Abstract
Callose is a polysaccharide in the form of β-1,3-glucan with some β-1,6-branches and it exists in the cell walls of a wide variety of higher plants. Callose plays important roles during a variety of processes in plant development and/or in response to multiple biotic and abiotic stresses. It is now generally believed that callose is produced by callose synthases and that it is degraded by β-1,3-glucanases. Despite the importance of callose in plants, we have only recently begun to elucidate the molecular mechanism of its synthesis. Molecular and genetic studies in Arabidopsis have identified a set of genes that are involved in the biosynthesis and degradation of callose. In this mini-review, we highlight recent progress in understanding callose biosynthesis and degradation and discuss the future challenges of unraveling the mechanism(s) by which callose synthase operate.
Key words: Arabidopsis thaliana, callose, callose synthase, glucan synthase-like, pollen, plasmodesmata, cell plate, stress
Introduction
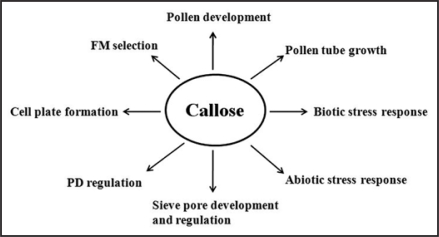
Callose is widespread in higher plants, in which it is a component of specialized cell walls or cell wall-associated structures at particular stages of growth and differentiation.1 Callose is involved at multiple stages of pollen development as a structural component.1,2 It is also deposited at cell plates during cytokinesis.3,4 In addition, callose can be deposited at plasmodesmata (PD) to regulate the cell-to-cell movement of molecules by controlling the size exclusion limit (SEL) of PD.5,6 Callose deposition can also be induced by wounding, infection of pathogens, aluminum, abscisic acid, and other physiological stresses1 (Fig. 1).
Figure 1.
Callose is involved in multiple aspects of plant growth and development and response to biotic and abiotic stress. FM, functional megaspore; PD, plasmodesmata.
Given the importance of callose, it may seem surprising that our understanding of its mechanism of synthesis is incomplete. Callose synthesis and deposition have been investigation for several decades, but many technical problems have hindered research studies. Over the last several years, however, significant progress has been made, in particular using the model plant Arabidopsis thaliana. Owing to progress in Arabidopsis and due to the lack of other recent review articles addressing callose in plants, this review focuses on recent advances in understanding the molecular mechanism by which callose is synthesized and deposited to regulate plant growth, development and stress responses.
Callose Structure and Callose Synthase
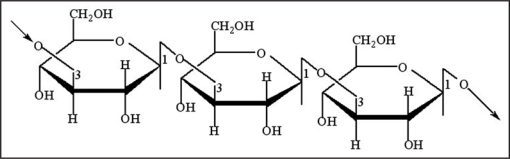
Callose is a linear homopolymer made up of β-1,3-linked glucose residue with some β-1,6-branches (Fig. 2) and callose biosynthesis uses UDP-glucose as a substrate. Biochemical evidence and molecular studies7–9 in several plant species indicate that callose is synthesized by a class of enzymes, termed callose synthases. In the model plant Arabidopsis thaliana, twelve genes encoding putative callose synthase have been identified by two independent research groups.7,10 Accordingly, two different nomenclatures have been adopted for the Arabidopsis genes. The group of Desh Verma uses the CalS (Callose synthase) system to name the twelve genes: AtCalS1-AtCalS12 based on their relative similarity to AtCalS1.4 The Somerville group refers to the twelve Arabidospsis genes as GSL (Glucan synthase-like) genes, and has designated them as AtGSL1 to AtGSL12.10 Due to its wide usage by the callose synthase research community, we have adopted the GSL nomenclature system.11–17
Figure 2.
A fragment of β-1,3-glucan showing how adjacent sugar residues are inverted.
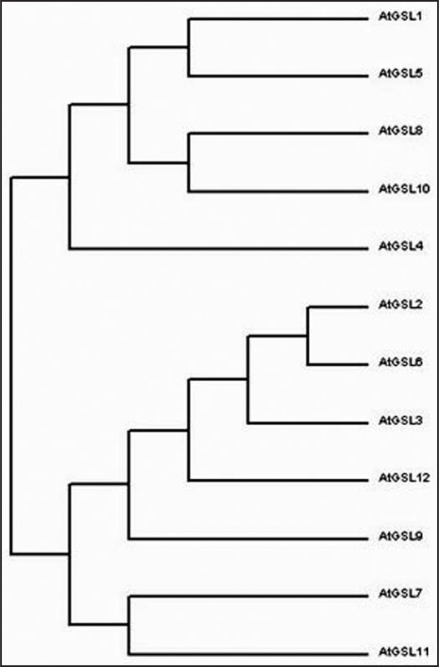
Phylogenetic analsysis of the AtGSL family suggests that the GSL family can be classified into four main subfamilies (Fig. 3). The first subfamily contains AtGSL1, AtGSL5, AtGSL8 and AtGSL10, the second subfamily contains AtGSL2, AtGSL3, AtGSL6 and AtGSL12, the third subfamily contains AtGSL7 and AtGSL11, and the last subfamily includes AtGSL4. According to previsously characterized functions of GSL genes, members belonging to different subfamilies exhibit partially redundant roles during pollen development or fertilization. A single GSL gene can also have diverse functions; for example, GSL5 is responsible for the synthesis of wound-and pathogen-inducible callose in leaf tissue; it also plays an important role in exine formation and pollen wall patterning.11–13
Figure 3.
Phylogenetic analysis of glucan synthase-like (GSL) genes in Arabidopsis thaliana. The composite tree was done using ClustalW2 by aligning deduced GSL amino acid sequences.
Based on gene structure modeling, most GSL genes have 40–50 exons; exceptions include GSL1 and GSL5, which have two and three exons, respectively. Most of the AtGSL genes encode proteins of around 2,000 amino acids, which is larger than most plant genes.7,13 All Arabidopsis GSL proteins contain multiple transmembrane domains that are clustered into two regions (N-terminal and C-terminal), leaving a large hydrophilic central loop that faces the cytoplasm. This loop contains the putative catalytic domain which has been further subdivided into two domains: the UDP-glucose binding domain and the glycosyltransferase domain. These domains are characterized by the presence of multiple aspartic acid triplets (D,D,D) and a QXXRW motif that is conserved in the CeSA superfamily.7,17,18
Over the last decade, most of our knowledge about callose in plants has been derived from analyses of a number of mutations that affect callose synthesis that were made by knocking out the individual callose synthase genes of Arabidopsis.11–18
Callose, a Multiple Player, in Plant Growth and Development: The Role of Callose during Pollen Development
In Arabidopsis, several studies demonstrated that multiple GSLs are involved in pollen development. Enns et al.13 reported that GSL1 and GSL5 are necessary and that they function redundantly in pollen development and fertility. GSL1 and GSL5 are responsible for the formation of the callose wall that separates the microspores of the tetrad; the genes also play a gametophytic role later in pollen grain germination.
Two other studies provided contradictory data about the function of the GSL2 (CalS5) gene in exine formation during microgametogenesis and pollen viability. Dong et al.18 identified two T-DNA knockout mutants of cals5 (cals5-1 and cals5-2) and found that the cals5 mutant displayed male sterility and lacked the normal callose wall which affected the exine pattern of the microspore. Tryphine, an extracellular pollen coat, was synthesized in the mutant, but was randomly deposited as aggregates on the surface of microspores. Upon release from the tetrad, mutant microspores did not survive and the pollen wall collapsed, demonstrating that callose is required both in the synthesis of pollen wall exine and in the viability of pollen. Contrary to the report described above, Nishikawa et al.14 isolated three additional T-DNA alleles of cals5 (cals5-3, cals5-4 and cals5-5) that similarly altered the exine deposition pattern, but which actually produced fertile pollen. Furthermore, mutation of cals5-3 resulted in formation of pollen tubes that lacked callose walls and plugs, however, these tubes could still perform fertilization, demonstrating that a structured exine is not essential for pollen development, viability or fertility. Additionally, this study demonstrated that callose is not required for pollen functions. Thus, together, these two studies highlight the importance of generation and analysis of an allelic series of a gene under study.
More recently, Töller et al.15 reported both gsl8 and gsl10 mutants to be male gametophytic lethal, and the authors failed to recover homozygous mutants for either genes. Further analysis of gsl8 and gsl10 mutants during development revealed specific malfunctions associated with asymmetric microspore division as well as failed entry of mutant microspores into mitosis. These authors proposed that GSL8 and GSL10 might exert indirect regulatory functions through interactions with other proteins, rather than through their catalytic activity alone.15 Interestingly, through yeast two-hybrid screening, Dong et al. found that the amino terminus of AtGSL6 could interact with a novel lectin-containing receptor-like kinase (LecRLK1).19 It will thus be very interesting to test whether GSL8 and GSL10 can also interact with receptor-like kinases (RLKs) to regulate pollen entry into mitosis. Recently, as described in more detail below, we also obtained a similar result from GSL10, but we successfully recovered homozygous gsl8 mutants.16
The Role of Callose in Cell Plate Formation
During cell division in higher plants, the first visible evidence of the new cell wall is deposition of the cell plate in an equatorial plane between daughter nuclei. Cell plate development is initiated with the fusion of Golgi-derived vesicles into a continuous membrane network in the center of the phragmoplast; this process defines where the new cell wall will be assembled.20 Immunolabeling data indicate that the callose is the main luminal component of forming cell plate and it forms a coat-like structure on the membrane surface.21 Based on electron micrograph studies, the maturation of tubular network into a fenestrated cell plate and then into a cell wall may be driven primarily by the synthesis of callose.20,21 This driving force of callose on the membranes might be increased by the polymerization of phragmoplastin and sequeezing of phragmoplastin polymers,7 since one of the subunits of the callose synthase complex interacts with phragmoplastin.22 Callose deposition is followed by the deposition and organization of cellulose and other cell wall components; at the same time, cell plate callose is degraded by β-1,3-glucanase.
Callose is reported to be formed by enzymes located in the forming plasma membrane since callose is not found in cell plate-targeting vesicles. Through the use of BY-2 cells, it has been demonstrated that one callose synthase, AtGSL6, is located at the developing cell plate where it interacts with two other cell plate-associated proteins, phragmoplastin and a UDP-glucose transferase. It is therefore possible that these three proteins form part of a larger cell plate callose synthase complex on cell plate.4,22 However, several T-DNA knockout mutants of gsl6 (cals1) exhibited no detectable cytokinesis phenotype and actually retained callose at the cell plate, suggesting that more than one callose synthase is involved in cell plate formation.23 (Chen XY, et al. unpublished data).
More recently, Thiele et al.17 cloned a putative callose synthase gene, GSL8/MASSUE. Mutants in gsl8 were found to be seedling lethal, and had typical cytokinesis-defective phenotypes, such as bi- or multi-nucleate cells with cell wall stubs. This finding indicates that callose is indeed essential for plant cytokinesis by synthesizing callose at cell plate. This result is also consistent with recent finding from our lab (Chen XY, et al. unpublished data).
Plasmodesmata Regulation by Callose
In higher plants, almost all cells are symplasmically connected through PD (dynamic plasma-membrane and ER-based intercellular channels) which regulate the trafficking of nutrients, signal molecules etc. PD have several functional states, such as open, closed and dilated.24 The exact functional state of PD depends on what plants require to respond to developmental and/or environmental cues. Callose is frequently found to deposit at PD, where it is generally believed to control the movement of molecules through plasmodesmata as a developmental regulator of symplasmic continuity. Callose can also deposit at PD in response to abiotic and/or biotic stresses. Here we will focus on its developmental roles.
Callose plugs at PD have been implicated to function in the maintenance of dormancy by isolating the meristem from symplasmic continuity with surrounding tissues. Rinne et al.25 found that a short photoperiod can induce the transition of two concentric symplasmic domains in the birch shoot apical meristem into completely symplasmically isolated cells by through the formation of a callose plug at corresponding PD. Interestingly, the same group also showed that the birch shoot apical meristem can restore its symplasmic organization to break bud dormancy after chilling treatment. And, furthermore, that this restoration is likely to be mediated by β-1,3-glucanase since β-1,3-glucan degraded from PD during chilling. These findings led the authors to propose a model for “dormancy cycling” that depicts the meristem as passing through sequential states of cellular communication with characteristic sensitivities to distinct environmental cues.25,26
Another example of a callose-regulated symplasmic domain is the cotton fiber cell. Each cotton fiber is a single cell that forms from the epidermis of the outer integument of the ovules at or just prior to anthesis.27 Previously, Ruan et al.27 demonstrated that the transient closure of PD facilitates elongation of the cotton fiber. The authors concluded that cotton fiber cells are symplamically isolated so as to maintain the high turgor pressure required for cotton fiber elongation. Furthermore, aniline blue staining and immunolocalization studies revealed that callose deposition and degradation at the cotton fiber base correlates with the closure and reopening of PD, respectively. In addition, the expression of a β-1,3-glucanase, GhGluc1, could play a role in this process by degrading callose and opening the PD.28 To date, however, no single callose synthase responsible for callose deposition at PD has been reported. It will be interesting to determine if blocking callose deposition at PD leads to developmental defects, in particular shoot apical meristem (SAM). Interestingly, it has been proposed that symplasmic domain formation in SAM is mediated by callose turnover at PD.25,26
Callose Deposition in Response to Stress
As described above, callose plays important roles in many aspects of plant growth and development. In addition, callose is deposited at the plasma membrane and cell wall interface as one of the many responses of plant tissue to a range of wound stresses. Observable callose deposition occurs within minutes of damage by mechanical, chemical or ultrasonic treatments and in physiological or biotic stress induced by plasmolysis, raised or lowered temperatures and microbial infection, respectively.1 Several independent research groups have reported GSL5/PMR4/CalS12 to be responsible for callose synthesis in sporophytic tissue in response to wounding and/or pathogen. Loss-of-function mutants of GSL5/PMR4/CalS12 failed to synthesize callose at papillae. Unexpectedly, depletion of callose in gsl5 mutants rendered the plants more resistant, not more susceptible to pathogens. These data indicate that callose exerts a negative effect on plant defense against pathogen infection. One possible explanation for this is that callose could hinder the plants' defense machinery against pathogen; removal of callose in the gsl5 mutant could therefore activate defense systems.11 Alternatively, pathogen-induced callose could negatively regulates the SA signaling pathway of plants, and the lack of callose in the pmr4 mutant may enhance the SA signaling, which results in increased resistance to pathogen.12 These studies demonstrated that multiple mechanisms, in addition to callose, are involved in plant pathogen response.11,12,29
Future Perspectives
Callose is involved in various biological processes associated with plant growth, development and stress responses. Although remarkable progress has been made over the last decade, a number of questions remain unanswered: What is the precise biochemical mechanism of callose synthesis? What are the functional components of the callose synthase complex? What is the function of callose during functional megaspore selection? Do GSL proteins interact with each other in homo-oligomers or in hetero-oligomers? Is there any cross-talk between callose-associated regulation mechanism(s) and other signal pathways? Answering these questions is essential for a deeper understanding of callose function in plants. In the near future, based on previous progress, multiple approaches such as biochemistry, cell biology, genetics and system biology will need to be employed to unravel these mysteries.
Acknowledgements
This work was supported by KOSEF/MEST grants to the National Research Laboratory Program (2009-0066339), the WCU program (R33-2008-000-10002-0), the Environmental Biotechnology National Core Research Center (R15-2003-01201003-0) and by the BK21 program.
Abbreviations
- CalS
callose synthase
- GSL
glucan synthase-like
- SEL
size exclusion limit
- PD
plasmodesmata
- FM
functional megaspore
- RLK
receptor-like kinase
- SAM
shoot apical meristem
- PMR4
powdery mildew resistant 4
Footnotes
Previously published online as a Plant Signaling & Behavior E-publication: http://www.landesbioscience.com/journals/psb/article/8359
References
- 1.Stone BA, Clarke AE. Chemistry and Biology of (1-3)-β-D-Glucans. Victoria, Australia: La Trobe University Press; 1992. [Google Scholar]
- 2.McCormick S. Male gametophyte development. Plant Cell. 1993;5:1265–1275. doi: 10.1105/tpc.5.10.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Samuels AL, Giddings TH, Staehelin LA. Cytokinesis in tobacco BY-2 and root tip cells: A new model of cell plate formation in higher plants. J Cell Biol. 1995;130:1345–1357. doi: 10.1083/jcb.130.6.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hong Z, Delauney AJ, Verma DPS. A cell plate-specific callose synthase and its interaction with phragmoplastin. Plant Cell. 2001;13:755–768. doi: 10.1105/tpc.13.4.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iglesias VA, Meins F., Jr Movement of plant viruses is delayed in a β-1,3-glucanase-deficient mutant showing a reduced plasmodesmatal size exclusion limit and enhanced callose deposition. Plant J. 2000;21:157–166. doi: 10.1046/j.1365-313x.2000.00658.x. [DOI] [PubMed] [Google Scholar]
- 6.Bucher GL, Tarina C, Heinlein M, Di Serio F, Meins F, Jr, Iglesias VA. Local expression of enzymatically active class I β-1,3-glucanase enhances symptoms of TMV infection in tobacco. Plant J. 2001;28:361–369. doi: 10.1046/j.1365-313x.2001.01181.x. [DOI] [PubMed] [Google Scholar]
- 7.Verma DPS, Hong Z. Plant callose synthase complexes. Plant Mol Biol. 2001;47:693–701. doi: 10.1023/a:1013679111111. [DOI] [PubMed] [Google Scholar]
- 8.Brownfield L, Ford K, Doblin MS, Newbigin E, Read S, Bacic A. Proteomic and biochemical evidence links the callose synthase in Nicotiana alata pollen tubes to the product of the NaGSL1 gene. Plant J. 2007;52:147–156. doi: 10.1111/j.1365-313X.2007.03219.x. [DOI] [PubMed] [Google Scholar]
- 9.Brownfield L, Wilson S, Newbigi E, Bacic A, Read S. Molecular control of the glucan synthase-like protein NaGSL1 and callose synthesis during growth of Nicotiana alata pollen tubes. Biochem J. 2008;414:43–52. doi: 10.1042/BJ20080693. [DOI] [PubMed] [Google Scholar]
- 10.Richmond TA, Somerville CR. The cellulose synthase superfamily. Plant Physiol. 2000;124:495–498. doi: 10.1104/pp.124.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jacobs AK, Lipka V, Burton RA, Panstruga R, Strizhov N, Schulze-Lefert P, Fincher GB. An arabidopsis callose synthase, GSL5, is required for wound and papillary callose formation. Plant Cell. 2003;15:2503–2513. doi: 10.1105/tpc.016097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nishimura MT, Stein M, Hou B, Vogel JP, Edwards H, Somerville SC. Loss of a callose synthase results in salicylic acid-dependent disease resistance. Science. 2003;301:969–972. doi: 10.1126/science.1086716. [DOI] [PubMed] [Google Scholar]
- 13.Enns LC, Kanaoka MM, Torii KU, Comai L, Okada K, Cleland RE. Two callose synthases, GSL1 and GSL5, play an essential and redundant role in plant and pollen development and in fertility. Plant Mol Biol. 2005;58:333–349. doi: 10.1007/s11103-005-4526-7. [DOI] [PubMed] [Google Scholar]
- 14.Nishikawa S, Zinkl GM, Swanson RJ, Maruyama D, Preuss D. Callose (β-1,3 glucan) is essential for Arabidopsis pollen wall patterning, but not tube growth. BMC Plant Biol. 2005;5:22. doi: 10.1186/1471-2229-5-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Töller A, Brownfield L, Neu C, Twell D, Schulze-Lefert P. Dual function of Arabidopsis glucan synthase-like genes GSL8 and GSL10 in male gametophyte development and plant growth. Plant J. 2008;54:911–923. doi: 10.1111/j.1365-313X.2008.03462.x. [DOI] [PubMed] [Google Scholar]
- 16.Huang L, Chen XY, Rim Y, Han X, Cho WY, Kim SW, Kim JY. Arabidopsis Glucan Synthase-Like 10 functions in male gametogenesis. J Plant Physiol. 2008 doi: 10.1016/j.jplph.2008.06.010. In press. [DOI] [PubMed] [Google Scholar]
- 17.Thiele K, Wanner G, Kindzierski V, Jürgens G, Mayer U, Pachl F, Assaad FF. The timely deposition of callose is essential for cytokinesis in Arabidopsis. Plant J. 2008 doi: 10.1111/j.1365-313X.2008.03760.x. [DOI] [PubMed] [Google Scholar]
- 18.Dong X, Hong Z, Sivaramakrishnan M, Mahfouz M, Verma DPS. Callose synthase (CalS5) is required for exine formation during microgametogenesis and for pollen viability in Arabidopsis. Plant J. 2005;42:315–328. doi: 10.1111/j.1365-313X.2005.02379.x. [DOI] [PubMed] [Google Scholar]
- 19.Dong X. Functional investigation of Arabidopsis callose synthase and the signal transduction pathway. Ohio State University; 2005. The PhD thesis. [Google Scholar]
- 20.Staehelin LA, Hepler PK. Cytokinesis in higher plants. Cell. 1996;84:821–824. doi: 10.1016/s0092-8674(00)81060-0. [DOI] [PubMed] [Google Scholar]
- 21.Samuels AL, Giddings TH, Staehelin LA. Cytokinesis in tobacco BY-2 and root tip cells: A new model of cell plate formation in higher plants. J Cell Biol. 1995;130:1345–1357. doi: 10.1083/jcb.130.6.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hong Z, Zhang Z, Olson JM, Verma DPS. A novel UDP-glucose transferase is part of the callose synthase complex and interacts with phragmoplastin at the forming cell plate. Plant Cell. 2001;13:769–779. doi: 10.1105/tpc.13.4.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hong Z, Verma DPS. Molecular analysis of the cell plate forming machinery in cell division control in plants. Heidelberg: Springer Berlin; 2007. [Google Scholar]
- 24.Lucas WJ, Lee J. Plasmodesmata as a supracellular control network in plants. Nat Rev Mol Cell Biol. 2004;5:712–726. doi: 10.1038/nrm1470. [DOI] [PubMed] [Google Scholar]
- 25.Rinne PLH, Van der Schoot C. Symplasmic fields in the tunica of the shoot apical meristem coordinate morphogenetic events. Development. 1998;125:1477–1485. doi: 10.1242/dev.125.8.1477. [DOI] [PubMed] [Google Scholar]
- 26.Rinne PLH, Kaikuranta PM, Van Schoot CD. The shoot apical meristem restores its symplasmic organization during chilling-induced release from dormancy. Plant J. 2001;26:249–264. doi: 10.1046/j.1365-313x.2001.01022.x. [DOI] [PubMed] [Google Scholar]
- 27.Ruan Y, Llewellyn DJ, Furbank RT. The control of single-celled cotton fiber elongation by developmentally reversible gating of plasmodesmata and coordinated expression of sucrose and K+ transporters and expansin. Plant Cell. 2001;13:47–60. doi: 10.1105/tpc.13.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ruan Y, Xu S, White R, Furbank RT. Genotypic and developmental evidence for the role of plasmodesmatal regulation in cotton fiber elongation mediated by callose turnover. Plant Physiol. 2004;136:4104–4113. doi: 10.1104/pp.104.051540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dong X, Hong Z, Chatterjee J, Kim S, Verma DPS. Expression of callose synthase genes and its connection with Npr1 signaling pathway during pathogen infection. Planta. 2008;229:87–98. doi: 10.1007/s00425-008-0812-3. [DOI] [PubMed] [Google Scholar]