plays a central role in regulating macrophage signaling pathways; (ii) Cn acts physiologically as a negative regulator of gene expression downstream of Ca2+ flux and IFN-γ; (iii) acidic pHi (controlled by V-ATPase) induces effector gene expression, strongly synergizes with IFN-γ for IL-12 production, and prolongs the TNF-α response to LPS; and (iv) Cn and V-ATPase inhibitors increase macrophage gene expression by activating NF-κB.
A central role for Cn is strongly suggested by the opposite effects of Cn inhibitors and constitutively active Cn on NF-κB activation and cytokine production. Support for the involvement of Cn comes from the similar effects of Cn inhibitors FK506, CsA, and cypermethrin, which act by different mechanisms (37, 38). Rapamycin, which binds to FK506-binding proteins and blocks their peptidylprolyl-isomerase activity but does not inhibit Cn (39), did not augment production of TNF-α and IL-12 induced in macrophages by Ca2+ flux. Thus, blocking immunophilin peptidylprolyl-isomerase activity is not responsible for the induction of gene expression by Cn inhibitors. Final proof that the observed activation of NF-κB and gene expression is due solely to the inhibition of Cn activity will require testing cells without functional Cn.
The concentration of FK506 required to activate NF-κB and effector gene expression in macrophages is higher than that required to inhibit NFAT activation and gene expression in T cells. This difference could result from a variety of factors, including higher cytoplasmic volume and/or vesicle abundance in macrophages, or differences in the levels of Cn isoforms or immunophilins. The mechanism of NFAT activation by Cn is also different from that of NF-κB inhibition, as these factors are regulated by distinct pathways. More complete and prolonged inhibition of Cn might be required for the activation of NF-κB [known to induce its own inhibitor, IκB (40)], whereas only partial inhibition of Cn may suffice for the inhibition of NFAT [which is actively exported from a nucleus (41)]. Evidence for different requirements for Cn inhibition for NF-κB activation vs. NFAT inactivation comes from the finding that in macrophages 0.1–1 μg/ml FK506 induces nuclear localization of NF-κB, whereas 0.01–1 μg/ml inhibits nuclear localization of NFATc3 (Fig. 4). The nuclear localization of NFATc3 in normal untreated macrophages indicates that Cn is active, possibly reflecting low intracellular Ca2+ concentration which may be induced by adherence (40).
In T cells no cytokine gene expression is induced by inhibitors of V-ATPase or Cn, and the effects of these inhibitors on the nuclear vs. cytoplasmic localization of NF-κB are less prominent than in other cell types. NF-κB activation induced via the protein kinase C pathway was previously shown to be dependent on Cn in Jurkat cells (42). We have confirmed this in our T cell line; when both the Ca2+ and the protein kinase C pathways are activated by CD3 ligation, induction of NF-κB is blocked by FK506 (not shown). However, inhibition of the basal Cn activity induces NF-κB in all tested cell types (albeit to a different extent), and inhibition of Cn in cells activated by Ca2+ flux activates NF-κB in several cell types with weakly inhibitory or no effect in T cells (Fig. 4). These results indicate that the mechanism of NF-κB regulation by Cn is cell-type specific and may depend on the particular NF-κB activating stimulus. Interestingly, although T cells treated with thapsigargin + FK506 have significantly more nuclear NF-κB than untreated cells, TNF-α mRNA is not expressed. Thus, in agreement with previous results (43, 44), even in the presence of nuclear NF-κB T cells are absolutely dependent on Cn-mediated NFAT activation for the expression of cytokine genes, including TNF-α. The different roles of Cn may reflect the fact that T cells can be activated only by specific antigen. Distinct signaling pathways may have evolved to mediate induction of cytokine genes after T cell receptor/CD3 ligation (45, 46).
On the basis of these findings we propose that the physiological roles of V-ATPase and Cn include preventing acidic pHi, Ca2+ flux, or IFN-γ from inappropriately activating NF-κB in cells where this transcription factor is sufficient to drive effector-function gene expression. V-ATPase and Cn appear to modulate a key signaling pathway(s) regulating expression of several macrophage effector genes. Inhibition of V-ATPase or Cn enhances levels of nuclear NF-κB and TNF-α mRNA induced by thapsigargin and synergistically increases production of the secreted protein. Post-transcriptional regulation of TNF-α has been previously demonstrated (47).
The ability to recognize acidic pHi may have developed as a signal for activating macrophage effector functions in response to bacteria (similar to LPS). Macrophages responding to a bacterial infection may be exposed to both LPS and low pH. As LPS blocks the function of V-ATPase (24), bacteria would provide positive feedback for the innate response. Conversely, as IL-1 enhances V-ATPase activity (25), increasing levels of this inflammatory cytokine may feed back to down-regulate the innate response.
The finding that Cn inhibitors do not synergize with LPS for TNF-α expression (Fig. 2C) appears to contrast with the activating effects of Cn. As LPS normally activates NF-κB and induces effector gene expression (13), it may act by inhibiting or overcoming the negative effect of Cn. Lower levels of nuclear NFATc3 after LPS treatment of macrophages or WEHI-3 cells support this idea. The contrasting inhibition by constitutively active Cn of LPS-induced NF-κB activation and TNF-α production suggests that LPS may normally suppress the activity of endogenous Cn by the autoinhibitory peptide that is absent from cCn.
No direct effect of Cn or V-ATPase/pHi on NF-κB activation has been described previously. NF-κB translocates to the nucleus when IκB is phosphorylated and degraded (34, 48, 49); therefore, maintenance of inactive NF-κB potentially could directly result from the phosphatase activity of Cn. Rescue of IκB from degradation by serine-dephosphorylation has been reported (50), but not linked to Cn. Alternatively, Cn and/or low pHi may activate NF-κB indirectly, by controlling other factors affecting the IκB dissociation/degradation pathway.
Effects of Cn inhibitors on gene expression have been previously studied in a variety of systems and in nearly all cases have been found to be suppressive (28). Conflicting results have been obtained in studies of the effects of CsA and/or FK506 on macrophage and other nonlymphoid cells (51, 52). FK506 was reported to induce NF-κB-linked IL-6 expression by L cell fibroblasts and mesangial cells (53). However, in those studies rapamycin, but not CsA, produced similar results, implicating the peptidylprolyl isomerase activity of immunophilins and not Cn in this response.
V-ATPase/pHi and Cn have important consequences at multiple physiological levels, negatively regulating cytokine gene expression and modulating inflammation in a variety of cell types and tissues. Cn inhibitors are commonly used as immunosuppressants in organ transplantation, reflecting their inhibitory effect on cytokine gene expression in T cells. However, Cn inhibitors may have pro-inflammatory effects on other cells, such as macrophages, if present in sufficiently high levels and/or combined with activating stimuli such as IFN-γ or activators of Ca2+ fluxes. Specific targeting of Cn (and/or V-ATPase) inhibitors to T cells, macrophages, or other cells could optimize their beneficial effects in suppressing T cell responses or in augmenting macrophage effector functions.
Acknowledgments
We acknowledge Drs. Gerald Crabtree, Luika Timmerman, and Martha Cyert for providing reagents and for helpful discussion; Irving Weissman, Ron Kopito, and Richard Lewis for critical evaluation of our ideas; and Raymond Swanson and Becky Stein for supplying primary astrocytes. This work was supported by funding from Stanford University. I.C. was supported by National Institutes of Health Training Grants T32GM07276 and T32HD07249.
ABBREVIATIONS
- LPS
lipopolysaccharide
- TNF-α
tumor necrosis factor α
- pHi
intracellular pH
- V-ATPase
vacuolar-type H+-ATPase
- Cn
calcineurin
- CsA
cyclosporin A
- RT
reverse transcriptase
- EMSA
electrophoretic mobility-shift assay
References
- 1.Remick D G. J Clin Care. 1995;10:198–212. doi: 10.1016/0883-9441(95)90011-x. [DOI] [PubMed] [Google Scholar]
- 2.Unanue E R. Adv Immunol. 1981;31:1–121. doi: 10.1016/s0065-2776(08)60919-0. [DOI] [PubMed] [Google Scholar]
- 3.Mosman T R, Coffman R L. Immunol Today. 1987;8:223–226. doi: 10.1016/0167-5699(87)90171-X. [DOI] [PubMed] [Google Scholar]
- 4.Mosman T R, Sad S. Immunol Today. 1996;17:138–146. doi: 10.1016/0167-5699(96)80606-2. [DOI] [PubMed] [Google Scholar]
- 5.Seder R A, Gazzinelli R, Sher A, Paul W E. Proc Natl Acad Sci USA. 1993;90:10188–10192. doi: 10.1073/pnas.90.21.10188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fiorentino D F, Zlotnik A, Viera P, Mosmann T R, Howard M, Moore K W, O’Garra A. J Immunol. 1991;146:3444–3451. [PubMed] [Google Scholar]
- 7.Rincin M, Anguita J, Nakamura T, Fikrig E, Flavel R A. J Exp Med. 1997;185:461–469. doi: 10.1084/jem.185.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cope A P, Liblau R S, Young X-D, Congia M, Laudanna C, Schreiber R D, Probert L, Kollias G, McDevitt H O. J Exp Med. 1997;185:1573–1584. doi: 10.1084/jem.185.9.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Young N S. Eur J Haematol Suppl. 1996;60:55–59. doi: 10.1111/j.1600-0609.1996.tb01646.x. [DOI] [PubMed] [Google Scholar]
- 10.Feldman M, Brennan F M, Maini R N. Cell. 1996;85:307–310. doi: 10.1016/s0092-8674(00)81109-5. [DOI] [PubMed] [Google Scholar]
- 11.Steinman L. Cell. 1996;85:299–302. doi: 10.1016/s0092-8674(00)81107-1. [DOI] [PubMed] [Google Scholar]
- 12.Liblau R S, Singer S M, McDevitt H O. Immunol Today. 1995;16:34–38. doi: 10.1016/0167-5699(95)80068-9. [DOI] [PubMed] [Google Scholar]
- 13.Keilian T L, Blecha F. Immunopharmacology. 1995;29:187–205. doi: 10.1016/0162-3109(95)00003-c. [DOI] [PubMed] [Google Scholar]
- 14.Bancroft G J, Kelly J P. Immunobiology. 1994;191:424–431. doi: 10.1016/S0171-2985(11)80448-1. [DOI] [PubMed] [Google Scholar]
- 15.Tripp C S, Unanue E R. Res Immunol. 1995;146:515–520. doi: 10.1016/0923-2494(96)83025-2. [DOI] [PubMed] [Google Scholar]
- 16.Trinchieri G, Gerosa F. J Leukoc Biol. 1996;59:505–511. doi: 10.1002/jlb.59.4.505. [DOI] [PubMed] [Google Scholar]
- 17.Ma X, Chow J M, Gri G, Carra G, Cerosa F, Wolf S F, Dzialo R, Trinchieri G. J Exp Med. 1996;183:147–157. doi: 10.1084/jem.183.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murphy T L, Cleveland M G, Kulesza P, Magram J, Murphy K M. Mol Cell Biol. 1995;15:5258–5267. doi: 10.1128/mcb.15.10.5258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Steeg P S, Moore R N, Johnson M, Oppenheim J J. J Exp Med. 1982;156:1780–1787. doi: 10.1084/jem.156.6.1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.West M A, Clair L, Bellingham J. J Trauma. 1996;41:647–652. doi: 10.1097/00005373-199610000-00009. [DOI] [PubMed] [Google Scholar]
- 21.Raddassi K, Berton B, Petit J F, Lemaire G. Cell Immunol. 1994;153:443–455. doi: 10.1006/cimm.1994.1041. [DOI] [PubMed] [Google Scholar]
- 22.Watanabe N, Suzuki J, Kobayashi Y. J Biochem (Tokyo) 1996;120:1190–1195. doi: 10.1093/oxfordjournals.jbchem.a021540. [DOI] [PubMed] [Google Scholar]
- 23.Sutterwala F S, Noel G J, Clynes R, Mosser D M. J Exp Med. 1997;185:1977–1985. doi: 10.1084/jem.185.11.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Swallow C J, Grinstein S, Rotstein O D. J Biol Chem. 1990;265:7645–7654. [PubMed] [Google Scholar]
- 25.Brisseau G F, Grinstein S, Hackam D J, Nordstrom T, Manolson M F, Khine A A, Rotstein O D. J Biol Chem. 1996;271:2005–2011. doi: 10.1074/jbc.271.4.2005. [DOI] [PubMed] [Google Scholar]
- 26.Bidani A, Heming T A. J Leukoc Biol. 1995;57:275–281. doi: 10.1002/jlb.57.2.275. [DOI] [PubMed] [Google Scholar]
- 27.Swallow C J, Grinstein S, Rotstein O D. J Leukoc Biol. 1992;52:395–399. doi: 10.1002/jlb.52.4.395. [DOI] [PubMed] [Google Scholar]
- 28.Rao A, Chun L, Hogan P G. Annu Rev Immunol. 1997;15:707–747. doi: 10.1146/annurev.immunol.15.1.707. [DOI] [PubMed] [Google Scholar]
- 29.Conboy I M, De-Kruyff R H, Tate K M, Cao Z A, Moore T A, Umetsu D T, Jones P P. J Exp Med. 1997;185:439–451. doi: 10.1084/jem.185.3.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Branch D R, Shan A, Guilbert L J. J Immunol Methods. 1991;143:251–261. doi: 10.1016/0022-1759(91)90050-p. [DOI] [PubMed] [Google Scholar]
- 31.Swanson R A, Choi D W. J Cereb Blood Flow Metab. 1993;13:162–169. doi: 10.1038/jcbfm.1993.19. [DOI] [PubMed] [Google Scholar]
- 32.Huang D Y, Prystowsky M B. J Biol Chem. 1996;271:1218–1225. doi: 10.1074/jbc.271.2.1218. [DOI] [PubMed] [Google Scholar]
- 33.Warner N L, Moore M A, Metcalf D. J Natl Cancer Inst. 1969;43:963–982. [PubMed] [Google Scholar]
- 34.Baldwin A S., Jr Annu Rev Immunol. 1996;14:649–681. doi: 10.1146/annurev.immunol.14.1.649. [DOI] [PubMed] [Google Scholar]
- 35.Clipstone N A, Crabtree G R. Nature (London) 1992;357:695–597. doi: 10.1038/357695a0. [DOI] [PubMed] [Google Scholar]
- 36.Hashimoto Y, Perrino B A, Soldering T R. J Biol Chem. 1990;265:1924–1927. [PubMed] [Google Scholar]
- 37.Ho S, Clipstone N, Timmerman L, Northrop J, Graef I, Fiorentino D, Nourse J, Crabtree G R. Clin Immunol Immunopathol. 1996;80:s40–s45. doi: 10.1006/clin.1996.0140. [DOI] [PubMed] [Google Scholar]
- 38.Wang J, Stelzer A. NeuroReport. 1994;5:2377–2380. doi: 10.1097/00001756-199411000-00041. [DOI] [PubMed] [Google Scholar]
- 39.Dumont F J, Su Q. Life Sci. 1996;58:373–395. doi: 10.1016/0024-3205(95)02233-3. [DOI] [PubMed] [Google Scholar]
- 40.Lofquist A K, Mondal K, Morris J S, Haskill J S. Mol Cell Biol. 1995;15:1737–1746. doi: 10.1128/mcb.15.3.1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beals C R, Sheridan C M, Turck C W, Gardener P, Crabtree G R. Science. 1997;275:1930–1934. doi: 10.1126/science.275.5308.1930. [DOI] [PubMed] [Google Scholar]
- 42.Frantz B, Nordby E E, Bren G, Steffan N, Paya C V, Kinaid R L, Tocci M J, O’Keefe S J, O’Neill E A. EMBO J. 1994;13:861–870. doi: 10.1002/j.1460-2075.1994.tb06329.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McCaffry P G, Goldfeld A E, Rao A. J Biol Chem. 1994;269:30445–30450. [PubMed] [Google Scholar]
- 44.Timmerman L, Clipstone N A, Ho S N, Northrop J P, Crabtree G R. Nature (London) 1996;383:837–840. doi: 10.1038/383837a0. [DOI] [PubMed] [Google Scholar]
- 45.Weismann I. Cell. 1994;76:207–218. [Google Scholar]
- 46.Nossal G J V. Cell. 1994;76:229–239. doi: 10.1016/0092-8674(94)90331-x. [DOI] [PubMed] [Google Scholar]
- 47.Solomon K A, Covington M B, DeCicco C P, Newton R C. J Immunol. 1997;159:4524–4531. [PubMed] [Google Scholar]
- 48.Maniatis T. Science. 1997;278:818–819. doi: 10.1126/science.278.5339.818. [DOI] [PubMed] [Google Scholar]
- 49.Baeuerle P, Baltimore D. Cell. 1996;87:13–20. doi: 10.1016/s0092-8674(00)81318-5. [DOI] [PubMed] [Google Scholar]
- 50.Link E, Kerr L D, Schreck R, Zabel U, Vermat I, Baeuerle P A. J Biol Chem. 1992;267:239–246. [PubMed] [Google Scholar]
- 51.Svensson U, Holst E, Sundler R. Mol Immunol. 1995;32:157–165. doi: 10.1016/0161-5890(94)00107-c. [DOI] [PubMed] [Google Scholar]
- 52.Durez P, Abramowicz D, Gerald C, van Mechelen M, Amraoui Z, Duboi D, Leo O, Velu T, Goldman M. J Exp Med. 1993;177:551–555. doi: 10.1084/jem.177.2.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Muraoka K, Fujimoto K, Sun X, Yoshioka K, Shimizu K, Yagi M, Bosse H, Jr, Miyazaki T, Yamamoto K. J Clin Invest. 1996;97:2433–2439. doi: 10.1172/JCI118690. [DOI] [PMC free article] [PubMed] [Google Scholar]