Abstract
The shoot apical meristem is maintained by the intercellular factor, CLV3, a dodecapeptide in Arabidopsis. CLV3 belongs to the CLE family and putative CLE genes have been found in various plants, even in the moss Physcomitrella patens. Here, we report that a pteridophyte, Selaginella moelendorffii, also has 15 putative CLE genes in its genome. On the other hand, CLV1 is reported to function as a receptor for the CLV3 peptide, and other CLE peptides might be recognized by CLV1 homologues in various plants. Recent genetic studies of the crn and sol2 mutants of Arabidopsis have revealed that SOL2/CRN encodes a receptor-like kinase protein. SOL2/CRN functions together with CLV2 independently of CLV1 in the CLE signaling pathway. Phylogenetic analysis of CLV1, CLV2 and SOL2/CRN revealed that Arabidopsis, rice, Populus trichocarpa and Vitis vinifera have one copy of the SOL2/CRN and CLV2 homologues, and Selaginella moelendorffii and Physcomitrella patens have no homologues. In contrast, a number of CLV1 homologues were identified in the genomic databases of Arabidopsis, rice, Populus trichocarpa, Vitis vinifera, and even a pteridophyte, Selaginella moelendorffii, and a moss, Physcomitrella patens. These results indicate that CLV1 and its homologues play multiple roles in plant development and environmental responses, whereas SOL2/CRN and CLV2 have more specific roles in vascular plants.
Key words: SOL2, CLE, CLV, CRN, meristem, receptor kinase
In multicellular organisms, cell-to-cell communication is a fundamental mechanism in coordinating cellular proliferation and differentiation. In vascular plants, the CLV signaling pathway regulates the stem cell fate in the SAM in a noncell-autonomous manner. CLV3 encodes a peptide ligand that functions as an intercellular signaling molecule to suppress the expression of the homeobox transcription factor WUS in the organization center. In this signaling pathway, CLV3 is believed to recognize the receptor complexes CLV1 and CLV2. The suppression of WUS expression leads to the restriction of the stem cell population in the SAM and balances cell proliferation and differentiation.
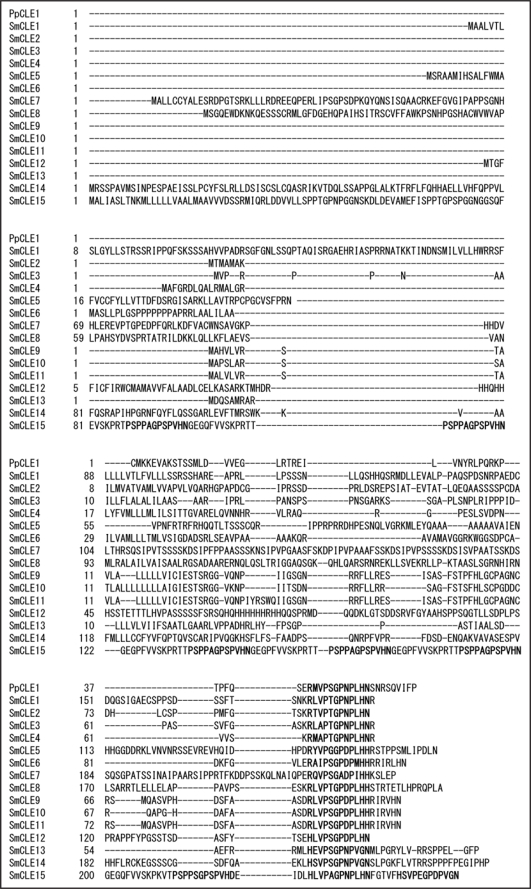
CLV3 is one of the 32 members of the CLV3/ESR-related (CLE) gene family in Arabidopsis, and CLV3 functions as a dodecapeptide.1,2 CLV3 and other CLE peptides are considered to function in plant morphogenesis as intercellular signaling molecules,3 ’9 and CLE genes have been found in various seed plants and even in the moss Physcomitrella patens.2,10,11 To identify putative CLEs in pteridophytes, we looked for genes encoding putative CLE domains in the Selaginella database (http://selaginella.genomics.purdue.edu/cgi-bin/blast_tmpl_s.cgi), using 12-amino-acid Arabidopsis CLE sequences as queries. The exon-intron structures were predicted with the FGENESH program (http://linux1.softberry.com/berry.phtml?topic=fgenesh&group=programs&subgroup=gfind). In total, 15 putative SmCLE genes were found in the Selaginella genome (Fig. 1). As has been noted previously,10 there is little sequence conservation among the CLE genes, except in the CLE domain. In Arabidopsis, the CLV3 and TDIF peptides have been identified in plant tissues, and their peptide sequences start from R and H, respectively. CLE peptides can be divided into two subclasses, the CLV3 class and the TDIF class. In rice, OsCLE502, OsCLE504 and OsCLE506 encode CLE proteins with multiple CLE domains, although no Arabidopsis genes encode such CLE proteins.2,12 In Selaginella, SmCLE15 encodes eight CLE domains. Six of these are class CLV3 and two of them are class TDIF. Thus, both Selaginella and rice have genes that encode multiple CLE domains, whereas Arabidopsis does not, and may have lost the genes encoding multiple CLE domains during its evolution.
Figure 1.
Alignment of the deduced polypeptides of the Selaginella CLE gene family and a Physcomitrella PpCLE1 gene. The conserved dodecapeptide CLE region is shown in boldface. The sequences of CLE genes are available in the DDBJ database (http://www.ddbj.nig.ac.jp/index-e.html). Accession numbers: SmCLE1, AB465350; SmCLE2, AB465351; SmCLE3, AB465352; SmCLE4, AB465353; SmCLE5, AB465354; SmCLE6, AB465355; SmCLE7, AB465356; SmCLE8, AB465357; SmCLE9, AB465358; SmCLE10, AB465359; SmCLE11, AB465360; SmCLE12, AB465361; SmCLE13, AB465362; SmCLE14, AB465363; SmCLE15, AB465364.
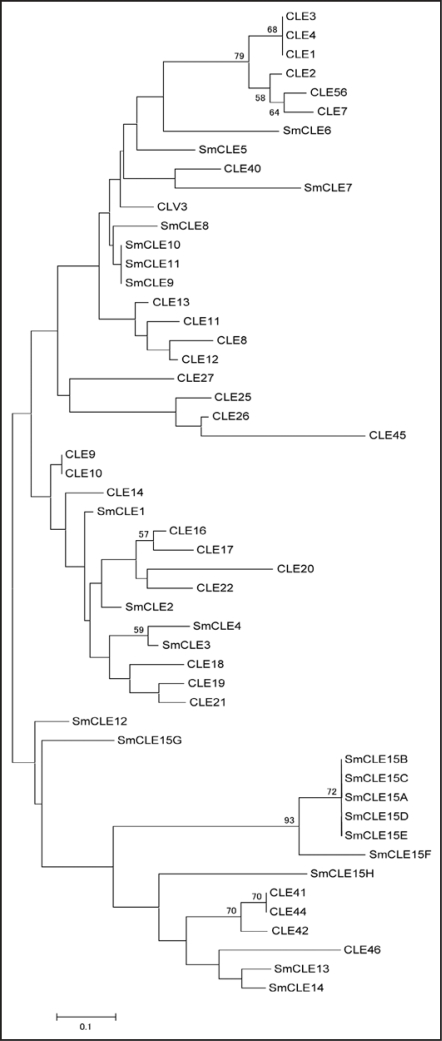
A phylogenetic tree was constructed with 12-amino-acid sequences corresponding to functional CLE dodecapeptide sequences (Fig. 2).1,5 It reveals that some of the Selaginella CLE peptides can be classified in a similar fashion to Arabidopsis CLE peptides (Fig. 2). However, no Selaginella CLEs were classified in the Arabidopsis CLE8, CLE11, CLE12, CLE13, CLE25, CLE26, CLE27 or CLE45 clades. Furthermore, SmCLE8, SmCLE9, SmCLE10, SmCLE11 and SmCLE15 were not included in any Arabidopsis CLE clade, so these proteins may be responsible for Selaginella specific physiological functions. We also identified the PpCLE1 gene from the moss Physcomitrella patens, so the CLE proteins seem to have functioned very early in plant evolution.
Figure 2.
An unrooted neighbor-joining tree of CLE dodecapeptides. Twelve-amino-acid sequences of the CLE peptide regions (Fig. 1) encoded by 15 Selaginella genes and 31 Arabidopsis CLE genes were used to construct the phylogenic tree. Bootstrap values of 30% and above, from the neighbor-joining method with Kimura's correction, are shown. The scale bar indicates the number of amino acid substitutions per site.
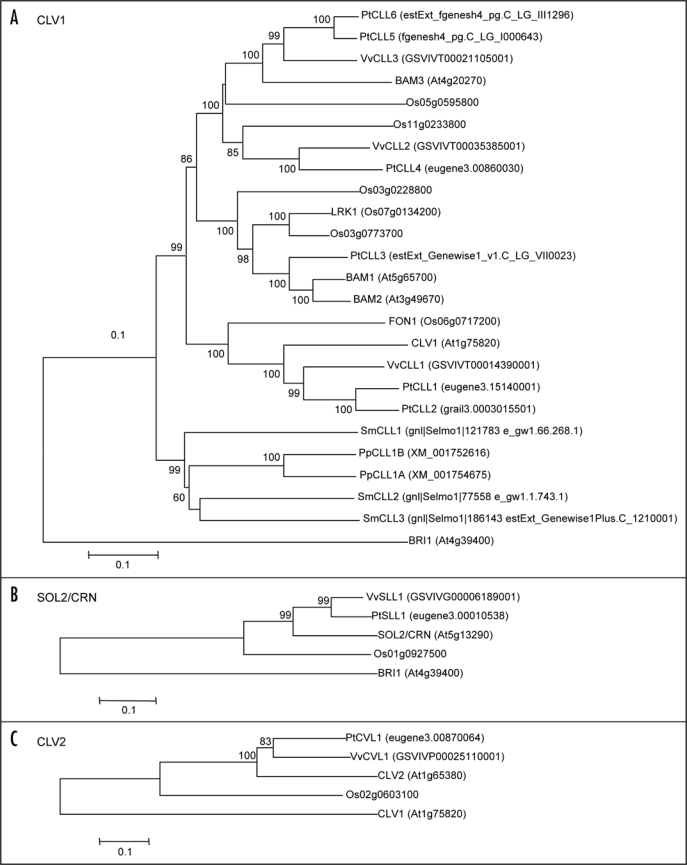
The CLE peptides are suggested to function through some receptors.13 CLV3 is reported to bind to the CLV1 protein in vitro.14 CLV1 encodes an LRR receptor-like kinase and CLV2 encodes an LRR receptor-like protein that lacks a kinase domain.15,16 Recently, genetic and physiological studies have demonstrated that SOL2/CRN, which encodes receptor-like kinase that lacks an extracellular domain,17,18 and CLV2 function together in the CLV3 pathway, independent of the CLV1 pathway, in SAM homeostasis.17,18 To understand the evolutionary context of these receptor-like proteins, we performed a phylogenetic analysis of CLV1, CLV2 and CRN/SOL2 from the Arabidopsis, rice, Populus trichocarpa, Vitis vinifera, Selaginella moelendorffii and Physcomitrella patens databases (Fig. 3).
Figure 3.
Comparison of SOL2/CRN, CLV2, CLV1, and their homologues. Phylogenetic relationships of SOL2/CRN, CLV2 and CLV1 to their counterparts from other plant species, Arabidopsis, rice, Populus trichocarpa, Vitis vinifera, Selaginella moelendorffii and Physcomitrella patens. The tree was calculated and drawn with the MEGA 3.1 program from an alignment of complete protein sequences, with gap deletion. Bootstrap values of 50% and above, from the neighbor-joining method with Kimura's correction, are shown. The amino-acid sequences of AtBRI1 (A and C) and AtCLV1 (B) were used as an outgroup. The scale bar indicates the number of amino acid substitutions per site. The cut-off of E-value for SOL2/CRN and CLV2 homologs is e−100. The cut-off of E-value for Selaginella CLV1 homologs is e−184.
The full amino-acid sequences of Arabidopsis CLV1, CLV2 and SOL2/CRN were used as queries for BLASTP search in the protein databases (http://blast.ncbi.nlm.nih.gov/Blast.cgi; http://gorgonzola.cshl.org/multi/blastview; http://xselaginella.genomics.purdue.edu/cgi-bin/blast_tmpl.cgi). Based on the protein structure and similarity, the receptor proteins were selected and used for the construction of the phylogenetic tree. CLV1 homologues were identified not only in vascular plants but also in the bryophyte Physcomitrella. CLV1 orthologues have also been identified in maize (THICK TASSEL DWARF 1 [TD1]) and in rice (FLORAL ORGAN NUMBER 1 [FON1]). The td1 mutant exhibits massive overproliferation of female inflorescence meristems, leading to the fasciated ear. The td1 mutant shows an abnormality in floral meristems and an increased number of floral organs.19 Similarly, the fon1 mutant displays enlarged floral meristems, resulting in an increased number of floral organs.20,21 These results suggest that some CLE signaling pathways may be conserved in bryophytes, pteridophytes and seed plants. Because a number of CLV1 homologues have been identified, CLV1 homologues may play central roles in the regulation of plant development and/or physiology.
In contrast, CRN/SOL2 and CLV2 have only a single homologue in the genomes of Arabidopsis, rice, Populus trichocarpa and Vitis vinifera, indicating that these genes might play more specific roles in higher plant development. Interestingly, we found no CRN/SOL2 or CLV2 homologues in the genomes of Selaginella moelendorffii or Physcomitrella patens. Because Physcomitrella and Selaginella have a single apical cell in the shoot apical region, SOL2/CRN and CLV2 might have specific functions in the multicellular organization of stem cells. It is postulated that CLV2 and SOL2/CRN are required for the evolution of the multicellular SAM from the single apical cell. The relative positions of the CLV2 and CRN/SOL2 homologues of Arabidopsis, rice, Populus trichocarpa and Vitis vinifera are well conserved on the phylogenetic tree (Fig. 3). This result also suggests the conservation of critical CLV2 and CRN/SOL2 functions in higher plant development.
Our phylogenetic analysis indicates that SOL2/CRN and CLV2 have evolved together recently, probably functioning in a meristem maintenance system. However, the functional diversity of CLV1 homologues remains to be resolved. The existence of a number of CLV1 homologues among seed plants implies a functional diversity among them. Except for the functions of CLV3 and TDIF, the functions of most CLE peptides are largely unknown. The direct interaction between CLE peptides and their cognate receptors will be examined with a biochemical approach because it is possible that these receptors bind other ligands. Further genetic and biochemical studies of these receptor-like proteins in various plant species will provide further evidence for CLE peptide and receptor functions in various aspects of plant development.
Acknowledgement
Grant-in Aid for Creative Scientific Research; Grant-in-Aid for Young Scientists S (19677001) from Japan Society of the Promotion of Science; Grant-in-Aid for Scientific Research for Priority Areas from the Ministry of Education, Culture, Sports, Science (19060009 to H.F., 20061004, and 19060016); Technology, and a Program of Basic Research Activities for Innovative Biosciences from Bio-oriented Technology Research Advancement Institution.
Abbreviations
- CLE
clavata3/ESR-related
- SAM
shoot apical meristem
- RAM
root apical meristem
- WUS
wuschel
- CLV
clavata
- LRR
leucin rich repeat
- CRN
coryne
- SOL2
suppressor of LLP1 2
- Vv
Vitis vinifera
- Pt
Populus trichocarpa
- Os
Oryza sativa
- Sm
Selaginella moelendorffii
- Pp
Physcomytrella patens
- CLL
CLV1-like protein
- CVL
CLV2-like protein
- SLL
SOL2-like protein
Footnotes
Previously published online as a Plant Signaling & Behavior E-publication: http://www.landesbioscience.com/journals/psb/article/8391
References
- 1.Kondo T, Sawa S, Kinoshita A, Mizuno S, Kakimoto T, Fukuda H, Sakagami Y. A plant peptide encoded by CLV3 identified by in situ MALDI-TOF MS analysis. Science. 2006;313:845–848. doi: 10.1126/science.1128439. [DOI] [PubMed] [Google Scholar]
- 2.Kinoshita A, Nakamura Y, Sasaki E, Kyozuka J, Fukuda H, Sawa S. Gain-of-function phenotypes of chemically synthetic CLAVATA3/ESR-related (CLE) peptides in Arabidopsis thaliana and Oryza sativa. Plant Cell Physiol. 2007;48:1821–1825. doi: 10.1093/pcp/pcm154. [DOI] [PubMed] [Google Scholar]
- 3.Chu H, Qian Q, Liang W, Yin C, Tan H, Yao X, et al. The floral organ number4 gene encoding a putative ortholog of Arabidopsis CLAVATA3 regulates apical meristem size in rice. Plant Physiol. 2006;142:1039–1052. doi: 10.1104/pp.106.086736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fiers M, Golemiec E, van der Schors R, van der Geest L, Li KW, Stiekema WJ, et al. The CLAVATA3/ESR motif of CLAVATA3 is functionally independent from the non-conserved flanking sequences. Plant Physiol. 2006;141:1284–1292. doi: 10.1104/pp.106.080671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ito Y, Nakanomyo I, Motose H, Iwamoto K, Sawa S, Dohmae N, et al. Dodeca-CLE peptides as suppressors of plant stem cell differentiation. Science. 2006;313:842–845. doi: 10.1126/science.1128436. [DOI] [PubMed] [Google Scholar]
- 6.Ni J, Clark SE. Evidence for functional conservation, sufficiency and proteolytic processing of the CLAVATA3 CLE domain. Plant Physiol. 2006;140:726–733. doi: 10.1104/pp.105.072678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sawa S, Kinoshita A, Nakanomyo I, Fukuda H. CLV3/ESR-related (CLE) peptides as intercellular signaling molecules in plants. Chem Rec. 2006;6:303–310. doi: 10.1002/tcr.20091. [DOI] [PubMed] [Google Scholar]
- 8.Strabala TJ, O'donnell PJ, Smit AM, Ampomah-Dwamena C, Martin EJ, Netzler N, et al. Gain-of-function phenotypes of many CLAVATA3/ESR genes, including four new family members, correlate with tandem variations in the conserved CLAVATA3/ESR domain. Plant Physiol. 2006;140:1331–1344. doi: 10.1104/pp.105.075515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suzaki T, Toriba T, Fujimoto M, Tsutsumi N, Kitano H, Hirano HY. Conservation and diversification of meristem maintenance mechanism in Oryza sativa: Function of the FLORAL ORGAN NUMBER2 gene. Plant Cell Physiol. 2006;47:1591–1602. doi: 10.1093/pcp/pcl025. [DOI] [PubMed] [Google Scholar]
- 10.Sharma VK, Ramirez J, Fletcher JC. The Arabidopsis CLV3-like (CLE) genes are expressed in diverse tissues and encode secreted proteins. Plant Mol Biol. 2003;51:415–425. doi: 10.1023/a:1022038932376. [DOI] [PubMed] [Google Scholar]
- 11.Oelkers K, Goffard N, Weiller GF, Gresshoff PM, Mathesius U, Frickey T. Bioinformatic analysis of the CLE signaling peptide family. BMC Plant Biol. 2008;8:1. doi: 10.1186/1471-2229-8-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sawa S, Kinoshita A, Betsuyaku S, Fukuda H. A large family of genes that share homology with CLE domain in Arabidopsis and rice. Plant Signaling Behavior. 2008;3:337–339. doi: 10.4161/psb.3.5.5344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miwa H, Kinoshita A, Fukuda H, Sawa S. Plant meristems: CLAVATA3/ESR-related signaling in the shoot apical meristem and the root apical meristem. J Plant Res. 2009;122:31–39. doi: 10.1007/s10265-008-0207-3. [DOI] [PubMed] [Google Scholar]
- 14.Ogawa M, Shinohara H, Sakagami Y, Matsubayashi Y. Arabidopsis CLV3 peptide directly binds CLV1 ectodomain. Science. 2008;319:294. doi: 10.1126/science.1150083. [DOI] [PubMed] [Google Scholar]
- 15.Clark SE, Williams RW, Meyerowitz EM. CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell. 1997;89:575–585. doi: 10.1016/s0092-8674(00)80239-1. [DOI] [PubMed] [Google Scholar]
- 16.Jeong S, Trotochaud AE, Clark SE. The Arabidopsis CLAVATA2 gene encodes a receptor-like protein required for the stability of the CLAVATA1 receptor-like kinase. Plant Cell. 1999;11:1925–1934. doi: 10.1105/tpc.11.10.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miwa H, Betsuyaku S, Iwamoto K, Kinoshita A, Fukuda H, Sawa S. The receptor-like kinase SOL2 mediates CLE signaling in Arabidopsis. Plant Cell Physiol. 2008;49:1752–1757. doi: 10.1093/pcp/pcn148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Müller R, Bleckmann A, Simon R. The receptor kinase CORYNE of Arabidopsis transmits the stem cell-limiting signal CLAVATA3 independently of CLAVATA1. Plant Cell. 2008;20:934–946. doi: 10.1105/tpc.107.057547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bommert P, Lunde C, Nardmann J, Vollbrecht E, Running M, Jackson D, et al. thick tassel dwarf1 encodes a putative maize ortholog of the Arabidopsis CLAVATA1 leucine-rich repeat receptor-like kinase. Development. 2005;132:1235–1245. doi: 10.1242/dev.01671. [DOI] [PubMed] [Google Scholar]
- 20.Nagasawa N, Miyoshi M, Kitano H, Satoh H, Nagato Y. Mutations associated with floral organ number in rice. Planta. 1996;198:627–633. doi: 10.1007/BF00262651. [DOI] [PubMed] [Google Scholar]
- 21.Suzaki T, Sato M, Ashikari M, Miyoshi M, Nagato Y, Hirano HY. The gene FLORAL ORGAN NUMBER1 regulates floral meristem size in rice and encodes a leucine-rich repeat receptor kinase orthologous to Arabidopsis CLAVATA1. Development. 2004;131:5649–5657. doi: 10.1242/dev.01441. [DOI] [PubMed] [Google Scholar]