Abstract
Plants use pattern recognition receptors (PRRs) to perceive pathogen-associated molecular pattern (PAMPs) and initiate defence responses. PAMP-triggered immunity (PTI) plays an important role in general resistance, and constrains the growth of most microbes on plants. Despite the importance of PRRs in plant immunity, the vast majority of them remain to be identified. We recently showed that the Arabidopsis LysM receptor kinase CERK1 is required not only for chitin signalling and fungal resistance, but plays an essential role in restricting bacterial growth on plants. We proposed that CERK1 may mediate the perception of a bacterial PAMP, or an endogenous plant cell wall component released during infection, through its extracellular carbohydrate-binding LysM-motifs. Here we report reduced activation of a PAMP-induced defence response on plants lacking the CERK1 gene after treatment with crude bacterial extracts. This demonstrates that CERK1 mediates perception of an unknown bacterial PAMP in Arabidopsis.
Key words: PAMP, PRR, PTI, LysM, chitin, bacteria, carbohydrate
Introduction
A key feature of active defence mechanisms is the ability to discriminate between self and nonself upon microbial infection. In higher eukaryotes, microbes are detected directly by perception of conserved pathogen-associated molecular patterns called PAMPs, or indirectly by sensing wound-or injury-related molecules released during the infection process. PAMPs constitute highly conserved molecules typical of whole classes of pathogens that are indispensable for the microbial lifestyle. The sensory function for PAMPs is provided by pattern recognition receptors (PRRs) which are typically localized in the plasma membrane. Plant PRRs belong to a large family of receptor kinases containing at least 600 members in Arabidopsis.1 Some well-studied examples are the flagellin receptor FLAGELLIN-SENSING 2 (FLS2) and the Arabidopsis EF-Tu receptor (EFR),2,3 both of which recognise bacterial proteins. Signalling through these receptors requires rapid association with a second receptor-like kinase called BAK1 (BRI1-ASSOCIATED KINASE 1) to integrate perception events into downstream signalling responses.4,5 Perception of PAMPs by PRRs leads to generation of reactive oxygen species (ROS), induction of defence gene expression, and callose deposition into cell walls at infected sites, together restricting the growth of most microbes.6,7 However, the vast majority of PRRs involved in microbe sensing and the microbial molecules that are perceived are largely unknown. Identification of such components represents an exciting challenge to understand fully the molecular basis of innate immunity.
CERK1, a LysM Receptor Kinase Required for Fungal and Bacterial Resistance
The bacterial pathogen Pseudomonas syringae pv. tomato DC3000 (Pto DC3000) uses a specialized type III secretion system (TTSS) to deliver more than 30 virulence effector molecules into the plant cell.8 Effectors contribute to pathogenesis by acting on host molecular targets to suppress PTI and defeat plant defences.9,10 Suppression of PAMP responses by effectors is crucial as Pseudomonas strains lacking the TTSS are not pathogenic.11 We recently identified the LysM receptor kinase CERK1 as a novel target for the Pseudomonas effector AvrPtoB.12 CERK1 is required for chitin signalling and fungal resistance in Arabidopsis.13,14 We demonstrated that CERK1 also constitutes an essential component that restricts bacterial growth on plants. Arabidopsis cerk1 mutants showed enhanced disease symptoms and supported higher bacterial growth when Pto DC3000 was sprayed onto leaves. Interestingly, bacterial growth assays indicated similar contributions of FLS2 and CERK1 to immunity, suggesting that CERK1 may play a role in bacterial recognition. Accordingly, a non-pathogenic Pto DC3000 strain unable to secrete effectors or suppress PAMP responses showed enhanced growth on cerk1 mutant plants. This implies that CERK1 is required for PAMP perception during bacterial pathogenesis. Our results show that AvrPtoB targets CERK1 directly to overcome bacterial perception and suppress innate immunity.
Does CERK1 Perceive a Bacterial PAMP or an Endogenous Host Molecule?
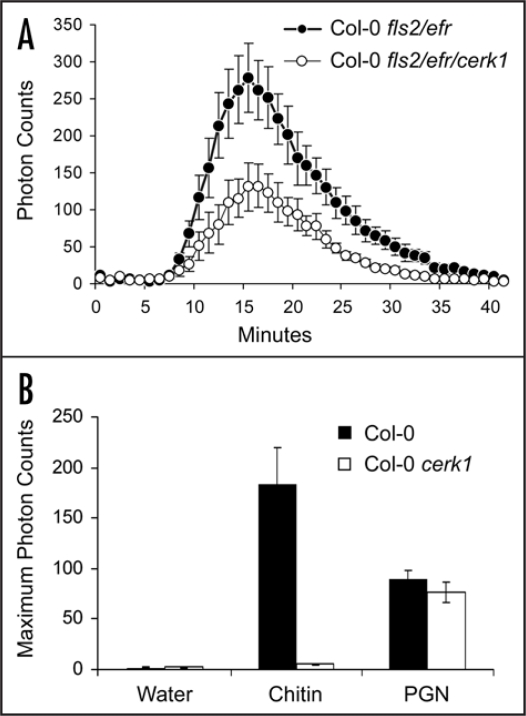
CERK1 is required for all defence responses induced by chitin, an N-acetyl-D-glucosamine polymer constituting the main component of fungal cell walls. CERK1 encodes a receptor kinase containing three extracellular LysM-domains and an intracellular kinase domain.13,14 LysM-motifs have been studied extensively and are generally regarded as carbohydrate-binding moieties.15 Although bacteria do not contain chitin per se, similar carbohydrate based structures are present in bacteria and may act as PAMPs on plant cells. However, similar structures are also present in plant cell walls and could potentially be released upon microbe invasion to act as danger signals. To test whether CERK1 perceives a molecule from Pto DC3000, we prepared crude bacterial extracts as reported previously7 and tested them for activation of defence responses in Arabidopsis plants lacking the FLS2 and EFR genes. These plants cannot perceive flagellin or EF-Tu and therefore constitute an ideal background to detect novel elicitors from bacteria. Treatment of these plants with crude Pto DC3000 bacterial extracts induced generation of ROS, indicating that as expected, Arabidopsis responds to bacterial PAMPs other than flagellin and EF-Tu (Fig. 1A). Interestingly, ROS production was strongly reduced in similar Arabidopsis plants containing an additional mutation in the cerk1 gene. Although indirect recognition of a plant cell wall fragment can not be completely excluded by this experiment, the result is consistent with recognition of a novel bacterial PAMP by CERK1 leading to induction of ROS.
Figure 1.
Generation of ROS by Pto DC3000 bacterial extracts in cerk1 mutant plants. (A) ROS burst in Arabidopsis Col-0 fls2/efr and Col-0 fls2/efr/cerk1 plants after treatment with 20 µl/ml of crude bacterial extracts from Pto DC3000. Crude bacterial extracts were prepared by boiling bacterial suspensions for 10 minutes and removing cell debris by centrifugation. Error bars indicate standard error of the mean (SEM). The results shown are representative of four independent experiments. (B) ROS burst in Arabidopsis Col-0 and Col-0 cerk1 plants upon treatment with water, 100 µg/ml chitin, or 30 µl/ml of Pto DC3000 peptidoglycan (PGN). Error bars indicate SEM. Similar results were obtained in two independent experiments.
Some LysM domains are known to bind peptidoglycan (PGN) from various bacteria.15 PGN is an essential structural component of the bacterial cell wall and acts as a PAMP on Arabidopsis.16,17 It is therefore plausible that PGN constitutes a ligand for any of the three LysM motifs of CERK1. To test this hypothesis, we purified Pto DC3000 PGN as described by Erbs et al.16 Both chitin and PGN induced ROS generation on Col-0 plants as expected (Fig. 1B). However, only chitin-induced defences were blocked by the cerk1 mutation. Thus, perception of PGN is independent of CERK1, and the bacterial molecule recognised by CERK1 remains unknown.
Carbohydrate Recognition by LysM-Containing Receptor Kinases
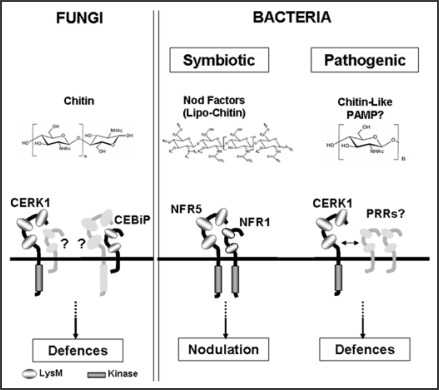
CERK1 is not the only LysM-containing receptor protein known to be involved in chitin perception. The receptor protein CEBiP of rice was previously identified as an essential component for chitin signalling.18 CEBiP encodes a transmembrane protein with two extracellular LysM-motifs that bind chitin directly. In contrast, direct binding of chitin to Arabidopsis CERK1 remains to be demonstrated. The lack of an intracellular signalling domain in the CEBiP protein provokes the hypothesis that it might associate with a receptor kinase to transduce the PAMP signal (Fig. 2). An analagous mechanism is perception of lipopolysacharides (LPS) by the mammalian TLR4/CD14 receptor complex.19 Both Arabidopsis and rice contain several CEBiP-and CERK1-like proteins, respectively. It is possible that CERK1 could act as an auxiliary component of multiple PRRs to transduce the signal, similar in concept to the requirement for BAK1 by FLS2 and other receptors.4,5 It is of great interest to determine whether CERK1 interacts with CEBiP.
Figure 2.
Schematic representation of microbial perception and signalling through LysM receptor kinases in plants. Plants sense fungi by recognition of chitin oligomers released from fungal cell walls through the LysM receptor proteins CERK1 and CEBiP in Arabidopsis and rice, respectively. The lack of an intracellular signaling domain in the CEBiP protein suggests that the receptor may associate with another protein such as a receptor kinase to transduce the signal. Chitin perception show many similarities with recognition of symbiotic bacteria in leguminous plants. CERK1 is homologous to the legume Nod factor receptors NFR1 and NFR5 mediating perception and signalling of lipo-chitin Nod factors, probably in a heterodimeric complex. The identification of CERK1 as a component required for bacterial immunity suggests that pathogenic bacteria also contain similar chitin-like PAMPs.
Fascinatingly, other LysM receptor kinases are known to mediate bacterial perception, but in an opposite role to plant defense. CERK1 is highly related to the receptor kinases NFR1 and NFR5 of legumes that control symbiotic interactions with rhizobial bacteria upon perception of Nod factors20–22 (Fig. 2). Nod factors are lipo-chitin oligosaccharide molecules.23 This core molecule carries additional decorations which determine the host specificity of each rhizobial strain,24 and may also function to avoid activation of defence responses elicited by the chitin oligomer core.25
Strikingly, Sinorhizobium meliloti Nod factors function not only as nodulation signals to initiate symbiosis, but also as key components to establish rhizobial biofilms.26 The role of Nod factors in biofilm formation is likely to be ancestral as N-acetylglucosamines function as adhesins promoting cell-to-cell and abiotic surface adhesion in a number of bacteria including Staphylococcus epidermidis, Staphylococcus aureus, Escherichia coli and Actinobacillus actinomycetemcomitans.27–30 This evolutionary relatedness suggests a potential common origin between the recognition of pathogenic and symbiotic bacteria molecules through diversified LysM-containing receptor proteins. Identification of the bacterial molecule conferring CERK1-mediated immunity is the first priority for understanding the role of LysM proteins in general bacterial recognition.
Acknowledgements
We are very grateful to Dr. Cyril Zipfel for providing the Col-0 fls2/efr seeds. We thank Dr. Dagmar Hann for many points of discussion and a critical review of the manuscript. The support of the Gatsby Charitable Foundation is gratefully acknowledged.
Footnotes
Previously published online as a Plant Signaling & Behavior E-publication: http://www.landesbioscience.com/journals/psb/article/8697
References
- 1.Shiu SH, Bleecker AB. Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc Natl Acad Sci USA. 2001;98:10763–10768. doi: 10.1073/pnas.181141598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gomez-Gomez L, Boller T. FLS2: An LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell. 2000;5:1003–1011. doi: 10.1016/s1097-2765(00)80265-8. [DOI] [PubMed] [Google Scholar]
- 3.Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones J, Boller T, Felix G. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell. 2006;125:749–760. doi: 10.1016/j.cell.2006.03.037. [DOI] [PubMed] [Google Scholar]
- 4.Chinchilla D, Zipfel C, Robatzek S, Kemmerling B, Nurnberger T, Jones JD, et al. A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature. 2007;448:497–500. doi: 10.1038/nature05999. [DOI] [PubMed] [Google Scholar]
- 5.Heese A, Hann DR, Gimenez-Ibanez S, Jones AM, He K, Li J, et al. The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc Natl Acad Sci USA. 2007;104:12217–12222. doi: 10.1073/pnas.0705306104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zipfel C, Robatzek S, Navarro L, Oakeley EJ, Jones JD, Felix G, Boller T. Bacterial disease resistance in Arabidopsis through flagellin perception. Nature. 2004;428:764–767. doi: 10.1038/nature02485. [DOI] [PubMed] [Google Scholar]
- 7.Kunze G, Zipfel C, Robatzek S, Niehaus K, Boller T, Felix G. The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cell. 2004;16:3496–3507. doi: 10.1105/tpc.104.026765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schechter LM, Vencato M, Jordan KL, Schneider SE, Schneider DJ, Collmer A. Multiple approaches to a complete inventory of Pseudomonas syringae pv. tomato DC3000 type III secretion system effector proteins. Mol Plant Microbe Interact. 2006;19:1180–1192. doi: 10.1094/MPMI-19-1180. [DOI] [PubMed] [Google Scholar]
- 9.Xiang T, Zong N, Zou Y, Wu Y, Zhang J, Xing W, et al. Pseudomonas syringae effector AvrPto blocks innate immunity by targeting receptor kinases. Curr Biol. 2008;18:74–80. doi: 10.1016/j.cub.2007.12.020. [DOI] [PubMed] [Google Scholar]
- 10.Shan L, He P, Li J, Heese A, Peck SC, Nurnberger T, et al. Bacterial effectors target the common signaling partner BAK1 to disrupt multiple MAMP receptor-signaling complexes and impede plant immunity. Cell Host Microbe. 2008;4:17–27. doi: 10.1016/j.chom.2008.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roine E, Wei W, Yuan J, Nurmiaho-Lassila EL, Kalkkinen N, Romantschuk M, He SY. Hrp pilus: an hrp-dependent bacterial surface appendage produced by Pseudomonas syringae pv. tomato DC3000. Proc Natl Acad Sci USA. 1997;94:3459–3464. doi: 10.1073/pnas.94.7.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gimenez-Ibanez S, Hann DR, Ntoukakis V, Petutschnig E, Lipka V, Rathjen JP. AvrPtoB targets the LysM receptor kinase CERK1 to promote bacterial virulence on plants. Curr Biol. 2009;19:423–429. doi: 10.1016/j.cub.2009.01.054. [DOI] [PubMed] [Google Scholar]
- 13.Miya A, Albert P, Shinya T, Desaki Y, Ichimura K, Shirasu K, et al. CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc Natl Acad Sci USA. 2007;104:19613–19618. doi: 10.1073/pnas.0705147104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wan J, Zhang XC, Neece D, Ramonell KM, Clough S, Kim SY, et al. A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell. 2008;20:471–481. doi: 10.1105/tpc.107.056754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buist G, Steen A, Kok J, Kuipers OP. LysM, a widely distributed protein motif for binding to (peptido)glycans. Mol Microbiol. 2008;68:838–847. doi: 10.1111/j.1365-2958.2008.06211.x. [DOI] [PubMed] [Google Scholar]
- 16.Erbs G, Silipo A, Aslam S, De Castro C, Liparoti V, Flagiello A, et al. Peptidoglycan and muropeptides from pathogens Agrobacterium and Xanthomonas elicit plant innate immunity: structure and activity. Chem Biol. 2008;15:438–448. doi: 10.1016/j.chembiol.2008.03.017. [DOI] [PubMed] [Google Scholar]
- 17.Gust AA, Biswas R, Lenz HD, Rauhut T, Ranf S, Kemmerling B, et al. Bacteria-derived peptidoglycans constitute pathogen-associated molecular patterns triggering innate immunity in Arabidopsis. J Biol Chem. 2007;282:32338–32348. doi: 10.1074/jbc.M704886200. [DOI] [PubMed] [Google Scholar]
- 18.Kaku H, Nishizawa Y, Ishii-Minami N, Akimoto-Tomiyama C, Dohmae N, Takio K, et al. Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc Natl Acad Sci USA. 2006;103:11086–11091. doi: 10.1073/pnas.0508882103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jerala R. Structural biology of the LPS recognition. Int J Med Microbiol. 2007;297:353–363. doi: 10.1016/j.ijmm.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 20.Zhang XC, Wu X, Findley S, Wan J, Libault M, Nguyen HT, et al. Molecular evolution of lysin motif-type receptor-like kinases in plants. Plant Physiol. 2007;144:623–636. doi: 10.1104/pp.107.097097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Radutoiu S, Madsen LH, Madsen EB, Felle HH, Umehara Y, Gronlund M, et al. Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature. 2003;425:585–592. doi: 10.1038/nature02039. [DOI] [PubMed] [Google Scholar]
- 22.Limpens E, Franken C, Smit P, Willemse J, Bisseling T, Geurts R. LysM domain receptor kinases regulating rhizobial Nod factor-induced infection. Science. 2003;302:630–633. doi: 10.1126/science.1090074. [DOI] [PubMed] [Google Scholar]
- 23.Lerouge P, Roche P, Faucher C, Maillet F, Truchet G, Prome JC, Denarie J. Symbiotic host-specificity of Rhizobium meliloti is determined by a sulphated and acylated glucosamine oligosaccharide signal. Nature. 1990;344:781–784. doi: 10.1038/344781a0. [DOI] [PubMed] [Google Scholar]
- 24.Cullimore JV, Ranjeva R, Bono JJ. Perception of lipo-chitooligosaccharidic Nod factors in legumes. Trends Plant Sci. 2001;6:24–30. doi: 10.1016/s1360-1385(00)01810-0. [DOI] [PubMed] [Google Scholar]
- 25.Muller J, Staehelin C, Xie ZP, Neuhaus-Url G, Boller T. Nod factors and chitooligomers elicit an increase in cytosolic calcium in aequorin-expressing soybean cells. Plant Physiol. 2000;124:733–740. doi: 10.1104/pp.124.2.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fujishige NA, Lum MR, De Hoff PL, Whitelegge JP, Faull KF, Hirsch AM. Rhizobium common nod genes are required for biofilm formation. Mol Microbiol. 2008;67:504–515. doi: 10.1111/j.1365-2958.2007.06064.x. [DOI] [PubMed] [Google Scholar]
- 27.Baldassarri L, Donnelli G, Gelosia A, Voglino MC, Simpson AW, Christensen GD. Purification and characterization of the staphylococcal slime-associated antigen and its occurrence among Staphylococcus epidermis clinical isolates. Inf Immun. 1996;64:3410–3415. doi: 10.1128/iai.64.8.3410-3415.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maira-Litran T, Kropec A, Abeygunawardana C, Joyce J, Mark G, 3rd, Goldmann DA, Pier GB. Immunochemical properties of the staphylococcal poly-N-acetylglucosamine surface polysaccharide. Inf Immun. 2002;70:4433–4440. doi: 10.1128/IAI.70.8.4433-4440.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang X, Preston JF, 3rd, Romeo T. The pgaABCD locus of Escherichia coli promotes the synthesis of a polysaccharide adhesin required for biofilm formation. J Bacteriol. 2004;186:2724–2734. doi: 10.1128/JB.186.9.2724-2734.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaplan JB, Velliyagounder K, Ragunath C, Rohde H, Mack D, Knobloch JK, Ramasubbu N. Genes involved in the synthesis and degradation of matrix polysaccharide in Actinobacillus actinomycetemcomitans and Actinobacillus pleuropneumoniae biofilms. J Bacteriol. 2004;186:8213–8220. doi: 10.1128/JB.186.24.8213-8220.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]