Abstract
MLOs belong to the largest family of seven-transmembrane (7TM) domain proteins found in plants. The Arabidopsis and rice genomes contain 15 and 12 MLO family members, respectively. Although the biological function of most MLO family members remains elusive, a select group of MLO proteins have been demonstrated to negatively regulate defence responses to the obligate biotrophic pathogen, powdery mildew, thereby acting as “susceptibility” genes. Recently we identified a family of 17 putative VvMLO genes in the genome of the cultivated winegrape species, Vitis vinifera. Expression analysis indicated that the VvMLO family members respond differently to biotic and abiotic stimuli. Infection of V. vinifera by grape powdery mildew (Erysiphe necator) specifically upregulates four VvMLO genes that are orthologous to the Arabidopsis and tomato MLOs previously demonstrated to be required for powdery mildew susceptibility. We postulate that one or more of these E. necator responsive VvMLOs may have a role in the powdery mildew susceptibility of grapevine.
Key words: MLO, powdery mildew, resistance, susceptibility, grapevine
The MLO family is only found in plants and its members are characterised by the presence of 7TM and C-terminal calmodulin-binding domains.1–3 MLO proteins share the same topology and localisation within the plasma membrane as animal 7TM proteins.2 Animal 7TM proteins commonly function as G-protein coupled receptors (GPCRs) that sense external stimuli and trigger downstream signalling through heterotrimeric G-proteins.4 However, some 7TM proteins have been found to transmit signals independently of heterotrimeric G-proteins and plant MLOs appear to fall into that group.3,5
Despite the identification of large MLO families in different plant species, the function of MLO proteins in plants cells are still unknown. However, there is now a large body of evidence suggesting that certain members of the MLO family pre-dispose plants to powdery mildew infection and therefore act as “susceptibility” factors. For example, mutations in specific members of the MLO gene family from barley, tomato and Arabidopsis (either through natural genetic variation or targeted gene silencing) have been found to result in broad spectrum resistance to compatible powdery mildew isolates.6–9 This function as a susceptibility factor is unlikely to be the primary role of MLOs, since there must be a selection pressure for retaining functional MLO genes in the plant genome. It has therefore been suggested that powdery mildew exploits MLOs to gain entry to their plant host.10,11 This is similar to the exploitation of 7TM GPCRs in animals, by pathogens such as Streptococcus pneumoniae, to gain access to host cells.12
MLO proteins are localised in the plasma membrane, but during powdery mildew attack become polarised at the site of attempted powdery mildew ingress.13 It is postulated that MLO negatively regulates two independent penetration resistance pathways.14 The first involves the syntaxin PEN1/ROR2 which is thought to mediate vesicle trafficking.15 Yeast two hybrid studies have provided evidence of a direct physical interaction between MLO and ROR2.11 The second penetration resistance pathway involves a glycosyl hydrolase (PEN2) and an ABC transporter (PEN3) which may produce and secrete fungal toxins respectively.16,17
The cultivated grapevine, V. vinifera, is highly susceptible to the powdery mildew species E. necator. As a result, grape production worldwide is highly dependent on the use of fungicides to control this pathogen. The development of new grapevine cultivars with enhanced genetic resistance to powdery mildew would be of significant economic and environmental benefit. We therefore set out to identify and characterise the MLO gene family in grapevine with the aim of isolating potential candidates involved in powdery mildew susceptibility. Using the recently sequenced V. vinifera genome sequence,18 we identified a family of 17 putative VvMLO genes.19 Phylogenetic analysis of the translated products of the VvMLO gene family together with other known plant MLO sequences identified six VvMLO genes (VvMLO3, VvMLO4, VvMLO6, VvMLO9, VvMLO13 and VvMLO17) which clustered within the same clade as the Arabidopsis and tomato MLO genes that have been demonstrated to be required for powdery mildew susceptibility.6,7,19 Of these, three (VvMLO3, VvMLO4 and VvMLO17) were found to be significantly induced in grape leaves within 8 h of E. necator inoculation, which coincided with the commencement of fungal penetration.19 The transient transcriptional response of VvMLO3, VvMLO4 and VvMLO17 to E. necator penetration is consistent with that previously reported for HvMLO in response to barley powdery mildew (Blumeria graminis) infection.20 VvMLO9 was also induced, but not until 24 h post inoculation suggesting that this MLO may not be involved in the first stages of pathogen infection.19 We have also subsequently shown these powdery mildew-induced VvMLO proteins are localised to the plasma membrane (Fig. 1). Thus, the similarities in both sequence homology, transcriptional regulation and cellular localization suggests that VvMLO3, VvMLO4 and VvMLO17 may have functionally similar roles to those MLO genes from barley, Arabidopsis and tomato which have been shown to modulate powdery mildew susceptibility.6–9 It may also indicate some level of functional redundancy between these three VvMLO genes as is the case in Arabidopsis.7
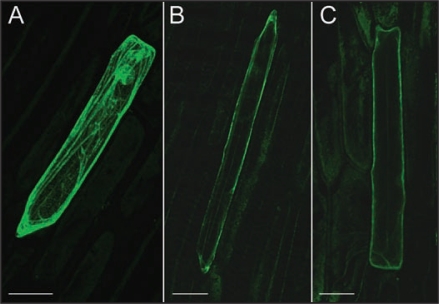
Figure 1.
Plasma membrane localisation of VvMLO. Onion epidermal cells expressing (A) ER-targeted mGFP (pCAMBIA1301 with mGFP5-ER introduced from pBIN mGFP5-ER) (B) VvMLO4-GFP (C-terminal GFP fusion of VvMLO4 in pN'GFP) (C) VvMLO17-GFP (C-terminal GFP fusion of VvMLO17 in pGWB5). Scale bar 100 µM.
It is not yet known which signalling molecules are responsible for the rapid induction of VvMLOs following powdery mildew infection. Despite the involvement of the signalling compounds H2O2 and salicylic acid in plant defence responses,21,22 these compounds were found to induce a different subset of VvMLOs to those induced by E. necator.19 On the other hand, wounding was found to induce the same subset of VvMLO transcripts (as well as VvMLO6), to those rapidly induced by E. necator infection, although the relative levels of induction differed.19 Gene-silencing studies are now underway to establish which of the E. necator induced VvMLOs are required for powdery mildew susceptibility in grapevine. It will also be important to determine if silencing of MLO gene(s) results in any pleiotropic effects on grapevine growth and development, as has been observed in barley and Arabidopsis mlo mutants.7,20,23 These include the spontaneous deposition of callose-containing cell wall appositions in pathogen-free conditions and the premature onset of leaf senescence. However, these were absent in powdery-mildew resistant tomato mlo mutants.6 The appearance of similar pleiotropic effects on grapevine leaves is unlikely to cause significant issues for grape-growers because photosynthetic capacity is not normally a limiting factor to grape production. Indeed, it is likely that most growers would tolerate some loss in maximum cropping capacity in exchange for durable resistance to powdery mildew, provided there are no significant effects on grape berry quality.
Footnotes
Previously published online as a Plant Signaling & Behavior E-publication: http://www.landesbioscience.com/journals/psb/article/8575
References
- 1.Buschges R, Hollricher K, Panstruga R, Simons G, Wolter M, Frijters A, et al. The barley Mlo gene: a novel control element of plant pathogen resistance. Cell. 1997;88:695–705. doi: 10.1016/s0092-8674(00)81912-1. [DOI] [PubMed] [Google Scholar]
- 2.Devoto A, Piffanelli P, Nilsson I, Wallin E, Panstruga R, von Heijne G, Schulze-Lefert P. Topology, subcellular localization and sequence diversity of the MLO family in plants. J Biol Chem. 1999;274:34993–35004. doi: 10.1074/jbc.274.49.34993. [DOI] [PubMed] [Google Scholar]
- 3.Kim MC, Panstruga R, Elliott C, Muller J, Devoto A, Yoon HW, et al. Calmodulin interacts with MLO protein to regulate defence against mildew in barley. Nature. 2002;416:447–451. doi: 10.1038/416447a. [DOI] [PubMed] [Google Scholar]
- 4.Bockaert J, Pin JP. Molecular tinkering of G protein-coupled receptors: an evolutionary success. EMBO J. 1999;18:1723–1729. doi: 10.1093/emboj/18.7.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brzostowski JA, Kimmel AR. Signaling at zero G: G-protein-independent functions for 7-TM receptors. Trends Biochem Sci. 2001;26:291–297. doi: 10.1016/s0968-0004(01)01804-7. [DOI] [PubMed] [Google Scholar]
- 6.Bai Y, Pavan S, Zheng Z, Zappel NF, Reinstadler A, Lotti C, et al. Naturally occurring broad-spectrum powdery mildew resistance in a Central American tomato accession is caused by loss of mlo function. Mol Plant Microbe Interact. 2008;21:30–39. doi: 10.1094/MPMI-21-1-0030. [DOI] [PubMed] [Google Scholar]
- 7.Consonni C, Humphry ME, Hartmann HA, Livaja M, Durner J, Westphal L, et al. Conserved requirement for a plant host cell protein in powdery mildew pathogenesis. Nature Gen. 2006;38:716–720. doi: 10.1038/ng1806. [DOI] [PubMed] [Google Scholar]
- 8.Jorgensen JH. Discovery, Characterization and exploitation of MLO powdery mildew resistance in barley. Euphytica. 1992;63:141–152. [Google Scholar]
- 9.Piffanelli P, Ramsay L, Waugh R, Benabdelmouna A, D'Hont A, Hollricher K, et al. A barley cultivation-associated polymorphism conveys resistance to powdery mildew. Nature. 2004;430:887–891. doi: 10.1038/nature02781. [DOI] [PubMed] [Google Scholar]
- 10.Panstruga R, Schulze-Lefert P. Corruption of host seven-transmembrane proteins by pathogenic microbes: a common theme in animals and plants? Microbes Infect. 2003;5:429–437. doi: 10.1016/s1286-4579(03)00053-4. [DOI] [PubMed] [Google Scholar]
- 11.Panstruga R. Serpentine plant MLO proteins as entry portals for powdery mildew fungi. Biochemical Soc Trans. 2005;33:389–392. doi: 10.1042/BST0330389. [DOI] [PubMed] [Google Scholar]
- 12.Cundell DR, Gerard NP, Gerard C, Idanpaan-Heikkila I, Tuomanen EI. Streptococcus pneumoniae anchor to activated human cells by the receptor for platelet-activating factor. Nature. 1995;377:435–438. doi: 10.1038/377435a0. [DOI] [PubMed] [Google Scholar]
- 13.Bhat RA, Miklis M, Schmelzer E, Schulze-Lefert P, Panstruga R. Recruitment and interaction dynamics of plant penetration resistance components in a plasma membrane microdomain. Proc Natl Acad Sci USA. 2005;102:3135–3140. doi: 10.1073/pnas.0500012102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Underwood W, Somerville SC. Focal accumulation of defences at sites of fungal pathogen attack. J Exp Bot. 2008;59:3501–3508. doi: 10.1093/jxb/ern205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Collins NC, Thordal-Christensen H, Lipka V, Bau S, Kombrink E, Qiu JL, et al. SNARE-protein-mediated disease resistance at the plant cell wall. Nature. 2003;425:973–977. doi: 10.1038/nature02076. [DOI] [PubMed] [Google Scholar]
- 16.Lipka V, Dittgen J, Bednarek P, Bhat R, Wiermer M, Stein M, et al. Pre- and post-invasion defenses both contribute to nonhost resistance in Arabidopsis. Science. 2005;310:1180–1183. doi: 10.1126/science.1119409. [DOI] [PubMed] [Google Scholar]
- 17.Stein M, Dittgen J, Sanchez-Rodriguez C, Hou BH, Molina A, Schulze-Lefert P, et al. Arabidopsis PEN3/PDR8, an ATP binding cassette transporter, contributes to nonhost resistance to inappropriate pathogens that enter by direct penetration. Plant Cell. 2006;18:731–746. doi: 10.1105/tpc.105.038372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jaillon O, Aury J-M, Noel B, Policriti A, Clepet C, Casagrande A, et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007;449:463–468. doi: 10.1038/nature06148. [DOI] [PubMed] [Google Scholar]
- 19.Feechan A, Jermakow AM, Torregrosa L, Panstruga R, Dry IB. Identification of grapevine MLO gene candidates involved in susceptibility to powdery mildew. Funct Plant Biol. 2008;35:1255–1266. doi: 10.1071/FP08173. [DOI] [PubMed] [Google Scholar]
- 20.Piffanelli P, Zhou FS, Casais C, Orme J, Jarosch B, Schaffrath U, et al. The barley MLO modulator of defense and cell death is responsive to biotic and abiotic stress stimuli. Plant Physiol. 2002;129:1076–1085. doi: 10.1104/pp.010954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alvarez ME, Pennell RI, Meijer PJ, Ishikawa A, Dixon RA, Lamb C. Reactive oxygen intermediates mediate a systemic signal network in the establishment of plant immunity. Cell. 1998;92:773–784. doi: 10.1016/s0092-8674(00)81405-1. [DOI] [PubMed] [Google Scholar]
- 22.Thomma BP, Penninckx IA, Broekaert WF, Cammue BP. The complexity of disease signaling in Arabidopsis. Curr Opin Immunol. 2001;13:63–68. doi: 10.1016/s0952-7915(00)00183-7. [DOI] [PubMed] [Google Scholar]
- 23.Peterhänsel C, Freialdenhoven A, Kurth J, Kolsch R, Schulze-Lefert P. Interaction analyses of genes required for resistance responses to powdery mildew in barley reveal distinct pathways leading to leaf cell death. Plant Cell. 1997;9:1397–1409. doi: 10.1105/tpc.9.8.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]