Abstract
Previously, we established that natural killer (NK) cells from C57BL/6 (B6), but not BALB/c, mice lysed Chinese hamster ovary (CHO) cells, and we mapped the locus that determines this differential CHO-killing capacity to the NK gene complex on chromosome 6. The localization of Chok in the NK gene complex suggested that it may encode either an activating or an inhibitory receptor. Here, results from a lectin-facilitated lysis assay predicted that Chok is an activating B6 NK receptor. Therefore, we immunized BALB/c mice with NK cells from BALB.B6–Cmv1r congenic mice and generated a mAb, designated 4E4, that blocked B6-mediated CHO lysis. mAb 4E4 also redirected lysis of Daudi targets, indicating its reactivity with an activating NK cell receptor. Furthermore, only the 4E4+ B6 NK cell subset mediated CHO killing, and this lysis was abrogated by preincubation with mAb 4E4. Flow cytometric analysis indicated that mAb 4E4 specifically reacts with Ly-49D but not Ly-49A, B, C, E, G, H, or I transfectants. Finally, gene transfer of Ly-49DB6 into BALB/c NK cells conferred cytotoxic capacity against CHO cells, thus establishing that the Ly-49D receptor is sufficient to activate NK cells to lyse this target. Hence, Ly-49D is the Chok gene product and is a mouse NK cell receptor capable of directly triggering natural killing.
Keywords: natural killer cell, cytotoxicity, tumor
The mechanisms by which natural killer (NK) cells recognize and kill certain tumor targets or some virus-infected cells are understood incompletely (1). In mice, many of the receptors that regulate NK cell activity are encoded within the NK gene complex (NKC) on chromosome 6. First defined by the Ly49 and Nkrp1 multigene families, the NKC is now known to span >2 megabases and also includes Cd69, Cd94, and the Nkg2 genes (2–4). These genes encode C type lectin type II integral membrane proteins that either inhibit or activate NK cell effector functions.
Mouse NK cells express two classes of inhibitory receptors specific for MHC class I, the Ly-49 family of homodimeric molecules and the recently characterized heterodimeric CD94/NKG2 molecule. Whereas members of the Ly-49 family have been shown to bind to classical MHC class I molecules and globally inhibit NK cells, the CD94/NKG2A receptor binds to the nonclassical MHC class I molecule Qa-1b, paralleling the interaction between the human CD94/NKG2A and HLA-E (5–11). Although both types of inhibitory receptors have distinct extracellular regions, they share conserved sequences in their cytoplasmic domains that mediate inhibitory activity. The NKG2A molecule and most of the Ly-49 receptors contain immunoreceptor tyrosine-based inhibitory motifs in their cytoplasmic domains. These motifs recruit the cytoplasmic tyrosine phosphatase SHP-1, resulting in inhibition of NK cell lytic activity (12–14). Thus, the molecular basis for enhanced NK cell activity against certain tumor or virus-infected cells that have down-regulated their MHC class I molecules is the absence of activity of inhibitory NK cell receptors specific to MHC class I (1, 15, 16).
Although significant advances have been made in understanding inhibitory NK cell receptors, very little is known regarding the receptors involved in target recognition and activation of NK cells. The mouse NKC encodes several activation receptors [including NK1.1 (musNKR-P1C), Ly-49D, Ly-49H, and CD94/NKG2C] that lack immunoreceptor tyrosine-based inhibitory motifs in their cytoplasmic domains (17–19). Instead, these molecules have a charged residue in their transmembrane regions that can facilitate the association with the chains containing immunoreceptor tyrosine-based activation motifs, such as the DAP12 molecule (20–22). These candidate activation receptors have been identified primarily by using an experimental assay system known as (antibody-induced) redirected lysis. This assay employs Fc receptor (FcR)-expressing target cells, which are relatively insensitive to spontaneous NK cell-mediated lysis. On addition of a mAb specific for an activating NK cell surface antigen, the FcR on target cells binds the Fc portion of the mAb, thereby bridging and crosslinking the NK cell receptor, which induces lysis of the targets. However, the role of the NKC-encoded activating receptors in natural killing and in vivo NK cell function remains to be determined.
In addition to genes for known molecules, the NKC encodes several phenotypically defined loci that have not been characterized structurally. These include the Cmv1 and Rmp1 genes, which regulate replication of mouse cytomegalovirus and ectromelia virus, respectively (23, 24). Recently, we also characterized and genetically mapped Chok to the NKC (25). This locus regulates NK cell activity against Chinese hamster ovary (CHO) cells, including natural killing in vitro and tumor elimination in vivo. In particular, C57BL/6 (B6) NK cells kill CHO targets in vitro and eliminate labeled CHO cells in vivo, whereas BALB/c NK cells do not. The localization of Chok to the NKC suggested that the Chok gene product, like other NKC-encoded molecules, may either activate or inhibit NK cell function. Chok may therefore encode an activation receptor, expressed by B6 NK cells, which binds a ligand on CHO cells and triggers cytolysis. BALB/c NK cells may fail to transduce a signal through such a receptor because of either structural alterations or dysregulated receptor expression. Alternatively, BALB/c NK cells may express an inhibitory receptor that recognizes a CHO cell ligand that is either lacking or sufficiently different in the B6 background such that it does not bind CHO cells or inhibit the NK cells.
In this report, we describe an experimental strategy to discriminate between these possibilities, one of which predicts that Chok encodes a B6 NK cell activation receptor. We then used the congenic BALB.B6–Cmv1r, a mouse strain that contains the B6 Cmv1r allele and other NKC-encoded loci back-crossed onto the BALB/c genomic background, to generate a mAb that could block killing of CHO cells by B6 NK cells. The derivation of this Chok-specific mAb facilitated identification of the Chok antigen as Ly-49D. Gene transfer experiments confirmed that Ly-49D is sufficient to confer functional CHO cell target specificity to BALB/c NK cells. This study identifies a mouse NK cell receptor that directly activates natural killing and validates a genetic approach to understanding NK cell specificity.
MATERIALS AND METHODS
Mice.
B6 and BALB/c mice were obtained from the National Cancer Institute (Frederick, MD). The BALB.B6–Cmv1r, a mouse strain that contains B6 alleles for Cmv1 and other NKC-encoded loci on the BALB/c genetic background, was derived as described (26). All mouse strains were maintained in a pathogen-free facility at Washington University.
Cells and Cell Lines.
CHO cells, a gift from P. Stanley (Albert Einstein College of Medicine, Bronx, NY), were maintained in MEM-α (GIBCO) and supplemented with ribonucleosides, deoxyribonucleosides, and 10% (vol/vol) FCS (Harlan Breeders, Indianapolis, IN) without antibiotics. YAC-1 and Daudi cells were obtained from the American Type Culture Collection and maintained in RPMI medium 1640 (GIBCO) or DMEM (GIBCO), respectively, each supplemented with l-glutamine (300 μg/ml), penicillin (100 units/ml), streptomycin (100 μg/ml), 50 μM β-mercaptoethanol, and 10% (vol/vol) FCS. DMEM was supplemented additionally with sodium pyruvate (1 mM) and nonessential amino acids (0.1 mM). Human 293T cells, provided by J. Vaage (University of Oslo, Oslo), and B cell hybridomas were maintained in complete DMEM with 10% (vol/vol) FCS. IL-2-activated NK cells were generated as described (25) with minor modifications. On day 4, adherent cells were washed with prewarmed Hanks’ balanced salt solution (GIBCO) with 10% (vol/vol) FCS and expanded for 4–6 days in complete RPMI medium 1640 supplemented with 1,000 units/ml of rIL-2. Functional assays were performed with adherent cells harvested on days 7–10. Naive splenocytes were prepared by first lysing red blood cells as described (25). Viable cells were collected by using a Lympholyte-M density gradient (Cedarlane Laboratories) and then stained for fluorescence-activated cell sorter (FACS) analysis.
Generation of mAb 4E4.
BALB.B6–Cmv1r IL-2-activated NK cells (n = 1 × 106 to 8 × 106) mixed with the formulation (monophosphoryl lipid A and synthetic trehalose dicorynomycolate in squalene and Tween 80) of the Ribi Adjuvant System were used to immunize BALB/c mice as described by the manufacturer (Ribi Immunochem). B cell fusions with the murine myeloma P3X63Ag8.653 were performed at the Washington University School of Medicine Hybridoma Center. Hybridoma supernatants were screened for ability to block CHO killing by B6, (BALB/c × B6)F1, or BALB.B6–Cmv1r NK cells. Candidate hybridoma supernatants then were screened for inhibition of B6 NK-mediated CHO killing, redirected lysis of Daudi cells, and anti-B6 NK cell reactivity by FACS analysis. Two hybridoma supernatants (4E4 and 5A9) met all three criteria, and these were cloned by limiting dilution. Only 4E4 subclones survived two additional rounds of subcloning and rescreening. mAb 4E4 was isotyped as IgG2a by using the clonotyping system-HRPP kit (Southern Biotechnology Associates) and then was purified from culture supernatants by using protein A affinity chromatography (Amersham Pharmacia).
Other Antibodies and Reagents.
mAbs 2B4 (anti-2B4), phycoerythrin-conjugated DX5 (anti-pan NK antigen), and phycoerythrin-conjugated 5E6 (anti-Ly-49C/I) were purchased (PharMingen). All other antibodies that were used were purified by using protein A or protein G affinity chromatography, including AF6–88.5.3 (anti-H-2Kb), A1 (anti-LY-49A), 4D11(anti-Ly-49G2/A), and 4E5 (anti-Ly-49D; ref. 27). Biotinylated A1, 4D11, and 4E5 were used with a streptavidin–phycoerythrin secondary detection reagent (PharMingen). mAb 4E4 was conjugated directly with FITC-Celite (Calbiochem).
Cell Surface Expression Analysis and Sorting.
FACS staining was performed as described (25). Stained cells were analyzed on a FACScan with cellquest software (Becton Dickinson). Dead cells were excluded by propidium iodide staining. For separation of 4E4+ and 4E4− NK cells, day 5 adherent IL-2-activated NK cells were incubated with FITC-conjugated 4E4 mAb (10 μg/ml) and sorted with a FACS into 4E4+ and 4E4− populations. (These cells were cultured for 3–5 h after sorting and then were harvested and replated.) Sorted cells were expanded for an additional 7 days before use in functional assays.
Natural Killing.
Standard 51Cr release assays were performed as described (25). For lectin-facilitated lysis assays, varying concentrations of conconavalin A (ConA) were added before the addition of the labeled targets. For experiments examining antibody effects, mAbs (10 μg/ml, unless indicated otherwise) were added to effector cells and incubated for 15–30 min at room temperature before the addition of labeled target cells.
Transient Transfections.
Transient transfections of 293T cells were carried out with Lipofectamine reagent (GIBCO) according to the manufacturer’s protocol by using B6 Ly-49C cDNA in pAX142 vector (a gift from F. Takei, University of British Columbia, Vancouver, BC, Canada) and Ly-49D cDNA cloned in a pcDNAI/Neo vector (Invitrogen). Cells were FACS analyzed 48 h after transfection.
Vaccinia Virus Infection of IL-2-Activated BALB/c NK cells.
Generation of recombinant vaccinia was performed as described (28). Briefly, the B6 Ly-49D cDNA was inserted into the pSport1 vector (GIBCO). This construct was expressed in a recombinant vaccinia vector, driven by the LacI-inducible early gene promoter that was inserted into a NotI site as described (28). Wild-type Western Reserve (WR) vaccinia strain and recombinant Ly-49D–vaccinia virus were purified by using a semipurification protocol (29), and the titer in plaque-forming units (pfu) was determined on TK− cells. Vaccinia infections of mouse IL-2-activated NK cells were performed as described (30, 31). Cells were used in standard 51Cr release assays and/or analyzed by FACS. Data presented in this study reflect results obtained from 6-h infections with 50 pfu of virus per cell. Expression of Ly-49D on BALB/c NK cells was confirmed by using mAb 4E4.
RESULTS
Chok Does Not Inhibit BALB/c NK Cell Cytotoxicity.
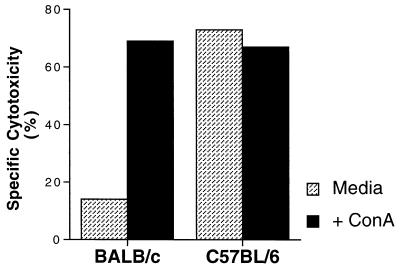
To determine whether the Chok product represented an inhibitory receptor on BALB/c NK cells or a stimulatory receptor on B6 NK cells, we examined BALB/c NK cells for the capacity to mediate lectin-facilitated lysis of CHO cells. Because the Ly-49A inhibitory signal has been shown to be dominant over all experimental assays of NK cell activation (5), we predicted that a putative BALB/c inhibitory receptor specific for CHO cells should similarly “protect” CHO targets even in the presence of lectin. Instead, IL-2-activated BALB/c NK cells displayed efficient CHO cell lysis in the presence of ConA (Fig. 1), suggesting that Chok does not transduce negative signals in BALB/c NK cells. These results support the hypothesis that the Chok gene product is an activating molecule that is present in B6 NK cells.
Figure 1.
Lectin-facilitated lysis of CHO targets by BALB/c NK cells. Cytotoxicity assays against CHO cells in the presence or absence of ConA (8 μg/ml) were performed by using BALB/c and B6 IL-2-activated NK cells at effector:target (E:T) ratios of 10:1 and 20:1, respectively.
Chok Activates B6 NK Cell Cytotoxicity.
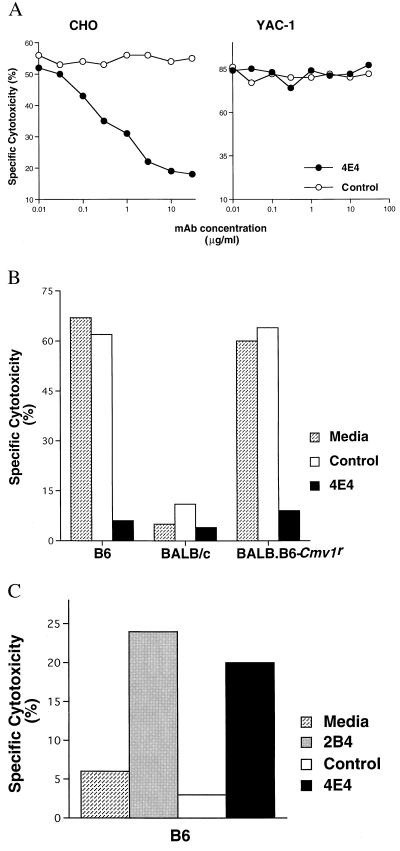
Based on this hypothesis, we immunized BALB/c mice with NK cells derived from the BALB.B6–Cmv1r congenic strain. This immunization scheme was designed to generate mAbs specific for B6 alleles of NKC-encoded molecules. To isolate an anti-ChokB6 allele-specific mAb, we implemented the following selection criteria: (i) blockade of B6-mediated CHO lysis; (ii) induction of redirected lysis of FcR+ (human) Daudi targets; and (iii) specificity for the ChokB6 allele expressed on NK cells by FACS analysis. One such candidate mAb, clone 4E4, was identified. mAb 4E4 specifically inhibits CHO lysis in a dose-dependent manner, with maximal inhibition achieved at a final concentration of 10 μg/ml (Fig. 2A). Abrogation of CHO lysis by mAb 4E4 is specific, judging from the fact that this mAb had no effect on YAC-1 lysis and that an isotype-matched control mAb had no effect on CHO lysis. Likewise, mAb 4E4 had no effect on BALB/c mediated CHO lysis, whereas 4E4 significantly reduced B6 and BALB.B6–Cmv1r NK cell-mediated CHO lysis (Fig. 2B). Taken together, these results indicate that mAb 4E4 specifically interferes with CHO killing by NK cells expressing B6 NKC molecules. Significantly, mAb 4E4 also increased B6 NK-mediated lysis of the FcR+ human Daudi targets (Fig. 2C) in a dose-dependent fashion (data not shown). This induction of redirected lysis of Daudi cells by mAb 4E4 is comparable to levels induced by other anti-activation receptor mAbs such as 2B4 (Fig. 2C), suggesting that mAb 4E4 reacts with a molecule capable of activating NK cell lysis.
Figure 2.
Establishment of anti-Chok mAb. (A) mAb 4E4 mediates dose-dependent inhibition of B6 NK-mediated CHO lysis. B6-derived IL-2-activated NK cells were assessed for their capacity to lyse CHO and YAC-1 targets in the presence of increasing concentrations of either mAb 4E4 or AF6–88.5.3, an isotype matched control (anti-H-2Kb), at an E:T ratio of 6.7:1. (B) mAb 4E4 blocks CHO killing by B6 and BALB.B6–Cmv1r NK cells. CHO cell lysis by IL-2-activated NK cells derived from B6, BALB/c, and BALB.B6–Cmv1r mouse strains was determined in medium alone or in the presence of 10 μg/ml of either mAb 4E4 or anti-H-2Kb control mAb (E:T of 6.7:1). (C) mAb 4E4 induces redirected lysis. mAbs 4E4, 2B4, and anti-H-2Kb control were evaluated for their capacity to influence B6 IL-2-activated NK cell-mediated lysis of FcR+ Daudi cells (E:T of 6.7:1).
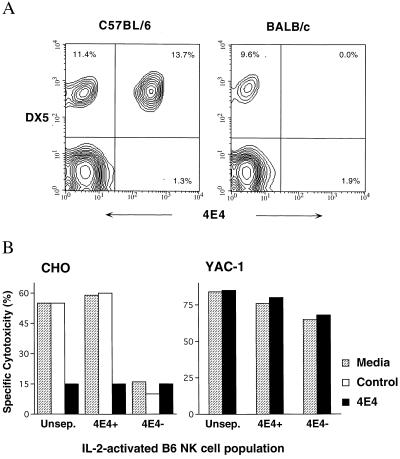
To characterize the cellular reactivity of mAb 4E4 further, flow cytometric analysis was performed on splenocytes and IL-2-activated NK cells derived from B6 and BALB/c mice. mAb 4E4 staining was evident on 55% of CD3− DX5+ B6 splenocytes, whereas there was no detectable reactivity among BALB/c splenocytes (Fig. 3A). Similar results were obtained on CD3− NK1.1+ B6 NK cells (data not shown). Furthermore, essentially no NK1.1− or DX5− cells were stained with mAb 4E4, suggesting restriction of the 4E4 epitope to the NK cell lineage. FACS analysis indicated that 37% of IL-2-activated B6 NK cells were 4E4+, whereas no 4E4+ BALB/c NK cells were detected (data not shown). These results are consistent with the CHO killing capacity of these strains. In addition, examination of NK cells from BALB.B6–Cmv1r mice, as well as from the seven BALB/cBy × B6By recombinant inbred mouse strains previously analyzed for CHO killing (25), indicated that the strain distribution pattern of 4E4 reactivity is concordant with the Chok phenotype (data not shown). This finding constitutes convincing genetic evidence that the Chok gene product is recognized by mAb 4E4.
Figure 3.
mAb 4E4 identifies a subset of B6 NK cells that mediate CHO cell lysis. (A) mAb 4E4 reacts with a population of B6 but not BALB/c NK cells. Two-color flow cytometric analysis of naïve B6 and BALB/c splenocytes was performed with 4E4 and DX5 mAbs. (B) mAb 4E4-reactive cells mediate CHO lysis. Unseparated as well as FACS-sorted 4E4+ and 4E4− IL-2-activated B6 NK cells were assessed for their capacity to lyse CHO and YAC-1 targets alone or in the presence of either mAb 4E4 or anti-H-2Kb control mAb (E:T of 5:1).
To confirm mAb 4E4 recognition of Chok, we sorted B6 IL-2-activated NK cells into 4E4+ and 4E4− populations and compared their capacity to mediate CHO and YAC-1 lysis. The 4E4+ population contained >80% 4E4+ NK cells, as determined by FACS analysis, whereas the 4E4− population contained no detectable 4E4+ NK cells (data not shown). Both NK cell subpopulations performed efficient YAC-1 cell lysis equivalent to unseparated B6 and BALB/c-derived NK cell cytotoxicity. However, only bulk B6 and 4E4+ NK cells efficiently lysed CHO cells (Fig. 3B), establishing that the 4E4+ cells alone are responsible for CHO lysis. Moreover, 4E4+ cell-mediated CHO lysis was blocked by mAb 4E4. Collectively, these results indicate that mAb 4E4 is specific for Chok, an activation receptor expressed by a subpopulation of B6 NK cells that mediates CHO killing.
Chok Is Ly-49D.
Biochemical analysis indicated that mAb 4E4 immunoprecipitates a ≈100 kDa species from B6 and BALB.B6–Cmv1r IL-2-activated NK cells (but not from BALB/c). This species reduces to ≈60 kDa, suggesting disulfide-linked subunits (data not shown). Expression as a disulfide-linked homodimer on a subpopulation of NK cells is characteristic of the Ly-49 family of receptors, suggesting that Chok may represent a member of this family (32).
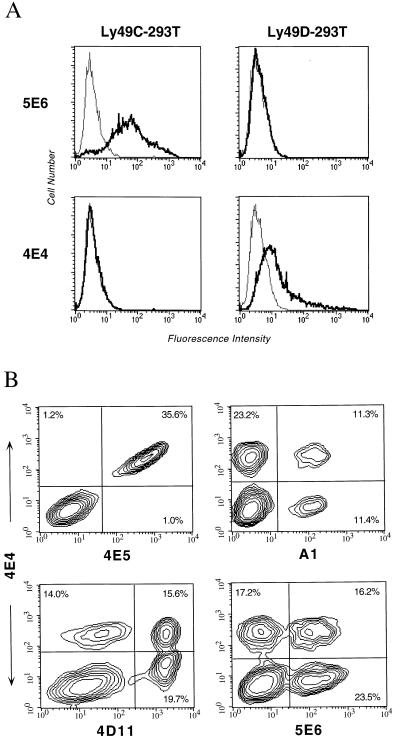
To test the reactivity of mAb 4E4 with Ly-49 molecules, cDNAs for Ly-49C and Ly-49D were transiently transfected into 293T cells, and expression levels were confirmed by using mAbs 5E6 and 4E5, respectively (Fig. 4A and data not shown). Significantly, mAb 4E4 reacted with only the Ly-49D-transfected cells. The low level of Ly-49D expression is consistent with its requirement for DAP12 for efficient expression on the cell surface in some transfected cell systems (22). To determine whether mAb 4E4 reacts with other Ly-49 molecules, we FACS analyzed cells stably transfected with a panel of Ly-49 cDNAs, including Ly-49A, Ly-49B, Ly-49E, Ly-49G, Ly-49A/H (a chimeric protein containing the cytoplasmic and transmembrane domain of Ly-49A and the extracellular domain of Ly-49H; H.R.C.S. and W.M.Y., unpublished results), or Ly-49I. (We have not been able to establish expression of our Ly-49F cDNA; H.R.C.S. and W.M.Y., unpublished results.) mAb 4E4 did not react with any of the transfectants, despite the expression of Ly-49 receptor, as determined by immunoprecipitation with an antipeptide antiserum crossreactive with all Ly-49 molecules or by FACS (data not shown). Thus, mAb 4E4 reacts with Ly-49D and does not recognize Ly-49A, Ly-49B, Ly-49E, Ly-49G, Ly-49H, or Ly-49I molecules.
Figure 4.
mAb 4E4 reacts with Ly-49D. (A) Flow cytometric analysis of 293T cells transfected with cDNA expression constructs for either Ly-49C or Ly-49D. (B) Two-color FACS analysis of B6 IL-2-activated NK cells by using 4E4 vs. 4E5 (anti-Ly-49D), A1 (anti-Ly-49A), 4D11 (anti-Ly-49G2/A), or 5E6 (anti-Ly-49C/I) mAbs.
To investigate the specificity of mAb 4E4 further, we performed two-color flow cytometric analysis with anti-Ly-49 mAbs (Fig. 4B). The expression patterns observed for 4E4 vs. A1, 4D11, and 5E6 indicated distinct populations in all four quadrants. Notably, two-color FACS analysis of B6 NK cells with 4E4 and 4E5, another monospecific anti-Ly-49D mAb, identified a significant lack of single-positive populations, which indicates that the epitopes for both of these mAbs are coexpressed and may be coincident. Thus, mAb 4E4 specifically recognizes Ly-49D.
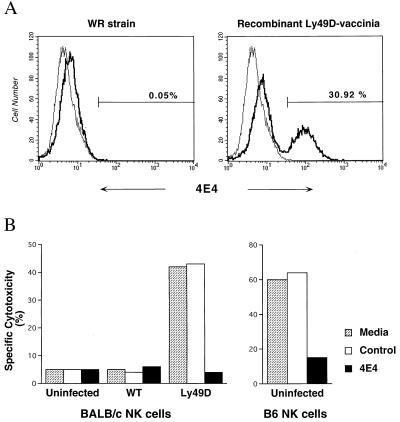
To establish the role of Ly-49D in recognition of CHO targets directly, we transferred expression of Ly-49D into IL-2-activated BALB/c NK cells by using the recombinant vaccinia virus expression system described for human killer inhibitory receptors (30, 31, 33). Infection conditions for a recombinant Ly-49D–vaccinia virus were optimized by using expression analysis with mAb 4E4 as well as killing assays against YAC-1 and CHO targets. In all conditions, infections with the wild-type WR vaccinia strain were performed in parallel as a specificity control. A 6-h infection time with a multiplicity of infection of 50 pfu per cell yielded Ly-49D cell surface expression on ≈31% of BALB/c IL-2-activated NK cells (Fig. 5A). These results also independently confirm the specificity of mAb 4E4 to the Ly-49D molecule. Moreover, cytotoxicity assays indicated that the Ly-49D-transfected BALB/c NK cells gained the capacity to kill CHO targets and that this killing was blocked by mAb 4E4 (Fig. 5B). Cells infected with the wild-type strain did not lyse CHO, and all populations tested displayed efficient YAC-1 lysis (data not shown). Infections at higher multiplicities of infection resulted in a larger percentage of Ly-49D-expressing cells as well as increased CHO lysis; however, the lytic capacity of these cells against YAC-1 cells was diminished. Thus, these results establish that the Ly-49D receptor is sufficient to confer target recognition and cytolytic activity against the CHO target.
Figure 5.
Ly-49D expression in BALB/c NK cells confers cytotoxic capacity against CHO targets. (A) IL-2-activated BALB/c NK cells that were uninfected or infected with 50 pfu per cell of either wild-type vaccinia WR strain or a recombinant Ly-49D–vaccinia virus were analyzed for Ly-49D expression by using mAb 4E4. (B) The NK cell populations shown in A, as well as uninfected B6 NK cells, were assessed for their capacity to lyse CHO targets alone or in the presence of either 4E4 or an isotype-matched control mAb, mouse anti-rat Ig. YAC-l lysis is shown for comparison and was unaltered. Data shown represent lysis at E:T of 10:1.
DISCUSSION
In the present study, we describe the elucidation of the Chok gene product by using a mAb generated by immunizing BALB/c mice with BALB.B6–Cmv1r congenic NK cells. The anti-Chok mAb specifically inhibits CHO lysis by B6 NK cells. This mAb was designed to block murine NK-mediated killing of a tumor target specifically. In addition to blocking NK lysis, the anti-Chok mAb also activates NK-mediated killing of Daudi cells in redirected lysis assays. Biochemical and expression analysis indicated that Chok is a disulfide-linked homodimer expressed on a subpopulation of B6 but not BALB/c NK cells, consistent with Chok belonging to the Ly-49 family of receptors. Transfection and two-color FACS analysis established that Chok is Ly-49D. Although we previously did not observe blockade of CHO killing with the 4E5 mAb (25), more recent analysis with newly purified mAb 4E5 suggests that there were technical artifacts in our previous studies. Retitration of the activity of the original batch of mAb 4E5 showed that it had much lower specific activity but not protein content than originally determined. The current capacity to block CHO killing with new 4E5 mAb preparations (data not shown) provides independent confirmation of the conclusion that Chok is Ly-49D. Functional transfer experiments indicated that the Ly-49D molecule is sufficient to confer cytolytic activity against CHO cells to BALB/c NK cells. Moreover, genetic mapping analysis by using intra-NKC recombinant congenic mice did not identify any recombinations between the Chok locus and the Ly49 gene cluster (34), suggesting that Chok may be encoded by a member of the Ly49 gene family. The identification of Ly-49D as the product of the Chok gene establishes a role for Ly-49D in natural killing of a tumor target. Thus, we have described a mouse NK cell activation receptor capable of directly mediating natural killing of a specific target.
Because there were at least two hypotheses to explain the differential CHO killing by B6 and BALB/c NK cells, it was imperative to determine whether Chok is a B6 activation receptor or a BALB/c inhibitory receptor to formulate a successful immunization strategy. Lectin-facilitated lysis of CHO cells by BALB/c NK cells lent support to the former hypothesis. Another assay that could have been employed is antibody-dependent cellular cytotoxicity, because ligation of inhibitory receptors is known also to abrogate FcR-mediated cytolysis by NK cells. However, this assay requires a target-specific mAb that was not readily available in this case. Thus, the lectin-facilitated assay system is a convenient means to discriminate between these possibilities.
In addition to in vitro cytotoxicity, our previous characterization of Chok indicated that this locus plays an important role in tumor elimination in vivo. BALB/c mice failed to clear CHO tumor target from their lungs, whereas B6 and BALB.B6–Cmv1r congenic mice efficiently cleared this target (25). These studies establish that the Chok gene product, Ly-49D, plays an important role in vivo and that allelic variation at this locus has functional consequences.
Although it is formally possible that BALB/c mice possess the same Ly-49D sequence as the B6 strain (but fail to express this receptor), evidence from studies of at least two other members of the Ly-49 family suggest that BALB/c mice possess an allelic form of the Ly-49D molecule. Ly-49AB6 differs from its putative Ly-49ABALB allelic form in five amino acid residues (four of which are in the extracellular domain; ref. 35). Similarly, only four amino acid differences distinguish the Ly-49C alleles in these strains (36, 37). Analogously, Ly-49D alleles may have limited polymorphism among these strains. Thus, the identification of the BALB/c Ly-49D allele will not only lend insight into NK cell allelic specificity but may also implicate the specific residues involved in Ly-49D–CHO cell interaction.
BALB/c and B6 mouse strains differ in their susceptibility to pathogens as well as predisposition to certain tumors. Because allelic differences in Ly-49D play a role in vivo (25), it is conceivable that they may explain the differential susceptibilities of these strains. For example, Ly49d may be a candidate for a previously described gene with a major effect on susceptibility to induced lung tumors (Pas 1; ref. 38). Using strains that are genetically resistant or susceptible to urethane-induced lung tumors, Gariboldi et al. (38) have mapped this locus to a 35-centimorgan region on chromosome 6 that includes D6Mit13, a marker closely linked to the NKC. It is possible that Ly49d and Pas1 are the same or closely related genes. Further evaluation of the NKC and its role in innate immunity to pathogens and developing tumors will be aided by the availability of NKC congenic mice such as BALB.B6–Cmv1r and related strains.
At the time of this analysis, no definitive ligand for Ly-49D had been identified. Although binding assays suggested that Ly-49D may be an H-2k–specific receptor, the relevance of this interaction in functional assays has not been determined (39). However, recent advances provide compelling evidence suggesting that the MHC class I molecule, H-2Dd, may be a ligand for Ly-49D. Bone marrow transplantation studies suggested that Ly-49D may interact with H-2d antigens, because rejection of H-2d donor bone marrow was abrogated by the prior injection of anti-Ly-49D mAbs (40). George et al. (41), using ConA blasts derived from MHC congenic mouse strains as target cells, established that Ly-49D interacts with target cells expressing H-2Dd, Dr, and Dsp2 but not H-2b or H-2k. Consistent with this study, Nakamura et al. (42) recently presented definitive evidence that Ly-49D can interact with mouse H-2Dd. Using Ly-49D-expressing RNK-16 effector cells, Nakamura et al. (42) showed that Ly-49D specifically stimulated lysis of target cells transfected with H-2Dd but not those expressing H-2b or H-2k MHC antigens. Inclusion of mAbs directed against either Ly-49D or H-2Dd blocked this enhanced cytotoxicity. These experiments indicate that Ly-49D can recognize mouse allo-MHC class I molecules.
Hence, the ligand for Ly-49D on CHO cells could be a hamster MHC molecule that resembles a mouse MHC molecule. Consistent with this hypothesis, we have observed reactivity of CHO cells with anti-mouse MHC class I mAbs; however, none of the tested anti-H-2d mAbs bound CHO cells (A.H.I. and W.M.Y., unpublished observation). Furthermore, ConA blasts derived from Chinese hamster splenocytes were lysed by B6 but not BALB/c IL-2-activated NK cells, suggesting that the ligand for Ly-49D on CHO cells is also expressed on ConA blasts, supporting the possibility that the CHO ligand is a hamster MHC molecule rather than a tumor-specific molecule (A.H.I. and W.M.Y., unpublished observation). Because NK cells do not typically kill normal cells, and the basis for NK cell reactivity with cellular targets is poorly understood, the identification of the putative ligand for Ly-49D on CHO cells will be important in understanding NK cell recognition.
Until recently, one theory of NK cell reactivity assumed that there would be a clonotypic or restricted NK cell receptor responsible for natural killing. Current evidence argues against this theory. Studies with the NKR-P1-deficient RNK-16 rat NK cell line transfected with NKR-P1A receptor established a role for rat NKR-P1A in the natural killing of the IC-21 mouse target (43). However, the addition of a mAb against the NKR-P1A receptor could not block lysis of this target by parental RNK-16, even though it abolishes killing by the reconstituted mutant. These results suggested that additional pathways by which this target is lysed as well as members of the mouse NKR-P1 family may also be target-specific receptors. Hence, NK cells may be equipped with multiple activation receptors, each mediating lysis of certain targets, rather than a pan natural killing receptor responsible for killing all targets. Some of these receptors are expressed by all NK cells, and others are expressed by subsets of NK cells; however, taken together these receptors provide for a heterogeneous population of NK cells with the capacity to lyse a variety of tumor targets. This hypothesis is supported by the capacity of BALB/c NK cells to kill many tumor targets, although these cells do not seem to express the B6 alleles of NK1.1 or Ly-49D (25). Our work with the CHO target has been fruitful, because the predominant mechanism for the lysis of this target is via Ly-49D for B6 NK cells. By exploiting targets that have such unique specificity, we can begin to reduce the complexities underlying natural killing.
Acknowledgments
We are grateful to Drs. P. Stanley and J. Vaage for providing reagents, Kim Marlotte for immunizing the mice, the staff of the Washington University Hybridoma facility for carrying out the fusion, Robin Winkler-Pickett and Dr. E. Long for technical advice, and Jonathan Heusel and Michael Brown for critical reading of this manuscript. This work was supported by grants from the National Institutes of Health and the Barnes–Jewish Hospital Research Foundation (to W.M.Y.), and by National Health and Medical Research Council of Australia Grant 961305 (to A.A.S.). W.M.Y. is an investigator of the Howard Hughes Medical Institute.
ABBREVIATIONS
- NK
natural killer
- NKC
NK gene complex
- B6
C57BL/6
- CHO
Chinese hamster ovary
- FcR
Fc receptor
- FACS
fluorescence-activated cell sorter
- pfu
plaque-forming unit
- ConA
conconavalin A
- E:T
effector:target
- WR
Western Reserve
References
- 1.Trinchieri G. Adv Immunol. 1989;47:187–376. doi: 10.1016/S0065-2776(08)60664-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown M G, Fulmek S, Matsumoto K, Cho R, Lyons P A, Levy E R, Scalzo A A, Yokoyama W M. Genomics. 1997;42:16–25. doi: 10.1006/geno.1997.4721. [DOI] [PubMed] [Google Scholar]
- 3.Vance R E, Tanamachi D M, Hanke T, Raulet D H. Eur J Immunol. 1997;27:3236–3241. doi: 10.1002/eji.1830271222. [DOI] [PubMed] [Google Scholar]
- 4.Ho E L, Heusel J W, Brown M G, Matsumoto K, Scalzo A A, Yokoyama W M. Proc Natl Acad Sci USA. 1998;95:6320–6325. doi: 10.1073/pnas.95.11.6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karlhofer F M, Ribaudo R K, Yokoyama W M. Trans Assoc Am Physicians. 1992;105:72–85. [PubMed] [Google Scholar]
- 6.Vance R E, Kraft J R, Altman J D, Jensen P E, Raulet D H. J Exp Med. 1998;188:1841–1848. doi: 10.1084/jem.188.10.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Salcedo M, Bousso P, Ljunggren H G, Kourilsky P, Abastado J P. Eur J Immunol. 1998;28:4356–4361. doi: 10.1002/(SICI)1521-4141(199812)28:12<4356::AID-IMMU4356>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 8.Braud V M, Allen D S J, O’Callaghan C A, Söderström K, D’Andrea A, Ogg G S, Lazetic S, Young N T, Bell J I, Phillips J H, et al. Nature (London) 1998;391:795–799. doi: 10.1038/35869. [DOI] [PubMed] [Google Scholar]
- 9.Lee N, Llano M, Carretero M, Ishitani A, Navarro F, López-Botet M, Geraghty D E. Proc Natl Acad Sci USA. 1998;95:5199–5204. doi: 10.1073/pnas.95.9.5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brooks A G, Borrego F, Posch P E, Patamawenu A, Scorzelli C J, Ulbrecht M, Weiss E H, Coligan J E. J Immunol. 1999;162:305–313. [PubMed] [Google Scholar]
- 11.Leibson P J. Immunity. 1998;9:289–294. doi: 10.1016/s1074-7613(00)80611-1. [DOI] [PubMed] [Google Scholar]
- 12.Carretero M, Palmieri G, Llano M, Tullio V, Santoni A, Geraghty D E, López-Botet M. Eur J Immunol. 1998;28:1280–1291. doi: 10.1002/(SICI)1521-4141(199804)28:04<1280::AID-IMMU1280>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 13.Nakamura M C, Niemi E C, Fisher M J, Shultz L D, Seaman W E, Ryan J C. J Exp Med. 1997;185:673–684. doi: 10.1084/jem.185.4.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burshtyn D N, Long E O. Trends Cell Biol. 1997;7:473–479. doi: 10.1016/S0962-8924(97)01167-7. [DOI] [PubMed] [Google Scholar]
- 15.Ljunggren H G, Karre K. Immunol Today. 1990;11:237–244. doi: 10.1016/0167-5699(90)90097-s. [DOI] [PubMed] [Google Scholar]
- 16.Wiertz E J H J, Jones T R, Sun L, Bogyo M, Beuze H J, Ploegh H L. Cell. 1996;84:769–779. doi: 10.1016/s0092-8674(00)81054-5. [DOI] [PubMed] [Google Scholar]
- 17.Kung S K, Miller R G. J Immunol. 1995;154:1624–1633. [PubMed] [Google Scholar]
- 18.Smith H R C, Karlhofer F M, Yokoyama W M. J Immunol. 1994;153:1068–1079. [PubMed] [Google Scholar]
- 19.Mason L H, Anderson S K, Yokoyama W M, Smith H R C, Winklerpickett R, Ortaldo J R. J Exp Med. 1996;184:2119–2128. doi: 10.1084/jem.184.6.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lanier L L, Cortiss B C, Wu J, Leong C, Phillips J H. Nature (London) 1998;391:703–707. doi: 10.1038/35642. [DOI] [PubMed] [Google Scholar]
- 21.Lanier L L, Corliss B, Wu J, Phillips J H. Immunity. 1998;8:693–701. doi: 10.1016/s1074-7613(00)80574-9. [DOI] [PubMed] [Google Scholar]
- 22.Smith K M, Wu J, Bakker A B, Phillips J H, Lanier L L. J Immunol. 1998;161:7–10. [PubMed] [Google Scholar]
- 23.Scalzo A A, Fitzgerald N A, Wallace C R, Gibbons A E, Smart Y C, Burton R C, Shellam G R. J Immunol. 1992;149:581–589. [PubMed] [Google Scholar]
- 24.Delano M L, Brownstein D G. J Virol. 1995;69:5875–5877. doi: 10.1128/jvi.69.9.5875-5877.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Idris A H, Iizuka K, Smith H R C, Scalzo A A, Yokoyama W M. J Exp Med. 1998;188:2243–2256. doi: 10.1084/jem.188.12.2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scalzo A A, Lyons P A, Fitzgerald N A, Forbes C A, Shellam G R. Immunogenetics. 1995;41:148–151. doi: 10.1007/BF00182328. [DOI] [PubMed] [Google Scholar]
- 27.Mason L H, Willette-Brown J, Anderson S K, Gosselin P, Shores E W, Love P E, Ortaldo J R, McVicar D W. J Immunol. 1998;160:4148–4152. [PubMed] [Google Scholar]
- 28.Earl P L, Moss B. In: Current Protocols in Molecular Biology. Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Vol. 2. New York: Wiley; 1988. pp. 16.17.1–16.17.16. [Google Scholar]
- 29.Ting A T, Dick C J, Schoon R A, Karnitz L M, Abraham R T, Leibson P J. J Biol Chem. 1995;270:16415–16421. doi: 10.1074/jbc.270.27.16415. [DOI] [PubMed] [Google Scholar]
- 30.Burshtyn D N, Lam A S, Weston M, Gupta N, Warmerdam P A, Long E O. J Immunol. 1999;162:897–902. [PubMed] [Google Scholar]
- 31.Wagtmann N, Rajagopalan S, Winter C C, Peruzzi M, Long E O. Immunity. 1995;3:801–809. doi: 10.1016/1074-7613(95)90069-1. [DOI] [PubMed] [Google Scholar]
- 32.Yokoyama W M, Daniels B F, Seaman W E, Hunziker R, Margulies D H, Smith H R. Semin Immunol. 1995;7:89–101. doi: 10.1006/smim.1995.0013. [DOI] [PubMed] [Google Scholar]
- 33.Buchholz D, Scott P, Shastri N. J Biol Chem. 1995;270:6515–6522. doi: 10.1074/jbc.270.12.6515. [DOI] [PubMed] [Google Scholar]
- 34.Idris, A. H., Scalzo, A. A. & Yokoyama, W. M. (1999) Immunogenetics, in press. [DOI] [PubMed]
- 35.Held W, Roland J, Raulet D H. Nature (London) 1995;376:355–358. doi: 10.1038/376355a0. [DOI] [PubMed] [Google Scholar]
- 36.Brennan J, Lemieux S, Freeman J D, Mager D L, Takei F. J Exp Med. 1996;184:2085–2090. doi: 10.1084/jem.184.6.2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takei F. Immunol Rev. 1997;155:67–77. doi: 10.1111/j.1600-065x.1997.tb00940.x. [DOI] [PubMed] [Google Scholar]
- 38.Gariboldi M, Manenti G, Canzian F, Falvella F S, Radice M T, Pierotti M A, Della Porta G, Binelli G, Dragani T A. Nat Genet. 1993;3:132–136. doi: 10.1038/ng0293-132. [DOI] [PubMed] [Google Scholar]
- 39.Brennan J, Mahon G, Mager D L, Jefferies W A, Takei F. J Exp Med. 1996;183:1553–1559. doi: 10.1084/jem.183.4.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raziuddin A, Longo D L, Mason L, Ortaldo J R, Bennett M, Murphy W J. J Immunol. 1998;160:87–94. [PubMed] [Google Scholar]
- 41.George T C, Mason L H, Ortaldo J R, Kumar V, Bennett M. J Immunol. 1999;162:2035–2043. [PubMed] [Google Scholar]
- 42.Nakamura M C, Linnemeyer P A, Niemi E C, Mason L H, Ortaldo J R, Ryan J C, Seaman W E. J Exp Med. 1999;189:493–500. doi: 10.1084/jem.189.3.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ryan J C, Niemi E C, Goldfien R D, Hiserodt J C, Seaman W E. J Immunol. 1991;147:3244–3250. [PubMed] [Google Scholar]