Abstract
In plants, the division of peroxisomes is mediated by several classes of proteins, including PEROXIN11 (PEX11), FISSION1 (FIS1) and DYNAMIN-RELATED PROTEIN3 (DRP3). DRP3A and DRP3B are two homologous dynamin-related proteins playing overlapping roles in the division of both peroxisomes and mitochondria, with DRP3A performing a stronger function than DRP3B in peroxisomal fission. Here, we report the identification and characterization of the peroxisome division defective 2 (pdd2) mutant, which was later proven to be another drp3A allele. The pdd2 mutant generates a truncated DRP3A protein and exhibits pale green and retarded growth phenotypes. Intriguingly, this mutant displays much stronger peroxisome division deficiency in root cells than in leaf mesophyll cells. Our data suggest that the partial GTPase effector domain retained in pdd2 may have contributed to the distinct mutant phenotype of this mutant.
Key words: peroxisome division, dynamin-related protein, arabidopsis
In eukaryotic cells, peroxisomes are surrounded by single membranes and house a variety of oxidative metabolic pathways such as lipid metabolism, detoxification and plant photorespiration.1,2 To accomplish multiple tasks, the morphology, abundance and positioning of peroxisomes need to be highly regulated. Three families of proteins, whose homologs are present across different kingdoms, have been shown to be involved in peroxisome division in Arabidopsis. The PEX11 protein family is composed of five integral membrane proteins with primary roles in peroxisome elongation/tubulation, the initial step in peroxisome division.3–5 Although the exact function of PEX11s has not been demonstrated, these proteins are believed to participate in peroxisome membrane modification.6,7 The FIS1 family consists of two isoforms, which are C-terminal tail-anchored membrane proteins with rate limiting functions at the fission step.8,9 DRP3A and DRP3B belong to a superfamily of dynamin-related proteins, which are large and self-assembling GTPases involved in the fission and fusion of membranes by acting as mechanochemical enzymes or signaling GTPases.10 The function of PEX11 seems to be exclusive to peroxisomes, whereas DRP3 and FIS1 are shared by the division machineries of both peroxisomes and mitochondria in Arabidopsis.8,9,11–16 FIS1 proteins are believed to tether DRP proteins to the peroxisomal membrane,17,18 but direct evidence has not been obtained from plants. DRP3A and DRP3B share 77% sequence identity at the protein level and are functionally redundant in regulating mitochondrial division; however, DRP3A's role on the peroxisome seems stronger and cannot be substituted by DRP3B in peroxisome division.8,13,15
In a continuous effort to identify components of the plant peroxisome division apparatus from Arabidopsis, we performed genetic screens in a peroxisomal marker background expressing the YFP (yellow fluorescent protein)-PTS1 (peroxisome targeting signal 1, containing Ser-Lys-Leu) fusion protein. Mutants with defects in the morphology and abundance of fluorescently labeled peroxisomes are characterized. Following our analysis of the pdd1 mutant, which turned out to be a strong allele of DRP3A,8 we characterized the pdd2 mutant.
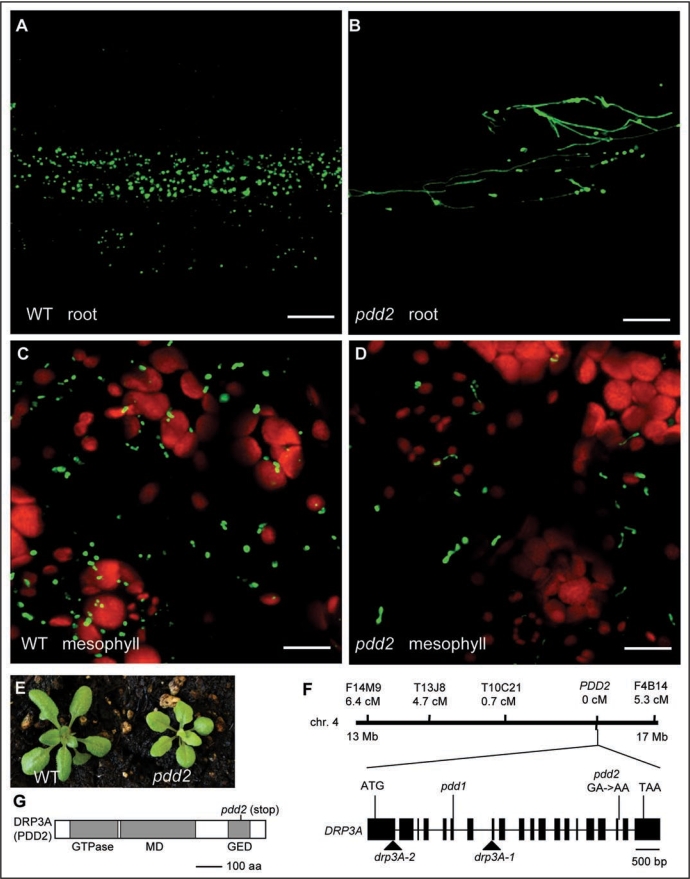
In root cells of the pdd2 mutant, extremely elongated peroxisomes and a beads-on-a-string peroxisomal phenotype are frequently observed (Fig. 1A and B). These peroxisome phenotypes resemble those of pdd1 and other strong drp3A alleles previously reported.8,15 However, the peroxisome phenotype seems to be less dramatic in leaf mesophyll cells. For instance, in addition to the decreased number of total peroxisomes, peroxisomes in leaf cells are only slightly elongated or exhibit a beads-on-a-string phenotype (Fig. 1C and D). Previously, we reported the phenotypes of three strong drp3A alleles, all of which contain a large number of peroxules, long and thin membrane extensions from the peroxisome,8 yet such peroxisomal structures are not observed in pdd2. On the other hand, pdd2 has a more severe growth phenotype than most drp3A alleles, as it is slow in growth and has pale green leaves (Fig. 1E). Genetic analysis showed that pdd2 segregates as a single recessive mutation (data not shown).
Figure 1.
Phenotypic analyses of pdd2 and identification of the PDD2 gene. (A–D) Confocal micrographs of root and mesophyll cells in 3-week-old wild type and pdd2 mutant plants. Green signals show peroxisomes; red signals show chloroplasts. Scale bars = 20 µm. (E) Growth phenotype of 3-week-old mutants. (F) Map-based cloning of the PDD2 gene. Genetic distance from PDD2 is shown under each molecular marker. Positions for mutations in previously analyzed drp3A alleles and pdd2 are indicated in the gene schematic. drp3A-1 and drp3A-2 are T-DNA insertion mutants, whereas pdd1 is an EMS mutant containing a premature stop codon in exon 6. (G) A schematic of the DRP3A (PDD2) protein with functional domains indicated. The pdd2 allele encodes a truncated protein lacking part of the GED domain.
The unique combination of peroxisomal and growth phenotypes of pdd2 prompted us to use map-based cloning to identify the PDD2 gene, with the hope to discover novel proteins in the peroxisome division machinery. A population of approximately 6,000 F2 plants (pdd2 × Ler) was generated. After screening 755 F2 mutants, the pdd2 mutation was mapped to the region between markers T10C21 and F4B14 on the long arm of chromosome 4 (Fig. 1F). Since this region contains DRP3A, we sequenced the entire DRP3A gene in pdd2 and identified a G→A transition at the junction of the 18th exon and intron (Fig. 1F). Further analysis revealed that the point mutation at this junction caused mis-splicing of intron 18, introducing a stop codon in the GTPase effector domain GED near the C terminus (Fig. 1G).
DRPs share with the classic dynamins an N-terminal GTPase domain, a middle domain (MD), and a regulatory motif named the GTPase effector domain (GED) (Fig. 1G). To date, a total of 26 drp3A mutant alleles carrying missense or nonsense mutations along the length of the DRP3A gene have been isolated.8,15 The combined peroxisomal and growth phenotype of pdd2 and the nature of the mutation in this allele are unique among all the drp3A alleles, indicating that the partial GED domain retained in pdd2 may have created some novel function for this protein. Further analysis of the truncated protein may be necessary to test this prediction.
Footnotes
Previously published online as a Plant Signaling & Behavior E-publication: http://www.landesbioscience.com/journals/psb/article/8699
References
- 1.Beevers H. Microbodies in higher plants. Annu Rev Plant Physiol. 1979;30:159–193. [Google Scholar]
- 2.Purdue PE, Lazarow PB. Peroxisome biogenesis. Annu Rev Cell Dev Biol. 2001;17:701–752. doi: 10.1146/annurev.cellbio.17.1.701. [DOI] [PubMed] [Google Scholar]
- 3.Lingard MJ, Trelease RN. Five Arabidopsis Peroxin 11 homologs individually promote peroxisome elongation, duplication or aggregation. J Cell Sci. 2006;119:1961–1972. doi: 10.1242/jcs.02904. [DOI] [PubMed] [Google Scholar]
- 4.Orth T, Reumann S, Zhang X, Fan J, Wenzel D, Quan S, Hu J. The PEROXIN11 protein family controls peroxisome proliferation in Arabidopsis. Plant Cell. 2007;19:333–350. doi: 10.1105/tpc.106.045831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nito K, Kamigaki A, Kondo M, Hayashi M, Nishimura M. Functional classification of Arabidopsis peroxisome biogenesis factors proposed from analyses of knockdown mutants. Plant Cell Physiol. 2007;48:763–774. doi: 10.1093/pcp/pcm053. [DOI] [PubMed] [Google Scholar]
- 6.Fagarasanu A, Fagarasanu M, Rachubinski RA. Maintaining peroxisome populations: a story of division and inheritance. Annu Rev Cell Dev Biol. 2007;23:321–344. doi: 10.1146/annurev.cellbio.23.090506.123456. [DOI] [PubMed] [Google Scholar]
- 7.Thoms S, Erdmann R. Dynamin-related proteins and Pex11 proteins in peroxisome division and proliferation. FEBS J. 2005;272:5169–5181. doi: 10.1111/j.1742-4658.2005.04939.x. [DOI] [PubMed] [Google Scholar]
- 8.Zhang X, Hu J. Two small protein families, DYNAMIN-RELATED PROTEIN3 and FISSION1, are required for peroxisome fission in Arabidopsis. Plant J. 2009;57:146–159. doi: 10.1111/j.1365-313X.2008.03677.x. [DOI] [PubMed] [Google Scholar]
- 9.Zhang X, Hu J. FISSION1A and FISSION1B proteins mediate the fission of peroxisomes and mitochondria in Arabidopsis. Mol Plant. 2008;1:1036–1047. doi: 10.1093/mp/ssn056. [DOI] [PubMed] [Google Scholar]
- 10.Praefcke GJ, McMahon HT. The dynamin superfamily: universal membrane tubulation and fission molecules? Nat Rev Mol Cell Biol. 2004;5:133–147. doi: 10.1038/nrm1313. [DOI] [PubMed] [Google Scholar]
- 11.Arimura S, Aida GP, Fujimoto M, Nakazono M, Tsutsumi N. Arabidopsis dynamin-like protein 2a (ADL2a), like ADL2b, is involved in plant mitochondrial division. Plant Cell Physiol. 2004;45:236–242. doi: 10.1093/pcp/pch024. [DOI] [PubMed] [Google Scholar]
- 12.Arimura S, Tsutsumi N. A dynamin-like protein (ADL2b), rather than FtsZ, is involved in Arabidopsis mitochondrial division. Proc Nat Acad Sci USA. 2002;99:5727–5731. doi: 10.1073/pnas.082663299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fujimoto M, Arimura SI, Mano S, Kondo M, Saito C, Ueda T, et al. Arabidopsis dynamin-related proteins DRP3A and DRP3B are functionally redundant in mitochondrial fission, but have distinct roles in peroxisomal fission. Plant J. 2009 doi: 10.1111/j.1365-313X.2009.03786.x. In press. [DOI] [PubMed] [Google Scholar]
- 14.Logan DC, Scott I, Tobin AK. ADL2a, like ADL2b, is involved in the control of higher plant mitochondrial morphology. J Exp Bot. 2004;55:783–785. doi: 10.1093/jxb/erh073. [DOI] [PubMed] [Google Scholar]
- 15.Mano S, Nakamori C, Kondo M, Hayashi M, Nishimura M. An Arabidopsis dynaminrelated protein, DRP3A, controls both peroxisomal and mitochondrial division. Plant J. 2004;38:487–498. doi: 10.1111/j.1365-313X.2004.02063.x. [DOI] [PubMed] [Google Scholar]
- 16.Scott I, Tobin AK, Logan DC. BIGYIN, an orthologue of human and yeast FIS1 genes functions in the control of mitochondrial size and number in Arabidopsis thaliana. J Exp Bot. 2006;57:1275–1280. doi: 10.1093/jxb/erj096. [DOI] [PubMed] [Google Scholar]
- 17.Kobayashi S, Tanaka A, Fujiki Y. Fis1, DLP1 and Pex11p coordinately regulate peroxisome morphogenesis. Exp Cell Res. 2007;313:1675–1686. doi: 10.1016/j.yexcr.2007.02.028. [DOI] [PubMed] [Google Scholar]
- 18.Koch A, Yoon Y, Bonekamp NA, McNiven MA, Schrader M. A role for Fis1 in both mitochondrial and peroxisomal fission in mammalian cells. Mol Biol Cell. 2005;16:5077–5086. doi: 10.1091/mbc.E05-02-0159. [DOI] [PMC free article] [PubMed] [Google Scholar]