Abstract
The KcsA potassium channel from Streptomyces lividans is one of the most actively studied ion channels. However, there are still unresolved issues about its gating mechanism in vivo because the channel is only activated by highly acidic intracellular pH, meaning that it will be mostly inactive in its host environment. In this study we have used a genetic complementation assay of K+-auxotrophic E. coli (TK2420) and S. cerevisiae (SGY1528) to identify activatory or ‘gain-of-function’ mutations which allow functional activity of KcsA in the physiological environment of two markedly different expression systems. These mutations clustered at the helix-bundle-crossing in both TM1 and TM2 (residues H25, L105, A108, T112, W113, F114, E118 and Q119), and include residues previously implicated in the pH-gating mechanism. We discuss how these gain-of-function mutations may result in their activatory phenotype, the relative merits of the E. coli and S. cerevisiae genetic complementation approaches for the identification of gating mutations in prokaryotic K+ channels, and ways in which this assay may be improved for future use in screening protocols.
Keywords: KcsA, K+ channel, potassium channel, genetic complementation, ion channel gating, K+ auxotrophic
Introduction
The three-dimensional structure of the prokaryotic potassium channel KcsA was first solved by X-ray crystallography in 1998. Since then, KcsA has not only become the principal model for K+ ion selectivity and permeation, but has also provided a major insight into the mechanism of K+ channel gating, toxin inhibition and channel inactivation.1-4
Yet despite the intense interest that KcsA has generated over the last ten years there are still several unresolved issues about the physiological role of this channel in its host organism (Streptomyces lividans) and the mechanisms which might gate the channel in vivo.5,6 In particular, although the channel is activated by intracellular acidification, activation only occurs at highly acidic levels (below pH 5.0) and the intracellular pH of Streptomyces is fairly neutral.6,7 Consequently the channel is expected to be closed most of the time in its host environment. Furthermore, a mutant strain of S. lividans with a deletion of the KcsA gene has failed to shed any further light on channel activity in vivo due to the absence of any obvious phenotype.8 We therefore sought to investigate the mechanism of KcsA gating in vivo by searching for mutations, which would permit KcsA channel activity under normal ‘physiological’ conditions.
An approach is therefore required to search for mutations which permit the functional activity of KcsA when expressed in a prokaryotic or microbial host. Several previous studies have used a genetic complementation approach to screen for activatory or ‘gain-of-function’ mutations in prokaryotic potassium channels.9-11 Such assays are based on the fact that in wild type E. coli, K+ uptake is mediated by three principal transport pathways: kdp, trk and kup.12 The TK2420 strain has mutations in these genes and consequently exhibits a K+-auxotrophic phenotype i.e., no growth in media containing low concentrations of K+ (<10 mM). However, this K+-uptake deficiency can be complemented by recombinant expression of a functional K+ channel that provides an alternative pathway for the entry of K+. The assay can therefore be used to examine the functional properties of prokaryotic K+ channels,13 and as an unbiased screen for activatory mutations.9 A similar approach using K+-auxotrophic S. cerevisiae has also been used to identify gating mutations in a wide range of prokaryotic as well as eukaryotic K+ channels.14-17
In this study we randomly mutated KcsA and screened these mutants by genetic complementation in K+-auxotrophic E. coli. We identified a number of novel activatory mutations in both TM1 and TM2 of KcsA that demonstrated functional activity in both E. coli and S. cerevisiae. Mapping these mutations onto the structure of wild-type KcsA demonstrates that they all cluster at the helix-bundle crossing and highlight several residues now known to be involved in the pH-gating mechanism. We discuss how these mutations may result in their activatory phenotype and the relative merits of this genetic complementation approach for the identification of gating mutations in prokaryotic K+ channels.
Results and Discussion
Genetic selection of KcsA mutations
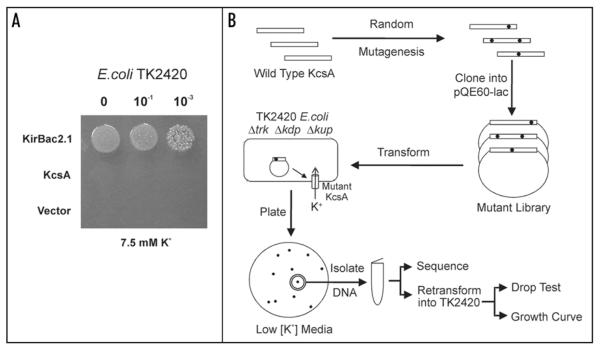
As can be seen in Figure 1A, wild-type KcsA does not complement the K+-uptake deficiency of TK2420 E. coli, presumably because KcsA can only be activated at highly acidic pH and the intracellular pH in E. coli is near neutral.18 By contrast, as a positive control, a different prokaryotic K+ channel, KirBac2.1 complements growth well.13
Figure 1.
Screen for Activatory Mutations in KcsA. (A) Wild-type KcsA does not complement the growth of K+-auxotrophic E. coli (TK2420) on low [K+] media (7.5 mM KCl). As a positive control KirBac2.1 complements growth well. The vector is pQE60-Lac. (B) Overall scheme for the random mutagenesis protocol.
A similar complementation assay was previously used to examine the functional consequences of tryptophan substitutions in TM1 and TM2 of KcsA.11 Several residues at the base of TM2 (in particular A108) were identified which affected channel gating and allowed functional expression in E. coli. However, only certain segments of the KcsA transmembrane domains were mutated and the mutagenesis approach was not fully randomized. We therefore decided to take a completely unbiased approach to the selection of activatory mutations in KcsA by randomly mutating the entire open reading frame of KcsA and selecting for activatory mutations on low [K+] media (Fig. 1B).
The mutant library was transformed into TK2420 E. coli and selected on K0 plates containing 7.5 mM KCl. After false positives were eliminated by retransformation into TK2420 and confirmation of the functional phenotype, 84 independent clones were sequenced. Where clones had more than one mutation, site-directed mutagenesis and/or subcloning was used to identify single mutants. Eight unique residues were identified which gave rise to an activatory phenotype (Fig. 2 and Table 1). These included two mutations identical to those found previously (A108T, T112N),11 confirming the validity of this approach. However, the rest were novel mutations and included one residue in TM1 (H25).
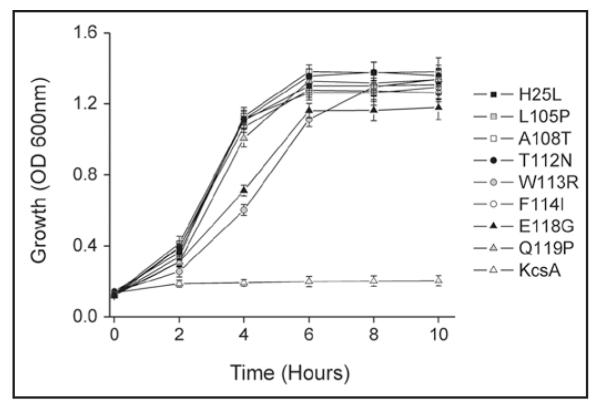
Figure 2.
Activatory mutations complement growth of TK2420 E. coli. The activatory mutations identified in this screen complement the K+-uptake growth defect of TK2420. Cultures were grown in 7.5 mM K0 media and induced with 0.5 mM IPTG at time zero. Wild-type KcsA does not complement this growth defect but the mutations permit growth in low [K+]. Data shown are mean ± S.D, n = 3.
Table 1.
Activatory mutations found in KcsA
| Mutation | Number found |
|---|---|
| H25L | 15 |
| H25P | 4 |
| H25Y | 1 |
| L105P | 5 |
| A108T | 18 |
| A108S | 2 |
| T112N | 6 |
| W113R | 10 |
| F114I | 9 |
| F114S | 2 |
| E118G | 7 |
| Q119P | 5 |
| Total No | 84 |
The occurrence for each of the activatory mutations found in KcsA out of a total number of 84 mutations that were sequenced.
Expression of KcsA in K+-auxotrophic yeast
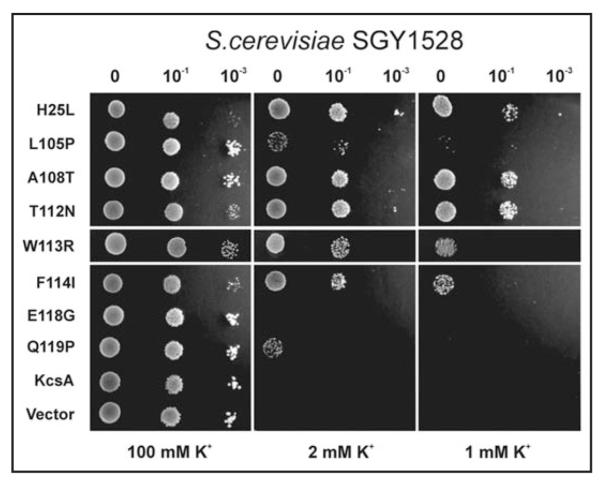
The SGY1528 strain of S. cerevisiae has mutations in both the trk1 and trk2 transport pathways that prevent growth on low [K+] media. We therefore tested the ability of wild-type and mutant KcsA to rescue the growth of this K+-uptake deficient yeast. The ability to express KcsA in S. cerevisiae may be of particular interest as their intracellular pH can drop as low as pH 5.0—pH 5.5 during certain parts of the cell cycles and may therefore be more appropriate for the functional expression of KcsA.19 However, despite this acidic intracellular pH, wild-type KcsA was not able to complement growth of this yeast strain (Fig. 3). Nevertheless, all of the activatory mutants identified in the E. coli screen above, with the exception of E118G, allowed complementation of SGY1528 growth on either 1 mM or 2 mM K+ media (Fig. 3).
Figure 3.
Activatory mutations complement growth of SGY1528 S. cerevisiae. With the exception of E118G, all of the activatory mutations identified in the E. coli random mutagenesis screen complement the growth of the K+-uptake deficient yeast strain, SGY1528. Some complement growth well on 1 mM K+, others only at 2 mM K+. The dilutions used in each drop test are shown above each column. As a control, all grow well on non-selective 100 mM K+ used for propagation of this strain. The vector is pYES-2m.
Activatory mutations cluster at helix-bundle crossing
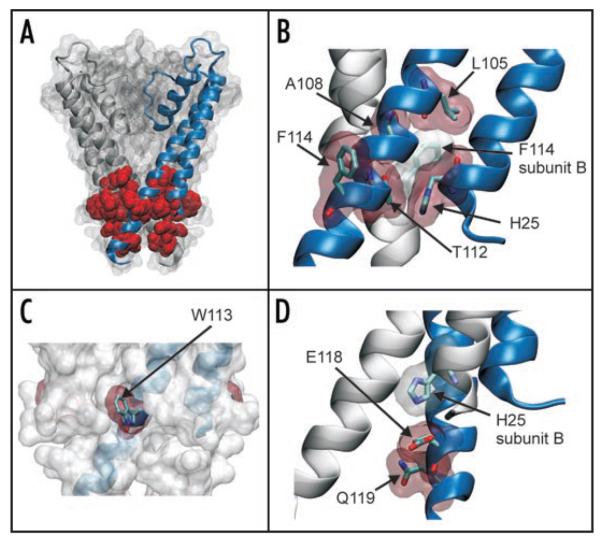
We used a high-resolution crystal structure of KcsA20 to examine the location of the 8 residues highlighted in this screen (Fig. 4A). It is of particular interest that they are only found in the transmembrane domains and all cluster at the helix-bundle crossing which is known to be an important structural component of the K+ channel gating mechanism.2
Figure 4.
Location of activatory mutations in the structure of KcsA. (A) The mutations identified in this study are mapped onto the structure of KcsA (PDB: 1K4C). For clarity the structure is shown as a surface map with only two subunits highlighted. The 8 residues (H25, L105, A108, T112, W113, F114, E118 and Q119) on all four subunits are shown in red as vdw spheres. All of these residues are located at the helix-bundle crossing. (B) Expanded view showing that residues H25, L105, A108, T112 and F114 all pack tightly at the helix-bundle crossing. Residues are shown as sticks with a vdw outline. In particular it can be seen how these residues pack tightly with F114 from an adjacent subunit. (C) The helix-bundle crossing is show as a vdw outline with one subunit highlighted. All four W113 residues are shown in red and can be seen to protrude from the surface of the channel. (D) Residues E118 and Q119 are below the helix-bundle-crossing. As shown previously,25 H25 H-bonds with E118 on an adjacent subunit forming part of the pH-gating mechanism. Q119 resides below and the side chain faces into the pore of the channel.
This closed-state structure reveals quite clearly that H25 in TM1 packs tightly in an intrasubunit interaction with several of the residues identified in TM2, in particular T112 and F114, as well as with E118 on TM2 of an adjacent subunit. Likewise, L105 and A108 in TM2 pack closely with F114 on an adjacent subunit (Fig. 4B). W113 is also close (<3.5 Å) to its neighboring residue F114, but points away from the TM helices into the lipid membrane (Fig. 4C); a role consistent with the frequent occurrence of aromatic belts at the membrane interface of ion channels.21 The final residue identified (Q119) is slightly lower down at the base of the helix-bundle crossing and all four side chains point in towards the centre of the channel (Fig. 4D).
However, despite the fact that these residues all cluster together at the helix-bundle crossing there are several potential mechanisms by which they may allow functional expression of KcsA in S. cerevisiae and/or E. coli. The initial presumption is that wild-type KcsA does not complement the growth defect in either the E. coli or yeast strains because their intracellular pH is not acidic enough to activate KcsA and the channel remains closed. These mutations may disrupt tight packing of the TM helices at the helix-bundle crossing and destabilize the closed state of the channel, even at neutral pH. A previous study has shown that the A108T mutation has a higher open-probability than wild-type KcsA.11 It is possible that similar effects on gating are observed for the F114I mutation, which also packs tightly at the bundle crossing and L105P, which may induce a kink in the TM2 helix. However, the effects of the Q119P mutation are more difficult to predict, because as well as inducing a potential kink in the helix, Q119 has also been proposed to form a hydration ‘basket’ at the intracellular mouth of the pore.22
The consequences of the W113R mutation are probably due to the dramatic charge substitution; tryptophan residues at the membrane interface are thought to form hydrophobic girdles stabilizing the channel in the membrane,21 and the fact that this residue faces the lipid in the available crystal structures would support this idea. It is surprising that this extreme substitution is even tolerated (the ∆∆G in solvation energy between Trp and Arg is >18 kcal/mol), but a positive charge at this position may result in novel interactions between the arginine and negatively charged lipid headgroups at the interface producing lateral forces on the helix-bundle crossing which destabilize the closed state of the channel.23
Activatory mutations target KcsA pH-sensing mechanism
During the course of this study two reports were published which also help explain the activatory effect of some of the mutations we have identified. A solution-state NMR study of KcsA gating demonstrated that mutation of H25 abolished a pH-dependent conformational rearrangement.24 Expanding on this, Nimigean and colleagues25 demonstrated that H25 forms part of a complex network of inter- and intra-subunit salt bridges and hydrogen bonds at the helix-bundle crossing with residues E118, E120 and R121, and that these pH-dependent interactions stabilize the closed state of the channel. A similar role for TM-TM interactions at the helix-bundle crossing in the control of pH-sensitivity has also been proposed for mammalian Kir channels.26-28 Thus it is likely that any shift in the pH-sensitivity of KcsA produced by mutation of these residues would allow a higher level of channel activity at more neutral pH and thus complementation of the growth defect.
However, our genetic screen only revealed mutations at positions H25 and E118, and did not uncover any of the other residues within the proposed pH-gating mechanism.25 The reason for this may be twofold. Firstly, the biological significance of a pH-gating mechanism is not clear, and is still controversial given that intracellular pH in Streptomyces is closely controlled.7 The extreme pH changes which induce channel opening may simply mimic the action of an unidentified cellular effector(s) which bind to the cytoplasmic domains of the channel to regulate channel activity, and the mutations we have identified may affect this ‘in vivo’ gating mechanism rather than the pH-sensing mechanism per se. Alternatively, it may be that only a modest shift in pH-sensitivity is required to allow enough channel activity and K+ influx to complement growth. Yet if small changes in pH-sensitivity were solely responsible then this would not explain why the E118G mutation fails to function in yeast, but does complement in E. coli, because the lower intracellular pH in yeast would be more likely to reveal subtle shifts in KcsA pH-sensitivity. Nevertheless, other similar studies demonstrate that there is clear correlation between the ability of channels to complement the K+-uptake deficiency and channel activity.9 However, the direct relation between complementation and channel open-probability is less clear, especially when changes in regulation by factors such as pH are taken into account.
Two separate gates
KcsA inactivates rapidly once it has opened, but this inactivation can be prevented by mutations within the pore (e.g., E71A).3,29 These mutations produce channels with a high open-probability at both negative and positive potentials. However, our random mutagenesis screen did not reveal any activatory mutations within the pore or selectivity filter, only mutations at the helix-bundle crossing.
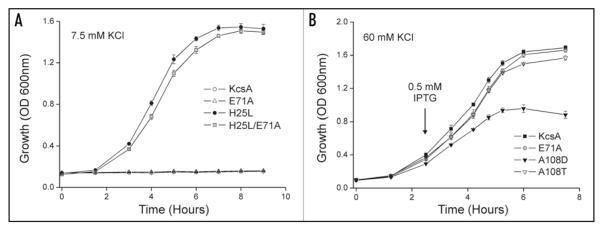
To examine whether this was due to incomplete mutagenesis, or whether these inactivation mutants do not complement the K+-uptake deficiency in our assay, we examined the ability of the E71A mutant to complement the growth of TK2420 E. coli. However, as shown in Figure 5A, the E71A mutant does not complement the growth defect. To exclude the possibility that this mutation is so overactive that it generates too much K+ entry, which is toxic to E. coli, we examined the double mutant (H25L/E71A), but this complements growth well (Fig. 5A).
Figure 5.
Inactivation mutations and sensitivity of the assay. (A) The inactivation mutation E71A fails to complement growth of TK2420 in 7.5 mM K+ K0 media indicating that removal of inactivation in the selectivity filter gate is not enough to open the channel in vivo. The double mutant H25L/E71A complements growth well demonstrating that the E71A mutation does not fail to complement due to ‘toxicity’. (B) Mutant and wild-type KcsA were grown in high K+ K0 media (60 mM) and expression was induced as indicated by the arrow. Wild-type KcsA, E71A and A108T exhibited robust growth over this 8 hour period. By contrast A108D cultures started to die off. Consequently such overactive mutations would not be found in this gain-of-function screen due to their intrinsic toxicity. Data shown are mean ± S.D., n = 3.
This has two implications. Firstly, even though the E71A mutation creates a non-inactivating channel with a high open-probability, these changes do not permit complementation, indicating that inactivation of wild-type KcsA per se is not what prevents its functional activity in vivo. Secondly, it demonstrates that this assay can distinguish between the two separate gating mechanisms in KcsA; the gating mechanism within the pore, which is affected by the E71A mutation, and the pH-dependent gating mechanism at the helix-bundle crossing that is affected by the activatory mutations. These results are therefore consistent with the idea that even though the gate at the selectivity filter may be open, no K+ permeation can occur unless the lower gate is also open.
Use of both E. coli and S. cerevisiae to identify activatory mutations in K+ channels
The functional expression of prokaryotic channels in eukaryotic systems can be problematic due to differences in lipid requirements and/or the different intracellular environments. The ability to express KcsA in S. cerevisiae may not be surprising given that a codon-optimized version of KcsA has been expressed in cultured mammalian cells,30 but it does demonstrate that both the E. coli and S. cerevisiae complementation assays can be used flexibly and interchangeably for prokaryotic ion channels, and this may have important future implications for the development of high-throughput assays based on this technology.31 It may also prove useful in future cases where different screening protocols may be used to exploit the differences in lipid environment and intracellular physiology between E. coli and yeast, in particular the acidic intracellular environment of S. cerevisiae and/or the absence of phosphoinositides in E. coli.18,19,32
Sensitivity of the assay
A final question remains however as to the limit of ‘sensitivity’ of this assay i.e., could it ever be used to uncover mutations, which generate constitutively open channels? The problem is that large numbers of constitutively open channels in the membrane would eventually be toxic to the growth of both yeast and E. coli and so they would not grow in this assay. It is likely that there is an optimal level or ‘window’ of channel activity required to rescue growth; below this level no growth is observed, but continuous levels of K+ entry above this level would be toxic, even if they initially permit growth. Indeed, we have observed such effects before in our assay of KirBac expression in TK2420 E. coli where reduced levels of expression at lower temperatures were required for optimal complementation.13
This may explain why we did not uncover the A108D mutation which was previously shown to markedly increase channel open-probability and channel conductance.33 The A108D mutation by itself did not complement growth in this assay (data not shown). This is probably due to the fact that expression appears toxic to the growth of E. coli; Figure 5B shows that when A108D expression is induced in high [K+] media the cultures begin to die off. This effect is not seen in either wild-type KcsA, or E71A which presumably remain closed, or even with A108T which has a lower open-probability than A108D.33
It is therefore possible that by reducing expression levels (e.g., by using very low copy vectors and/or mutations in promoter sequences) mutations with a dramatically increased open-probability and/or single-channel conductance could be identified which do not prove toxic to the cell as they would not be present in sufficient numbers to generate excessive K+ influx.
Conclusions
By genetic complementation of a K+-uptake deficiency in TK2420 E. coli we have identified a number of novel KcsA gating mutations clustered in the helix-bundle crossing, including several residues within the pH-sensing mechanism. This indicates that these mutants probably increase channel activity in vivo by reducing channel inhibition by intracellular factors such as pH and/or a general structural impairment of the ability of the channel to fully close at neutral pH. A more detailed electrophysiological analysis is necessary to determine their precise effect on pH-sensitivity, but this relatively crude assay can clearly differentiate between gating mutations at the selectivity filter and mutations at the helix-bundle crossing. Future development of this genetic complementation assay could also include exploitation of the physiological and biochemical differences between S. cerevisiae and E. coli to screen for different types of gating mutations in other K+ channels, as well as manipulation of expression levels in order to identify mutations with maximal changes in channel activity.
Materials and Methods
Growth media and strains
TK2420 E. coli were grown in either high [K+] K115 media, or low [K+] K0 media (supplemented with KCl as described and isopropyl β-D-1-thiogalactopyranoside (IPTG) to 0.5 mM to induce channel expression); 1.5% bacteriological agar was added for all solid media. SGY1528 cells were grown in APKO drop-out media,34 composition (/L): 0.5 ml phosphoric acid, 2.1 g L-arginine base, 1 ml 1 M MgSO4, 0.1 ml 1 M CaCl2; 50 mM Glucose, adjusted to pH 6.0 with phosphoric acid, with additional amino acid drop-out supplement as required (Q-Biogene) and K+-free trace minerals and vitamins.34 High [K+] APKO-ura contained csm-ura dropout media (Q-Biogene) for propagation of pYES2m plasmids and KCl adjusted to 100 mM. Due to the trace levels of K+ in most forms of agar, purified electrophoresis-grade agarose (1%) was added to obtain solid media. APKO-met-ura contained csm-met-ura drop-out supplement (Q-Biogene) to induce channel expression and KCl adjusted as specified.
Molecular biology and random mutagenesis
Wild-type KcsA was cloned into the E. coli expression vector pQE60-lac between the Nco1 and HindIII restriction sites.13 For expression in yeast the methionine regulated pYES2m vector was used34 and the gene cloned between EcoR1 and Xho1 restriction sites, with a modified Kozak sequence of 6 adenine nucleotides (AAAAAAATG).35 A randomly mutated library was constructed for KcsA using the GeneMorph-II random mutagenesis system (Stratagene), which uses non-biased, error-prone PCR. The PCR reactions were quantified according to the manufacturers protocols to produce an error rate of ~1–3 mutations per open reading frame. PCR products were cloned into the pQE60-lac vector between the Nco1 and HindIII restriction sites. In order to maximise the transformation efficiency, ligations were purified by phenol:chloroform extraction and ethanol precipitation prior to transformation into Library Efficiency DH5α E. coli (Invitrogen) and growth in culture overnight. Analysis of the transformation efficiency prior to overnight growth indicated that the library contained approximately 1.3 × 104 independent clones. 25 independent clones were isolated on non-selective media and upon sequencing were found to contain an average of 1–3 mutations per open reading frame. All site-directed mutagenesis was performed using the QuikChange II system (Stratagene).
Screening mutant libraries in E. coli
200 ng of mutant plasmid DNA was transformed into 100 μl chemically competent TK2420 cells (1 × 106 cfu/μg) and plated out onto 90 mm Petri dishes containing K0 solid media supplemented to 5 mM KCl and 0.5 mM IPTG. Plates were incubated at 37°C, individual colonies picked, propagated in K115 and plasmid DNA isolated. The quality of plasmid DNA obtained from the TK2420 strain is relatively low and was therefore retransformed back into DH5α and repurified prior to sequencing. To eliminate false positives, potential activatory mutants were retransformed into TK2420 and confirmed by drop tests on low [K+] before sequencing the entire gene.
E. coli drop tests and growth curves
Drop tests and growth curves were done as previously described.13 Briefly, overnight cultures were spun down, washed and resuspended in an equal volume of K0, then 4 μl of undiluted, 1:10 and 1:1000 dilutions spotted onto the plates and allowed to dry before being grown overnight at 37°C. For growth curves, washed overnight cultures were diluted 20-fold into K0 at the indicated KCl concentration and growth monitored at OD600 after induction with 0.5 mM IPTG.
Yeast transformation and drop-tests
Wild-type and mutant KcsA were transformed into competent SGY1528 cells using a standard lithium acetate yeast transformation protocol. Transformants were grown on APKO-ura plates for ~48 hours before overnight growth in 4 ml APKO-ura (100 mM KCl). Cultures were washed in APKO-met-ura media (0.5 mM KCl) and 3.5 μl drops of either undiluted, 1:10 or 1:1000 dilutions made onto APKO-met-ura plates (with [KCl] as specified). Drop test plates were incubated at 30°C for ~72 hours.
Acknowledgements
This work was supported by grants from the British Heart Foundation, the Royal Society, the Wellcome Trust and the Physiological Society. We thank Mr. Guy Baker for his experimental help with the initial stages of this study, Prof. Wolf Epstein for the TK2420 E. coli strain and Dr. Blanche Schwappach for the SGY1528 yeast strain.
References
- 1.Cortes DM, Cuello LG, Perozo E. Molecular architecture of full-length KcsA: role of cytoplasmic domains in ion permeation and activation gating. J Gen Physiol. 2001;117:165–80. doi: 10.1085/jgp.117.2.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perozo E, Cortes DM, Cuello LG. Structural rearrangements underlying K+-channel activation gating. Science. 1999;285:73–8. doi: 10.1126/science.285.5424.73. [DOI] [PubMed] [Google Scholar]
- 3.Cordero-Morales JF, Cuello LG, Zhao Y, Jogini V, Cortes DM, Roux B, Perozo E. Molecular determinants of gating at the potassium-channel selectivity filter. Nat Struct Mol Biol. 2006;13:311–8. doi: 10.1038/nsmb1069. [DOI] [PubMed] [Google Scholar]
- 4.Ader C, Schneider R, Hornig S, Velisetty P, Wilson EM, Lange A, Giller K, Ohmert I, Martin-Eauclaire MF, Trauner D, Becker S, Pongs O, Baldus M. A structural link between inactivation and block of a K+ channel. Nat Struct Mol Biol. 2008;15:605–12. doi: 10.1038/nsmb.1430. [DOI] [PubMed] [Google Scholar]
- 5.Zakharian E, Reusch RN. Streptomyces lividans potassium channel KcsA is regulated by the potassium electrochemical gradient. Biochem Biophys Res Commun. 2004;316:429–36. doi: 10.1016/j.bbrc.2004.02.069. [DOI] [PubMed] [Google Scholar]
- 6.Negoda A, Xian M, Reusch RN. Insight into the selectivity and gating functions of Streptomyces lividans KcsA. Proc Natl Acad Sci USA. 2007;104:4342–6. doi: 10.1073/pnas.0700495104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corvini PF, Gautier H, Rondags E, Vivier H, Goergen JL, Germain P. Intracellular pH determination of pristinamycin-producing Streptomyces pristinaespiralis by image analysis. Microbiology. 2000;146:2671–8. doi: 10.1099/00221287-146-10-2671. [DOI] [PubMed] [Google Scholar]
- 8.Hegermann J, Overbeck J, Schrempf H. In vivo monitoring of the potassium channel KcsA in Streptomyces lividans hyphae using immuno-electron microscopy and energy-filtering transmission electron microscopy. Microbiology. 2006;152:2831–41. doi: 10.1099/mic.0.29002-0. [DOI] [PubMed] [Google Scholar]
- 9.Parfenova LV, Rothberg BS. Genetic screening for functionality of bacterial potassium channel mutants using K+ uptake-deficient Escherichia coli. Methods Mol Biol. 2006;337:157–65. doi: 10.1385/1-59745-095-2:157. [DOI] [PubMed] [Google Scholar]
- 10.Parfenova LV, Crane BM, Rothberg BS. Modulation of MthK potassium channel activity at the intracellular entrance to the pore. J Biol Chem. 2006;281:21131–8. doi: 10.1074/jbc.M603109200. [DOI] [PubMed] [Google Scholar]
- 11.Irizarry SN, Kutluay E, Drews G, Hart SJ, Heginbotham L. Opening the KcsA K+ channel: tryptophan scanning and complementation analysis lead to mutants with altered gating. Biochemistry. 2002;41:13653–62. doi: 10.1021/bi026393r. [DOI] [PubMed] [Google Scholar]
- 12.Epstein W. The roles and regulation of potassium in bacteria. Prog Nucleic Acid Res Mol Biol. 2003;75:293–320. doi: 10.1016/s0079-6603(03)75008-9. [DOI] [PubMed] [Google Scholar]
- 13.Sun S, Gan JH, Paynter JJ, Tucker SJ. Cloning and functional characterization of a superfamily of microbial inwardly rectifying potassium channels. Physiol Genomics. 2006;26:1–7. doi: 10.1152/physiolgenomics.00026.2006. [DOI] [PubMed] [Google Scholar]
- 14.Yi BA, Lin YF, Jan YN, Jan LY. Yeast screen for constitutively active mutant G protein-activated potassium channels. Neuron. 2001;29:657–67. doi: 10.1016/s0896-6273(01)00241-0. [DOI] [PubMed] [Google Scholar]
- 15.Bichet D, Lin YF, Ibarra CA, Huang CS, Yi BA, Jan YN, Jan LY. Evolving potassium channels by means of yeast selection reveals structural elements important for selectivity. Proc Natl Acad Sci USA. 2004;101:4441–6. doi: 10.1073/pnas.0401195101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chatelain FC, Alagem N, Xu Q, Pancaroglu R, Reuveny E, Minor DL., Jr The pore helix dipole has a minor role in inward rectifier channel function. Neuron. 2005;47:833–43. doi: 10.1016/j.neuron.2005.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sesti F, Rajan S, Gonzalez-Colaso R, Nikolaeva N, Goldstein SA. Hyperpolarization moves S4 sensors inward to open MVP, a methanococcal voltage-gated potassium channel. Nat Neurosci. 2003;6:353–61. doi: 10.1038/nn1028. [DOI] [PubMed] [Google Scholar]
- 18.Shechter E, Letellier L, Simons ER. Fluorescence dye as monitor of internal pH in Escherichia coli cells. FEBS Lett. 1982;139:121–4. doi: 10.1016/0014-5793(82)80501-2. [DOI] [PubMed] [Google Scholar]
- 19.Imai T, Ohno T. Measurement of yeast intracellular pH by image processing and the change it undergoes during growth phase. J Biotechnol. 1995;38:165–72. doi: 10.1016/0168-1656(94)00130-5. [DOI] [PubMed] [Google Scholar]
- 20.Zhou Y, Morais-Cabral JH, Kaufman A, MacKinnon R. Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 A resolution. Nature. 2001;414:43–8. doi: 10.1038/35102009. [DOI] [PubMed] [Google Scholar]
- 21.Sansom MS, Bond PJ, Deol SS, Grottesi A, Haider S, Sands ZA. Molecular simulations and lipid-protein interactions: potassium channels and other membrane proteins. Biochem Soc Trans. 2005;33:916–20. doi: 10.1042/BST20050916. [DOI] [PubMed] [Google Scholar]
- 22.Kariev AM, Znamenskiy VS, Green ME. Quantum mechanical calculations of charge effects on gating the KcsA channel. Biochim Biophys Acta. 2007 doi: 10.1016/j.bbamem.2007.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tikhonov DB, Zhorov BS. In silico activation of KcsA K+ channel by lateral forces applied to the C-termini of inner helices. Biophys J. 2004;87:1526–36. doi: 10.1529/biophysj.103.037770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takeuchi K, Takahashi H, Kawano S, Shimada I. Identification and characterization of the slowly exchanging pH-dependent conformational rearrangement in KcsA. J Biol Chem. 2007;282:15179–86. doi: 10.1074/jbc.M608264200. [DOI] [PubMed] [Google Scholar]
- 25.Thompson AN, Posson DJ, Parsa PV, Nimigean CM. Molecular mechanism of pH sensing in KcsA potassium channels. Proc Natl Acad Sci USA. 2008;105:6900–5. doi: 10.1073/pnas.0800873105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rapedius M, Paynter JJ, Fowler PW, Shang L, Sansom MS, Tucker SJ, Baukrowitz T. Control of pH and PIP2 gating in heteromeric Kir4.1/Kir5.1 channels by H-bonding at the helix-bundle crossing. Channels. 2007;1:327–30. doi: 10.4161/chan.5176. [DOI] [PubMed] [Google Scholar]
- 27.Rapedius M, Haider S, Browne KF, Shang L, Sansom MS, Baukrowitz T, Tucker SJ. Structural and functional analysis of the putative pH sensor in the Kir1.1 (ROMK) potassium channel. EMBO Rep. 2006;7:611–6. doi: 10.1038/sj.embor.7400678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rapedius M, Fowler PW, Shang L, Sansom MS, Tucker SJ, Baukrowitz T. H bonding at the helix-bundle crossing controls gating in Kir potassium channels. Neuron. 2007;55:602–14. doi: 10.1016/j.neuron.2007.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cordero-Morales JF, Cuello LG, Perozo E. Voltage-dependent gating at the KcsA selectivity filter. Nat Struct Mol Biol. 2006;13:319–22. doi: 10.1038/nsmb1070. [DOI] [PubMed] [Google Scholar]
- 30.Gao L, Mi X, Paajanen V, Wang K, Fan Z. Activation-coupled inactivation in the bacterial potassium channel KcsA. Proc Natl Acad Sci USA. 2005;102:17630–5. doi: 10.1073/pnas.0505158102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zaks-Makhina E, Kim Y, Aizenman E, Levitan ES. Novel neuroprotective K+ channel inhibitor identified by high-throughput screening in yeast. Mol Pharmacol. 2004;65:214–9. doi: 10.1124/mol.65.1.214. [DOI] [PubMed] [Google Scholar]
- 32.Nikawa J, Kodaki T, Yamashita S. Expression of the Saccharomyces cerevisiae PIS gene and synthesis of phosphatidylinositol in Escherichia coli. J Bacteriol. 1988;170:4727–31. doi: 10.1128/jb.170.10.4727-4731.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nimigean CM, Chappie JS, Miller C. Electrostatic tuning of ion conductance in potassium channels. Biochemistry. 2003;42:9263–8. doi: 10.1021/bi0348720. [DOI] [PubMed] [Google Scholar]
- 34.Minor DL, Jr, Masseling SJ, Jan YN, Jan LY. Transmembrane structure of an inwardly rectifying potassium channel. Cell. 1999;96:879–91. doi: 10.1016/s0092-8674(00)80597-8. [DOI] [PubMed] [Google Scholar]
- 35.Hamilton R, Watanabe CK, de Boer HA. Compilation and comparison of the sequence context around the AUG startcodons in Saccharomyces cerevisiae mRNAs. Nucleic Acids Res. 1987;15:3581–93. doi: 10.1093/nar/15.8.3581. [DOI] [PMC free article] [PubMed] [Google Scholar]