Abstract
Background
Voltage-gated Cav1.2 calcium channels play a crucial role in Ca2+ signaling. The pore-forming α1C subunit is regulated by accessory Cavβ subunits, cytoplasmic proteins of various size encoded by four different genes (Cavβ1 - β4) and expressed in a tissue-specific manner.
Methods and Results
Here we investigated the effect of three major Cavβ types, β1b, β2d and β3, on the structure of Cav1.2 in the plasma membrane of live cells. Total internal reflection fluorescence microscopy showed that the tendency of Cav1.2 to form clusters depends on the type of the Cavβ subunit present. The highest density of Cav1.2 clusters in the plasma membrane and the smallest cluster size were observed with neuronal/cardiac β1b present. Cav1.2 channels containing β3, the predominant Cavβ subunit of vascular smooth muscle cells, were organized in a significantly smaller number of larger clusters. The inter- and intramolecular distances between α1C and Cavβ in the plasma membrane of live cells were measured by three-color FRET microscopy. The results confirm that the proximity of Cav1.2 channels in the plasma membrane depends on the Cavβ type. The presence of different Cavβ subunits does not result in significant differences in the intramolecular distance between the termini of α1C, but significantly affects the distance between the termini of neighbor α1C subunits, which varies from 67 Å with β1b to 79 Å with β3.
Conclusions
Thus, our results show that the structural organization of Cav1.2 channels in the plasma membrane depends on the type of Cavβ subunits present.
Introduction
Voltage-gated Cav1.2 calcium channels react to membrane depolarization by creating a rapid and transient increase in intracellular free Ca2+ concentration, thereby playing an essential role in initiation of calcium signaling in a wide variety of cells [1]. In order to exhibit this function, Cav1.2 calcium channels require association of the pore-forming α1C subunit with accessory Cavβ and α2δ subunits as well as calmodulin. Calcium channels are clustered rather than evenly distributed along the surface membrane of neurons [2]–[4] and cardiac myocytes [5]–[7]. Single-molecule imaging of the functional recombinant EYFPN-α1C/β2a/α2δ channels revealed clusters composed of ∼40 channels [8]. In neuronal cell bodies and proximal dendrites in hippocampus and cerebral cortex, Cav1.2 clusters of 1.5–2 µm in diameter were observed with anti-α1C antibody [9]. Using electron microscopy in bird and amphibian cardiac muscle [5], [6] and immuno-gold labeling in mammalian ventricular myocytes [7], [10] it was shown that Cav1.2 clusters are loosely tethered to ryanodine receptors (RyR) of the sarcoplasmic reticulum. Although association of calcium channels and ryanodine receptors appears to be weaker in cardiac myocytes than in skeletal muscle [11] and may involve different mechanisms of coupling [12], Cav1.2 clustering is essential for excitation-contraction coupling [13], [14].
Little is known about the factors affecting the structure of Cav1.2 clusters or the mechanisms of their formation. Because the carboxyl-terminal “IQ” region of α1C mediates the calmodulin-dependent Ca2+-induced inactivation of the channel [15]–[18], it is reasonable to suggest that both calmodulin and the cytoplasmic 750-amino acid C-tail of α1C have a role in the formation and maintenance of the Cav1.2 clusters. Indeed, a splice variant of α1C (α1C,86) deprived of IQ does not show a distinct tendency to form clusters [19]. The role of IQ sequences in intermolecular interactions between neighboring α1C molecules was experimentally confirmed in recent diffraction study [20]. The impact of bulky cytoplasmic Cavβ subunits on Cav1.2 structure and clustering is not known. Cavβ subunits are important differential modulators of the electrophysiological properties of calcium channels [21]–[23]. These peripheral proteins of variable size are encoded by four different genes (Cavβ1 - β4), some of them being subject to alternative splicing [24]. They have a common binding site in the cytoplasmic linker between repeats I and II of α1C known as the α-interaction domain (AID) [25]. Here, we applied total internal fluorescence reflection (TIRF) and three-color FRET microscopy to assess the effects of Cavβ on cluster size and density of Cav1.2 as well as to measure inter- and intramolecular distances between the N- and C-termini of α1C and the N-tails of β1b, β2d and β3. Our results demonstrated that Cav1.2 channels form plasma membrane clusters and revealed the effect of the type of Cavβ present on molecular distances and packing of the channels.
Results
Differential effect of Cavβ subunits on cluster organization of Cav1.2 channels
Cav1.2 calcium channels retain functional activity after fusion of fluorescent proteins to the N- and C-termini of α1C and to the N-terminus of Cavβ. In our experiments, we labeled α1C at the N-tail with monomeric mVenus (Vα1C) and/or at the C-tail with monomeric mCerulean (α1C C) [26]. To investigate the effect of Cavβ subtype on size and density of Cav1.2 clusters, we chose three major Cavβ variants, neuronal/cardiac β1b [27], cardiac β2d [28], [29] and neuronal/cardiac/vascular β3 [30]–[32], which is the predominant Cavβ subunit in vascular smooth muscle cells [31], [33]. The more commonly used β2a was excluded from the experiments because its N-tail is palmytoylated and anchored to the inner leaflet of the plasma membrane.
Fluorescent microscopy is a convenient approach to detect clusters of recombinant calcium channels as fluorescent foci or groupings of labeled molecules [34]. In this study, we used TIRF microscopy to visualize Cav1.2 clusters on the basal plasma membrane. Wavelet transform was used for the detection of clusters (see Methods and Figure 1A ) to estimate the effect of the type of Cavβ present on the Cav1.2 clusters size (Figure 1B ) and density (defined here as number of clusters per µm2 of the plasma membrane, Figure 1C ). The smallest Cav1.2 clusters were observed with β1b present. Cav1.2 clusters were significantly (P<0.001) larger with β2d (by ∼20%) and β3 present (by ∼30%) (Figure 1B ). We also found that the average density of the Vα1C/β1b clusters in the plasma membrane was 2.5 times higher (P<0.01) than Vα1C/β3, with β2d again taking an intermediate value (Figure 1C ). Thus, Cavβ subunits differentially regulate the architecture of the Cav1.2 clusters.
Figure 1. Effect of Cavβ subunits on cluster organization of Cav1.2 channels.
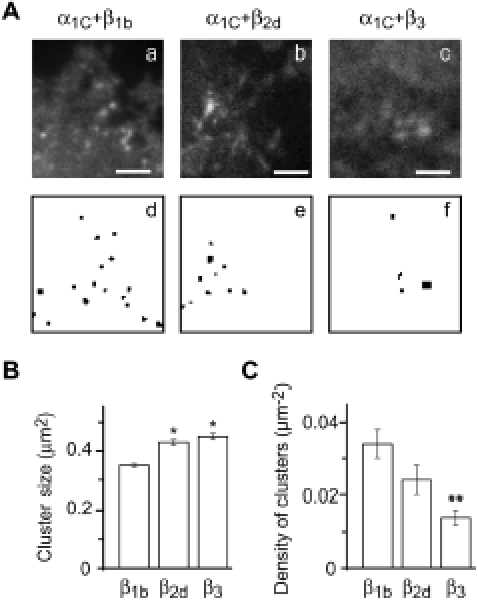
(A) TIRF images (a–c) and wavelet-derived clusters (d–f) of Cav1.2 channels containing β1b (a,d), β2d (b,e) or β3 (c,f). Scale, 4.5 µm. (B) Dependence of the average size of Cav1.2 clusters on the type of Cavβ present. β1b, mean size±SEM, 0.360±0.005 µm2 (number of clusters analyzed m = 1253); β2d, 0.430±0.013 mm2 (m = 270); β3, 0.450±0.017 µm2 (m = 205). *, P<0.001 relative to β1b. (C) Dependence of the number of Cav1.2 clusters (normalized to the area measured and defined as density) on the type of Cavβ present. β1b, mean number±SEM, 0.034±0.004 mm−2 (number of cells n = 27); β2d, 0.024±0.04 µm−2 (n = 30); β3, 0.014±0.02 µm−2 (n = 22). **, P<0.01 relative to β1b. Vα1C was co-expressed with α2δ and indicated Cavβ in COS1 cells.
In principle, the close proximity of channels within a cluster may generate intermolecular FRET between the V and C fluorophores of neighboring Vα1CC channels. This intermolecular FRET should be absent outside of clusters, where only intramolecular FRET should occur. The Vα1CC/α2δ/β3 channel was expressed in COS1 cells and two-color TIRF-FRET was measured inside and outside of clusters identified by wavelet transform. Based on FRET efficiency, a V-C distance (r)-frequency histogram of the total number of pixels within clusters revealed a possible bi-modal distribution, where a second (intermolecular) component of FRET is seen within clusters (Figure 2A ) but not outside of the clusters (Figure 2B ). Because TIRF microscopy captures only a small fraction of the cell plasma membrane, we used epifluorescent three-color FRET microscopy to quantitatively analyze the effect of Cavβ subtype on inter- and intra-molecular distance of Cav1.2 channels.
Figure 2. Intramolecular vs. intermolecular FRET in Vα1CC revealed in TIRF images.
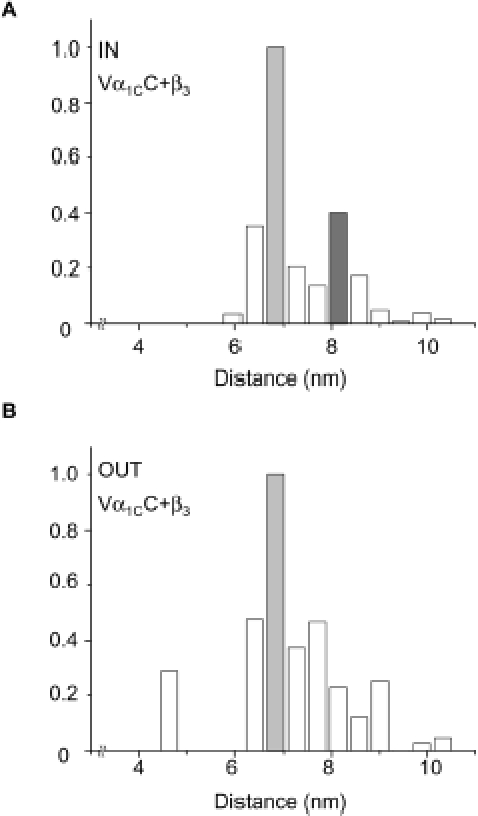
Vα1CC, α2δ and β3 were co-expressed in COS1 cells. Two-color FRET was measured in TIRF images and converted into distances r between V and C as described in Methods. Shown are normalized cumulative histograms (n = 11) for r calculated for ROI inside clusters (A, total number of pixels m = 231) and outside clusters (B, m = 3908) identified by wavelet transform. The same intramolecular (r V-C) distance ≈6.8 nm (light gray bars) was observed both inside and outside clusters, while intermolecular (r V∼C) distance ≈8.1 nm was observed only in clusters (dark gray).
The type of Cavβ present does not affect intramolecular distance between the N- and C-termini of the α1C subunit
We investigated the effect of Cavβ subtype on molecular distances in Cav1.2 channels by three-color FRET between Vα1CC and tagRFP (Rβ) fused to N-termini of β1b, β2d and β3. The advantage of three-color FRET cell microscopy applied to multisubunit complexes is that the method simultaneously detects the relative arrangement of the three different fluorophores (C, V and R) at a distance ≤2×R o, where R o is the Förster radius (R o(C-V) = 53 Å; R o(V-R) = 58 Å; R o(C-R) = 51 Å). Both mCerulean and mVenus are close analogs of GFP and can be approximated by a cylinder of 32×48 Å [35]. However, tagRFP [36], a monomeric analog of eqFP611, is larger in size and can be approximated by a cylinder of 34×54 Å [37]. Use of monomeric forms of fluorescent proteins excludes artifacts due to dimerization after expression [38]. The labeled constructs were co-expressed with α2δ in two different combinations as shown in Figure 3, and three-color FRET was measured using a multicube system [39]. Although membrane potential was not controlled during experiments, it was found to be on average −10.0±3.3 mV (n = 5) indicating that channels were predominantly in an inactivated state. In each fluorescent cell image, region of interest (ROI) was determined using a standard procedure as described earlier [40]. Within this ROI, only pixels with donor/acceptor ratio from 0.2 to 5 (Figure 4, left panels) were selected for further analysis [41]. FRET efficiency was determined according to [42] (Figure 4, middle panels) and then converted to the distance (r) between donor and acceptor (right panel) according to [43].
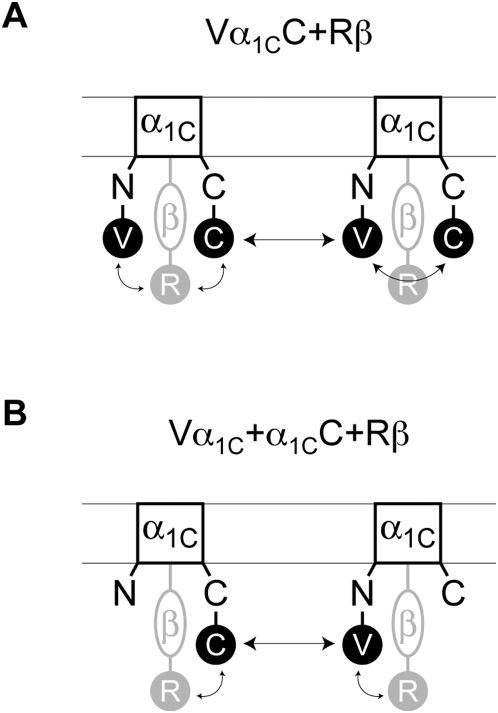
Figure 3. Investigated combinations of the labeled α1C and β subunits for three color FRET measurements.
Vα1CC and Rβ (A) and Vα1C, α1CC and Rβ (B) were co-expressed with α2δ (not shown). Arrows indicate revealed intramolecular and intermolecular distances.
Figure 4. Estimation of distance r between fluorophores fused to the N- and/or C-termini of the α1C subunit.
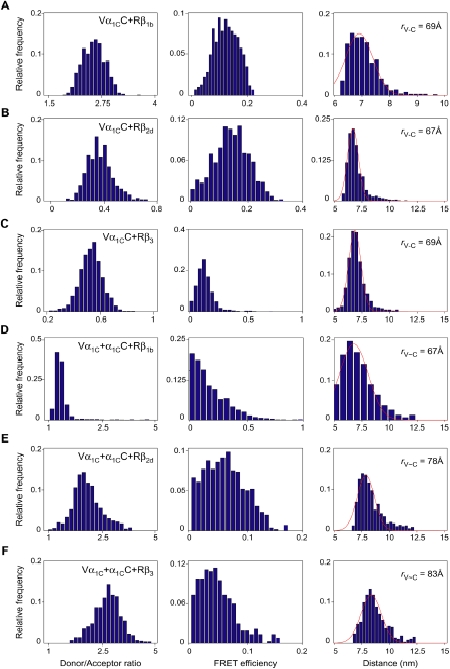
(A–C) Intramolecular FRET recorded with Vα1CC. (D–F) Intermolecular FRET recorded with Vα1C+α1CC. Channels were co-expressed in COS1 cells with α2δ and Rβ1b (A and D), Rβ2d (B and E) or Rβ3 (C and F). Shown are representative of histograms calculated from single exemplary cells for donor/acceptor ratio (left column), FRET efficiency (middle column) and distance (right column). Relative frequency was calculated for total number of pixels in ROI as described in Methods. The red solid line is the best fit to a Gaussian distribution with indicated means for r V-C and r V∼C.
Results of our measurements revealed that the tested Cavβ subunits did not affect intramolecular distance between the N- and C-termini of α1C. Measurement of FRET in the double-labeled Vα1CC co-expressed with α2δ and Rβ1b, Rβ2d or Rβ3 showed that the intramolecular distance r C-V between V and C did not vary significantly and was on average 68–69 Å, independent of the type of co-expressed Cavβ (Figure 4, A–C ; see Table 1 for statistics).
Table 1. Intra- and intermolecular distances between the Cav1.2 calcium channel α1C and β subunits measured by three-color FRET microscopy.
| Channel subunits | Measured distances (r) | β1b | β2d | β3 | ||
| r, Å | r, Å | c | r, Å | c | ||
| Vα1CC+Rβ | r C-V | 68±1 (17) | 68±2 (13) | 0.90±0.37 | 69±1 (19) | 0.60±0.05 |
| r C∼V | 72±3 (5) | 1.27±0.54 | 77±3 (6) | 1.16±0.17 | ||
| r V-R | 95±3 (13) | 99±3§ (8) | 1.70±0.27 | 90±2 (19) | 0.97±0.20 | |
| r V∼R | 107±1 (3) | 2.52±0.17 | 100±2 (15) | 1.72±0.23 | ||
| r C-R | 85±2‡ (13) | 84±2 (13) | 79±1 (14) | 0.70±0.10 | ||
| r C∼R | 85±1 (10) | 1.55±0.07 | ||||
| Vα1C+α1CC+Rβ | r C∼V | 67±1* (26) | 72±2 (13) | 79±4 (10) | ||
| r V-R | 90±2 (26) | 90±2 (13) | 90±5 (10) | |||
| r C-R | 78±1† (26) | 86±2 (6) | 80±4 (8) | |||
P<0.002 vs. β3.
P<0.05 vs. β2d.
P<0.05 vs. β3.
P<0.05 vs. r V-R in Vα1C+α1CC+Rβ2d.
FRET efficiency between the indicated fluorophores fused to the α1C and β1b, β2d or β3 subunits was measured in the plasma membrane of expressing COS1 cells and fitted to a Gaussian function. In cases when the routine curve fit showed two significantly different Gaussian distributions, the corresponding dispersion coefficients c (mean±SEM) are shown for both distances (see Experimental Procedures). V – mVenus, C- mCerulean, R – tagRFP. Shown values of r are mean±SEM. Number of cells is shown in parentheses.
Estimation of r C-V in the absence of Cavβ was not possible because of poor plasma membrane targeting by Vα1CC/α2δ under such conditions. To overcome this problem, we co-expressed Vα1CC and α2δ with tagRFP-labeled β2CED, a 42-amino acid fragment of β2 subunits which does not bind to AID, but interacts with the IQ region of the α1C subunit C-terminus, facilitates voltage gating and stimulates surface expression of the channel [44]. Results of FRET measurements showed r C-V = 68±1 Å (n = 22), essentially the same distance as that estimated when AID was occupied by Cavβ. Taken together, these results of our study suggest that type of Cavβ subunits present does not significantly affect the intramolecular distance between the N- and C-termini of α1C in Cav1.2 calcium channels.
Intermolecular distance between the α1C subunit N- and C-termini depends on the type of Cavβ present
Fitting of FRET data obtained with β2d and β3 to a sum of two Gaussian distributions (Table 1) revealed a statistically significant second component of Vα1CC FRET (Figure 5). Arising from neighboring Vα1CC molecules, this FRET provided estimates for the intermolecular distances (r C∼V) that were significantly different for β2d (72±3 Å, n = 5) and β3 (77±3 Å, n = 6). To verify our intermolecular distance measurements, we co-expressed a mixture of Vα1C and α1CC along with Rβ1b, Rβ2d or Rβ3 (Figure 4, D–F ). Any FRET between V and C in this recombinant system must be intermolecular FRET between termini of neighboring channels. Results, presented in Table 1, showed that intermolecular distances r C∼V measured in these complexes with β2d (72±2 Å, n = 13) and β3 (79±4 Å, n = 10) are not significantly different from the r C∼V values measured under the same conditions with Vα1CC. With β1b, the intermolecular distance r C∼V measured between Vα1C and α1CC was 67±1 Å (n = 26), a value not significantly different from the estimate for intramolecular Vα1CC distance (r V-C = 68±1 Å, n = 17). This explains why the data obtained in the presence of β1b were best fitted by a single Gaussian distribution. Thus, unlike β2d and β3, in the presence of β1b the inter- and intramolecular distances appear to be similar.
Figure 5. Intramolecular vs. intermolecular FRET in Vα1CC.
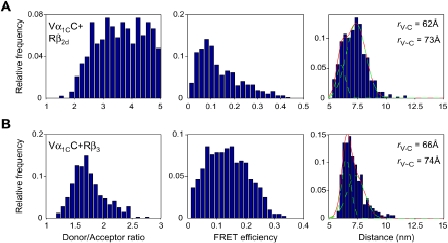
The Vα1CC subunit was co-expressed in COS1 cells with α2δ and Rβ2d (A) or Rβ3 (B). Shown are histograms of donor/acceptor ratio (left column), FRET efficiency (middle column) and distance (right column) determined in the plasma membrane region of two representative COS1 cells. The red solid line is the best fit to a sum of two Gaussian distributions with indicated means (green dotted lines) for intramolecular (r C-V) and intermolecular FRET (r C∼V).
The measurements with a mixture of Vα1C and α1CC confirm that Cav1.2 calcium channels containing β1b, β2d or β3 subunits are in close proximity to each other, thus supporting their clustering in the plasma membrane. The distance r C∼V between the N- and C-termini of the neighbor α1C subunits depends on the type of Cavβ. In the presence of β1b, the distance r C∼V (67±1 Å) is 1.2 nm smaller (P<0.002) than with β3 (79±4 Å), while r C∼V estimated in the presence of β2d (72±2 Å) is of an intermediate value. Subsequent measurements of three-color FRET between Rβ and the fluorophores of the α1C subunit added more certainty to this general picture (Figure 6A ).
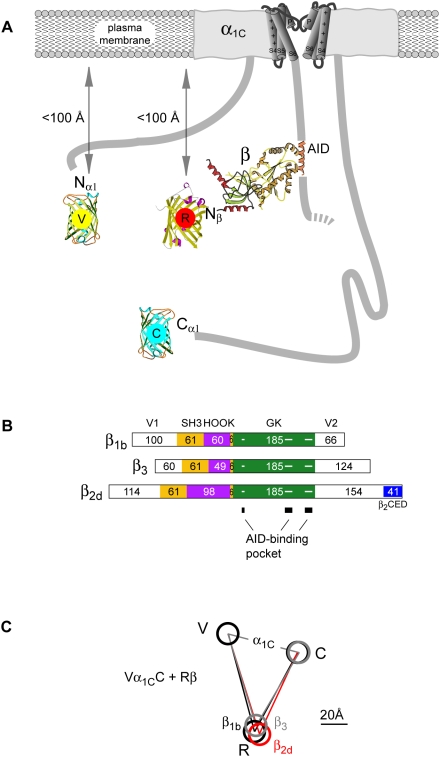
Figure 6. Molecular distances between the N- and C-termini of α1C and the Cavβ-subunit N-tail of β1b, β2d and β3.
(A) Schematic representation of Vα1CC with Rβ arranged under a vertically sliced α1C. The structures of TagRFP and Cavβ core MAGUK region were drawn based on PDB codes 1uisA [37] and 1t0j [62], respectively. FRET measurements with ECFP-labeled plekstrin homology domain in the inner leaflet of the plasma membrane [40], [63] showed that the N terminal tags of both the α1C and Cavβ subunits are located within the 2× Förster distance (<100 Å for ECFP/EYFP) from the plasma membrane. (B) Schematic representation of the domain organization of β1b, β2d and β3 aligned in regard to AID-binding guanylate kinase (GK) domain (green). Yellow box indicates the Src homology 3 (SH3) domain, purple the variable HOOK region, and blue the β2CED [44]. Number of amino acids is shown inside boxes. Amino acids involved in AID-binding pocket are marked in GK by three horizontal lines (for details see [62], [64], [65]). (C) Schematic representation of the results of simultaneous measurements of the molecular distances between three fluorophores shown in panel (A) in Vα1CC/α2δ/Rβ in the presence of Rβ1b (black lines), Rβ3 (gray lines) and Rβ2d (red lines).
FRET between tagRFP-labeled Cavβ and mCerulean/mVenus-labeled α1C
The three Cavβ subunits selected for our study vary in molecular mass (β1b, 53.2 kDa; β2d, 73.5 kDa; β3, 54.5 kDa) and in the size of the variable N-terminal (V1), central (HOOK) and C-terminal (V2) regions (see Figure 6B ). There are large differences between the three Cavβ subunits in variable regions on both sides of the AID-binding pocket, which anchors Cavβ to the I–II linker of α1C (Figure 6A ). In spite of that, the intramolecular distance r V-R between Rβ and Vα1C estimated in all tested three-color FRET combinations, including single- or double-labeled α1C (Vα1C+α1CC+Rβ, Vα1CC+Rβ), was not significantly different for all tested Cavβ subunits except for Rβ2d (see § in Table 1) Although the average distances r C-R between Rβ and Vα1CC were significantly different for Rβ1b (85±2 Å, n = 13) and Rβ3 (79±1 Å, n = 14), they were not significantly different between Rβ and α1CC. A superposition of all three simultaneously measured arrangements between Rβ and Vα1CC (Figure 6C ) illustrates differences in the positions of Rβ subunits as reflected by statistically significant differences in r V-R and r C-R (Table 1).
Fitting to a sum of two Gaussian distributions did not reveal the second (intermolecular) component of FRET between Vα1CC and Rβ1b (Table 1). However in the case of Rβ3 two intermolecular FRET components were clearly observed, one corresponding to the distance r V∼R = 100±2 Å (in 15 of 19 cells) and the other corresponding to r C∼R = 85±1 Å (in 10 of 14 cells). In the presence of Rβ2d, the latter component was not observed (n = 13), suggesting that the related distance r C∼R exceeded 102 Å. However, intermolecular FRET between Vα1C and Rβ2d was distinctly revealed in 3 out of 8 cells in a range close to the limits of resolution of the method with an estimate of r V∼R = 107±1 Å (Table 1). Taken together, FRET measurements between Rβ and the labeled tails of Vα1CC corroborated data on intermolecular FRET obtained with Vα1C+α1CC+Rβ and demonstrated that (a) calcium channels are in close proximity in the plasma membrane, and (b) both the intra- and intermolecular architecture of Cav1.2 channels depend on the type of Cavβ present.
Discussion
Cav1.2 calcium channels initiate Ca2+ signal transduction to many different downstream targets in wide variety of cells. Investigation of factors affecting structural organization of Cav1.2 channels is crucial for better understanding the mechanisms of Ca2+ signaling. The tendency of Cav1.2 channels to form clusters in the plasma membrane of different cell types has been poorly investigated. Here we studied effects of three major Cavβ subunits on structural organization of recombinant Cav1.2 channels expressed in COS1 cells. Because untransfected COS1 cells do not express endogenous calcium channels, they lack natural intracellular partners (e.g., cardiac RyR2) in proximity of exogenous Cav1.2 channels that might promote their clustering through “junctional” coupling [45]. However, recombinant Cav1.2 channels expressed in COS1 cells establish functional coupling to CREB-dependent transcriptional activation [46], pointing to a physiologically relevant integration of recombinant Cav1.2 into a naturally occurring signaling cascade with Ca2+/calmodulin-dependent protein kinase II mediating this activity in native cells [47].
TIRF microscopy revealed clusters of recombinant Cav1.2 channels in the plasma membrane of COS1 cells. The size and the plasma membrane density of the clusters significantly depend on the type of Cavβ present. This important observation suggests that the type of Cavβ present determines the structure of the Cav1.2 clusters. The average cluster size varies from 360 (β1b) to 450 nm2 (β3). Corroborating reasonable dimensions of these values, a mean size of the Cav1.2 cluster with the major cardiac β2d (430 nm2) is within the estimated size range (250–560 nm2) of rat ventricular RyR2 clusters [48].
Relative arrangement of α1C and Cavβ was estimated with sub-nanometer precision using three-color FRET microscopy in live cells with calcium channels in a stable, inactivated state. Our study revealed that in spite of substantial differences in molecular structure (Figure 6B ), the intramolecular distance between the α1C subunit tails does not significantly depend on the type of Cavβ present. Relative position of Rβ1b, Rβ2d and Rβ3 did not differ significantly. This is interesting because, unlike β1b and β3, β2d has a C-terminal β2CED domain, which interacts with the IQ region of the α1C C-tail [44].
Another important observation is that N- and C-termini of α1C and N-termini of Cavβ subunits of neighbor channels are in close (<120 Å) proximity to each other, which corroborates with the tendency of Cav1.2 to form clusters. Intermolecular distance between the α1C subunits significantly depends on the type of Cavβ and increases from 67 Å in the presence of β1b to 79 Å with β3. Measurements of FRET between Rβ and neighbor V/C-α1C supported this general picture and showed a significant effect of the type of Cavβ present on the relative position of neighbor channels.
Interestingly, freeze-fracture of the surface membrane revealed that distances between Cav1.2 channels trapped in cardiac junctions with RyR2 is variable and, on average, are larger than those identified by FRET [49]. It is known that the cytoskeleton and RyR2 associate with Cav1.2 plasma membrane clusters in heart cells [50]. Thus, one can not exclude that the distance between Cav1.2 channels in clusters in cardiac junctions is affected by RyR2. However, it is not clear whether clustering affects the ability of Cav1.2 channels to initiate Ca2+ signaling and whether every channel is responsive to depolarizing stimuli. In cardiac muscle cells, a single Cav1.2 opening triggers activity of 4–6 RyR2 [51]. The average size of a RyR2 cluster in ventricular myocytes plasma membrane is 250 nm2 (∼100 RyR2 molecules) [48] and interaction between Cav1.2 and RyR2 is weaker than that between Cav1.1 and RyR1 in skeletal muscle. Thus, activation of a RyR2 cluster may be mediated by random opening of few Cav1.2 channels in clusters located at a larger distance than that estimated by FRET.
Little is known about molecular determinants underlying physiologically important cluster organization of Cav1.2 channels in neurons [52]. It was shown recently that scaffolding proteins (AKAP79/150 and PDZ) participating in organizing plasma membrane signaling complexes in neurons were not responsible for organizing Cav1.2 channel clusters [53]. The involvement of Cavβ in Cav1.2 channel cluster organization, identified in our study, does not contradict the earlier report that the calmodulin-binding IQ region of α1C has a role in Cav1.2 clustering [19]. Because Cavβs interact with IQ [23], [44], it is possible that both act as concerted determinants in Cav1.2 channel clustering.
In conclusion, our study revealed effects of Cavβ subunits on the structural organization of Cav1.2 calcium channel in the plasma membrane in the absence of “junctional” interactions. It remains to be seen whether the observed differences in the cluster packing and arrangement of Cav1.2 contribute to the observed differences in calcium signaling among the cell types with preferential expression of a certain type of Cavβ [54]–[56].
Materials and Methods
Labeling α1C subunit with mVenus and/or mCerulean
To avoid dimerization, only monomeric forms of fluorescent proteins were used. The C-terminus of human Cav1.2 calcium channel α1C,77 subunit was amplified by PCR with sense 5′-ctattgaattcgatatcTGCCAGCAGCCTGGTGGAAGCG-3′ and antisense 5′-gtattaccggtggCAGGCTGCTGACGTAGACCCTGC-3′ primers. The PCR fragment was cleaved with ECoRI and AgeI and incorporated into an mCerulean-N1 [57] vector cleaved with the same enzymes, and the 5′-ECoRV/NotI-3′ fragment from the resulting plasmid was then incorporated into α1C,77-pCDNA3 cleaved with AleI and NotI, resulting in the mCeruleanC-α1C,77-pCDNA3 plasmid coding for α1CC. The 5′-NdeI/KpnI-3′ fragment from mVenus-C1 vector [26] was incorporated into α1C,77-pCDNA3 and mCeruleanC-α1C,77-pCDNA3 cleaved with the same enzymes to yield mVenusN-α1c,77-pCDNA3 and mVenusN-mCeruleanC-α1c,77-pCDNA3, respectively, coding for Vα1C and Vα1CC.
Labeling of Cavβ subunits with monomeric fluorescent tags
The cDNA of human β1b and β3 subunits was cloned from a human heart mRNA (Promega) by a nest RT-PCR strategy. For β1b, 5′-GACGGGCAGGGCGCCCACTAC-3′ was used as primer for the reverse transcription, sense 5′-GAGGCTCCTCTCCATGGTCCAG-3′ and antisense 5′-CCACTACATGGCATGTTCCTGC-3′ primers were used for the first round PCR, sense 5′-GCCACCATGGTCCAGAAGACCAG-3′ and antisense 5′-CACTACATGGCATGTTCCTGCTC-3′ primers were used for the second round PCR. For β3, primer 5′-CGCCTGTGCCTGCCAGGGTAGGGCAGCAGG-3′ was used for the reverse transcription, sense 5′-GACTCCCCCATGTATGACGAC-3′ and antisense 5′-GGCTGTCAGTAGCTATCCTTG-3′ primers were used for the first round PCR, sense 5′-GCCACCATGTATGACGACTCC-3′ and antisense 5′-TGTCAGTAGCTATCCTTGGGC-3′ primers were used for the second round PCR. The cDNA was cloned into a TA cloning vector pCR 2.1 (Invitrogen) and confirmed by DNA sequencing. The 5′-EcoRV/BamHI-3′ fragment of a β1b TA clone was incorporated into the pTagRFP-C vector (Evrogen, Moscow, Russia), which was cleaved with XhoI, filled in with Klenow and then cleaved with BamHI to generate RFP-β1b (Rβ1b). In a similar way the 5′-XhoI/HindIII-3′ fragment of a β3-TA clone was incorporated into the pTagRFP-C vector to generate monomeric Rβ3. To prepare RFP-β2d, β2d was amplified by PCR using mVenus-β2d [44] as template with sense primer 5′-CGGAGATCTATGGTCCAAAGGGACATGTC-3′ and antisense primer 5′-GGGGTCGACTCATTGGGGGATGTAAACATC-3′, and then the PCR product was cleaved with BglII and SalI, and incorporated into the pTagRFP-C vector cleaved with the same enzymes.
FRET calibration constructs
CTV, C5V, C39V, CVC and VCV were obtained from Drs. Ikeda and Vogel (NIAAA, NIH). The 5′-NheI/BsrGI:(Klenow-filled-in)-3′ fragments of mVenus-C1 and mCerulean-C1 were cloned into pTagRFP-C, which was cleaved with AgeI, filled in with Klenow and then cleaved with NheI, to make V4R and C4R respectively. The 5′-NheI/BamHI:(Klenow-filled-in)-3′ fragment of pTagRFP-C was cloned into mCerulean-N1, which was cleaved with EcoRI, filled in with Klenow and then cleaved with NheI, to make R39C. To prepare R17V and R17C, the 5′-NheI/XhoI:(Klenow-filled-in)-3′ fragment of pTagRFP-C was cloned, respectively, into mVenus-N1 and mCerulean-N1, which were cleaved with BamHI (Klenow filled in) and NheI. CTV was cleaved with BspEI, and the 0.7 Kb fragment was inserted into R17V and R17C to generate RTV and RTC, respectively. mCerulean was amplified by PCR using sense primer 5′-TATATCCGGAGATATCATGGTGAGCAAGGGCGAGGAG-3′ and antisense primer 5′-TATAGAATTCTTTGTACAGCTCGTCCATGCCGA-3′. After cleavage with BspEI and EcoRI, the PCR product of mVenus was inserted into pTagRFP-C to yield R5V; the PCR product of mCerulean was inserted into pTagRFP-C and C4R to yield R5C and CRC, respectively. RFP was amplified by PCR with sense primer 5′-TATAGAATTCGATATCATGGTGTCTAAGGGCGAAGAGCTG-3′ and antisense 5′-ATATGGTACCATTAAGTTTGTGCCCCAGTTTGCTAG-3′, cleaved with EcoRI and KpnI, and incorporated into R5V and R5C cleaved with the same enzymes to yield RVR and RCR, respectively.
Imaging
Images were recorded with a pixel size of ca. 200 nm using a 14-bit Hamamatsu C9100-12 digital camera (Hamamatsu City, Japan) mounted on a Nikon TE2000 epifluorescent microscope (Tokyo, Japan) equipped with a 60×1.45 numerical aperture (n.a.) oil objective and multiple filter sets (Chroma Technology, Rockingham, VT). Excitation light was delivered by a 175 W xenon lamp. Excitation filter sets were changed by a high-speed filter wheel system (Lambda 10-2, Sutter Instrument, Novato, CA). The Dual-View system (Optical Insights, Santa Fe, NM) was used for the simultaneous acquisition of two fluorescence images (donor and FRET). Images were collected and analyzed using C-Imaging (Compix, Cranberry Township, PA) and MATLAB v.7.0.4 (The Mathworks, Natick, MA).
Two-color FRET was quantified with three filter sets: for the yellow fluorescent protein (YFP) cube, excitation filter 500/20 nm, dichroic beam splitter 515 nm, emission filter 535/30 nm; for the cyan fluorescent protein (CFP) cube, excitation filter 436/20 nm, dichroic beam splitter 505 nm, emission filter 480/40 nm; for the FRET cube (CFP/YFP), excitation filter 436/20 nm, dichroic beam splitter 5051nm, emission filter 540/30 nm. For three-color FRET, the six-filter method described in [39] was used. All FRET images were acquired sequentially. For imaging mCerulean/mVenus pairs, the same filter arrangement as for two-color FRET was used. For the mCerulean/tagRFP combination, the following settings were used: for CFP cube, excitation filter 436/20 nm, dichroic beam splitter 505 nm, emission filter 480/40 nm; for RFP cube, excitation filter 555/28 nm, dichroic beam splitter 565 nm, emission filter 630/50 nm; for the FRET cube, excitation filter 436/20 nm, dichroic beam splitter 565 nm, emission filter 630/50 nm. With the mVenus/tagRFP combination, the following filter arrangement was used: for YFP cube, excitation filter 500/20 nm, dichroic beam splitter 515 nm, emission filter 535/30 nm; for RFP cube, excitation filter 555/28 nm, dichroic beam splitter 565 nm, emission filter 630/50 nm; for the FRET cube, excitation filter 484/15, dichroic beam splitter 565 nm, emission filter 630/50 nm. TIRF images were obtained with TIRF2 Nikon system mounted on Nikon TE2000 microscope and argon-ion laser with 514 nm line and diode laser with 440 nm line, dichroic beam splitter 505 nm, emission filters 470/30 nm and 550/30 nm.
Clusters within TIRF images were identified using 2D continuous wavelet transform similar to [58]. Images were analyzed using a two-dimensional mexican hat wavelet over scales 0.5 through 2 to identify ROI of locally increased signal fluorescence up to 5 µm2 in area. Similar approaches have been employed for cluster detection in clinical and cell biology imaging [46], [59], [60]. Corrected FRET intensity was calculated from data acquired using the three filter sets (CFP, YFP, and FRET) as described previously [40] using MATLAB. Briefly, corrected FRET values (FRETc) were calculated according to
where a and b are bleedthrough coefficients and IFRET, Id and Ia are FRET, donor and acceptor intensities.
Measurement of the G factor, which relates the increase in sensitized acceptor emission to the loss of donor fluorescence (quenching), is critical for calculating FRET efficiency (E) using the three-filter cube method. G factor is a constant for a particular fluorophore pair and imaging setup [42]. This method requires preparation of cDNA constructs encoding donor-acceptor fusion fluorescent proteins differing as widely as possible in FRET efficiency. This was accomplished by varying the length and composition of the linker residues connecting mCerulean and mVenus, mCerulean and tagRFP or mVenus and tagRFP. G factor was determined as
where Iaa1, Idd1 and Fc1 are acceptor, donor and corrected FRET intensity of the construct with the shortest linker between donor and acceptor, and Iaa2, Idd2 and Fc2 are acceptor, donor and corrected FRET intensity of the construct with the longest linker between donor and acceptor. Using this formula, we found G factors of 1.81 for the mCerulean/mVenus pair, 1.30 for the mVenus/tagRFP pair, and 0.38 for the mCerulean/tagRFP pair. These G factor values allowed us to calculate FRET efficiency according to [42] as follows:
The distance between two fluorophores was calculated in accordance with Förster theory:
The Förster distances (R0), the characteristic distance where the FRET efficiency is 50%, was calculated according to [61]:
where QD is the donor quantum yield, ε A is the maximal acceptor extinction coefficient, and J(λ) is the spectral overlap integral between the normalized donor fluorescence and the acceptor excitation spectra. All these parameters were calculated based on data obtained from Evrogen for tagRFP and reported for mCerulean and mVenus in [61]. Other parameters included coefficient C = 8.786×10−11 mol⌖L−1⌖cm⌖nm2, κ 2≤2/3 representing the angle between the two fluorophore dipoles assuming random orientation, and η≤1.4, the typical refractive index for biomolecules in aqueous solution [43]. The Förster distance estimated for mCerulean-mVenus was 5.3 nm, while R 0 for mCerulean-tagRFP was 5.1 nm and R 0 for mVenus-tagRFP was 5.8 nm. The k factor - the ratio of donor to acceptor (D/A) fluorescence intensity for equimolar concentrations in the absence of FRET, was determined for each construct in accordance with [42]:
The k factor for mCerulean/mVenus was calculated to be 0.41, while mVenus/tagRFP gave k = 1.60 and mCerulean/tagRFP gave k = 0.27.
D/A ratio for arbitrary concentrations of donor and acceptor was calculated according to [42]:
For corrected FRET efficiency measurements, this ratio should be in the range from 0.2 to 5.0 [41]. During analysis, the pixels with D/A ratio outside this range were eliminated from the FRET efficiency calculations.
Validation of G and k factors is presented in Figure S1 for two- and three-color FRET standards with different FRET efficiencies (linkers) and D/A stoichiometry. In our three-color FRET experiments, the major energy transfer was observed directly between mCerulean and tagRFP and not from cascade transfer through mVenus. If there would be a significant contribution of cascade FRET through mVenus, we would see a decrease in efficiency when we used two-color FRET (mCerulean/tagRFP) compared with three-color FRET, potentially including contributions from mCerulean/mVenus/tagRFP cascade. We did not observe a decrease in efficiency with two-color FRET, as experiments with Rβ3 and α1CC gave the same efficiency of 0.05 (r = 80 nm) as three-color FRET experiments with Rβ3, α1CC and Vα1C. Additional control experiments showed that the third fluorophore did not have a significant effect on mCerulean-mVenus FRET: we did not observe a significant difference between the distance between mCerulean/mVenus fluorophores (73±3, n = 10) measured by two-color FRET with Vα1CC and unlabeled β2d and that obtained with three-color FRET using Rβ2d and Vα1CC (68±2 nm, n = 13).
For each cell, we calculated FRET efficiency and distances (r) between fluorophores in each pixel of ROI. Gaussian fitting of the r distribution (20 bin histogram) was done in MATLAB using the fit function:
where b is the position of the center of the peak (mean) and c (dispersion coefficient) reflects the width of the distribution.
Supporting Information
FRET efficiency and donor/acceptor ratio of FRET standards. Shown are bar graphs summarizing the mean FRET efficiency (A) and the D/A ratio (B) for the indicated FRET calibration constructs. Data are presented as mean±SEM. (A) FRET efficiency values were: C4R (0.110±0.005), R39C (0.081±0.003), CTV (0.023±0.004), V4R (0.433±0.011), RTV (0.191±0.009), C5V (0.474±0.014), C39V (0.266±0.014) and CTV (0.179±0.006). (B) D/A ratios were: C4R (0.99±0.009), R39C (1.00±0.10), RTC (1.00±0.06), CRC (1.96±0.10), RCR (0.48±0.03), V4R (1.00±0.07), RTV (1.00±0.10), RVR (0.53±0.09), C5V (1.00±0.04), C39V (0.95±0.09), CTV (1.00±0.06), CVC (2.00±0.10) and VCV (0.54±0.02). The number of tested cells is shown in the bars. As one can see, increasing the length of the linker between the fluorophores significantly reduced FRET efficiency consistent with an increased distance between donor and acceptor. The measured mean D/A ratio corresponds well to the expected values of 1.0 (1∶1), 2.0 (2∶1) and 0.5 (1∶2). D/A ratios were also determined for the three-color construct CRV. D/A ratio and FRET efficiency were calculated in CRV independently for each pair of fluorophores. D/A ratio (EFRET) are: for CV, 1.00±0.07 (0.45±0.02); for VR, 1.10±0.10 (0.41±0.02), and for CR, 0.99±0.07 (0.20±0.01), n = 11. Thus, the D/A ratio of three-color standards well corresponds to the expected 1∶1 ratio.
(0.29 MB EPS)
Acknowledgments
The authors thank Dr. Stephen R. Ikeda and Dr. Steven S. Vogel for their gift of mVenus and mCerulean/mVenus fused constructs, and Dr. David W. Piston for his gift of mCerulean. The authors are grateful to Dr. Clara Franzini-Armstrong for critical discussion of the results and to Dr. Kenneth Fishbein for critically reading the manuscript.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by the NIA Intramural Research Program (Z01 AG000294-08 to NMS). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Clapham DE. Calcium signaling. Cell. 1995;80:259–268. doi: 10.1016/0092-8674(95)90408-5. [DOI] [PubMed] [Google Scholar]
- 2.Lipscombe D, Madison DV, Poenie M, Reuter H, Tsien RY, et al. Spatial distribution of calcium channels and cytosolic calcium transients in growth cones and cell bodies of sympathetic neurons. Proc Natl Acad Sci USA. 1988;85:2398–2402. doi: 10.1073/pnas.85.7.2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Silver RA, Lamb AG, Bolsover SR. Calcium hotspots caused by L-channel clustering promote morphological changes in neuronal growth cones. Nature. 1990;343:751–754. doi: 10.1038/343751a0. [DOI] [PubMed] [Google Scholar]
- 4.Westenbroek RE, Ahlijanian MK, Catterall WA. Clustering of L-type Ca2+ channels at the base of major dendrites in hippocampal pyramidal neurons. Nature. 1990;347:281–284. doi: 10.1038/347281a0. [DOI] [PubMed] [Google Scholar]
- 5.Franzini-Armstrong C, Protasi F, Ramesh V. Comparative ultrastructure of Ca2+ release units in skeletal and cardiac muscle. Ann NY Acad Sci. 1998;853:20–30. doi: 10.1111/j.1749-6632.1998.tb08253.x. [DOI] [PubMed] [Google Scholar]
- 6.Franzini-Armstrong C, Protasi F, Tijskens P. The assembly of calcium release units in cardiac muscle. Ann NY Acad Sci. 2005;1047:76–85. doi: 10.1196/annals.1341.007. [DOI] [PubMed] [Google Scholar]
- 7.Gathercole DV, Colling DJ, Skepper JN, Takagishi Y, Levi AJ, et al. Immunogold-labeled L-type calcium channels are clustered in the surface plasma membrane overlying junctional sarcoplasmic reticulum in guinea-pig myocytes-implications for excitation-contraction coupling in cardiac muscle. J Mol Cell Cardiol. 2000;32:1981–1994. doi: 10.1006/jmcc.2000.1230. [DOI] [PubMed] [Google Scholar]
- 8.Harms GS, Cognet L, Lommerse PHM, Blab GA, Kahr H, et al. Single-molecule imaging of L-type Ca2+ channels in live cells. Biophys J. 2001;81:2639–2646. doi: 10.1016/S0006-3495(01)75907-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hell JW, Westenbroek RE, Warner C, Ahlijanian MK, Prystay W, et al. Identification and differential subcellular localization of the neuronal class C and class D L-type calcium channel α1 subunits. J Cell Biol. 1993;123:949–962. doi: 10.1083/jcb.123.4.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takagishi Y, Yasui K, Severs NJ, Murata Y. Species-specific difference in distribution of voltage-gated L-type Ca2+ channels of cardiac myocytes. Am J Physiol Cell Physiol. 2000;279:C1963–1969. doi: 10.1152/ajpcell.2000.279.6.C1963. [DOI] [PubMed] [Google Scholar]
- 11.Di Biase V, Franzini-Armstrong C. Evolution of skeletal type e-c coupling: a novel means of controlling calcium delivery. J Cell Biol. 2005;171:695–704. doi: 10.1083/jcb.200503077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Woo S-H, Soldatov NM, Morad M. Modulation of Ca2+ signalling in rat atrial myocytes: possible role of the α1C carboxyl terminal. J Physiol (Lond) 2003;552:437–447. doi: 10.1113/jphysiol.2003.048330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shuai JW, Jung P. Optimal ion channel clustering for intracellular calcium signaling. Proc Natl Acad Sci U S A. 2003;100:506–510. doi: 10.1073/pnas.0236032100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Inoue M, Bridge JHB. Ca2+ sparks in rabbit ventricular myocytes evoked by action potentials: Involvement of clusters of L-type Ca2+ channels. Circ Res. 2003;92:532–538. doi: 10.1161/01.RES.0000064175.70693.EC. [DOI] [PubMed] [Google Scholar]
- 15.Pate P, Mochca-Morales J, Wu Y, Zhang J-Z, Rodney GG, et al. Determinants for calmodulin binding on voltage-dependent Ca2+ channels. J Biol Chem. 2000;275:39786–39792. doi: 10.1074/jbc.M007158200. [DOI] [PubMed] [Google Scholar]
- 16.Peterson BZ, DeMaria CD, Adelman JP, Yue DT. Calmodulin is the Ca2+ sensor for Ca2+-dependent inactivation of L-type calcium channels. Neuron. 1999;22:549–558. doi: 10.1016/s0896-6273(00)80709-6. [DOI] [PubMed] [Google Scholar]
- 17.Qin N, Olcese R, Bransby M, Lin T, Birnbaumer L. Ca2+-induced inhibition of the cardiac Ca2+ channel depends on calmodulin. Proc Natl Acad Sci USA. 1999;96:2435–2438. doi: 10.1073/pnas.96.5.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zühlke RD, Pitt GS, Deisseroth K, Tsien RW, Reuter H. Calmodulin supports both inactivation and facilitation of L-type calcium channels. Nature. 1999;399:159–162. doi: 10.1038/20200. [DOI] [PubMed] [Google Scholar]
- 19.Kepplinger KJF, Kahr H, Förstner G, Sonnleitner M, Schneider H, et al. A sequence in the carboxy-terminus of the α1C subunit important for targeting, conductance and open probability of L-type Ca2+ channels. FEBS Lett. 2000;477:161–169. doi: 10.1016/s0014-5793(00)01791-9. [DOI] [PubMed] [Google Scholar]
- 20.Fallon JL, Baker MR, Xiong L, Loy RE, Yang G, et al. Crystal structure of dimeric cardiac L-type calcium channel regulatory domains bridged by Ca2+·calmodulins. Proc Natl Acad Sci U S A published online before print. 2009;March 11, 2009:1–6. doi: 10.1073/pnas.0807487106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Colecraft HM, Alseikhan B, Takahashi SX, Chaudhuri D, Mittman S, et al. Novel functional properties of Ca2+ channel β subunits revealed by their expression in adult rat heart cells. J Physiol (Lond) 2002;541:435–452. doi: 10.1113/jphysiol.2002.018515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Varadi G, Lory P, Schultz D, Varadi M, Schwartz A. Acceleration of activation and inactivation by the β subunit of the skeletal muscle calcium channel. Nature. 1991;352:159–162. doi: 10.1038/352159a0. [DOI] [PubMed] [Google Scholar]
- 23.Zhang R, Dzhura I, Grueter CE, Thiel W, Colbran RJ, et al. A dynamic α-β inter-subunit agonist signaling complex is a novel feedback mechanism for regulating L-type Ca2+ channel opening. FASEB J. 2005;19:1573–1575. doi: 10.1096/fj.04-3283fje. [DOI] [PubMed] [Google Scholar]
- 24.Dolphin AC. β Subunits of voltage-gated calcium channels. J Bioenerg Biomembr. 2003;35:599–620. doi: 10.1023/b:jobb.0000008026.37790.5a. [DOI] [PubMed] [Google Scholar]
- 25.Pragnell M, De Waard M, Mori Y, Tanabe T, Snutch TP, et al. Calcium channel β-subunit binds to a conserved motif in the I-II cytoplasmic linker of the α1-subunit. Nature. 1994;368:67–70. doi: 10.1038/368067a0. [DOI] [PubMed] [Google Scholar]
- 26.Koushik SV, Chen H, Thaler C, Puhl HL, III, Vogel SS. Cerulean, Venus, and VenusY67C FRET reference standards. Biophys J. 2006;91:L99–101. doi: 10.1529/biophysj.106.096206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Powers PA, Liu S, Hogan K, Gregg RG. Skeletal muscle and brain isoforms of a β-subunit of human voltage-dependent calcium channels are encoded by a single gene. J Biol Chem. 1992;267:22967–22972. [PubMed] [Google Scholar]
- 28.Takahashi SX, Mittman S, Colecraft HM. Distinctive modulatory effects of five human auxiliary β2 subunit splice variants on L-type calcium channel gating. Biophys J. 2003;84:3007–3021. doi: 10.1016/S0006-3495(03)70027-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herzig S, Khan IFY, Grundemann D, Matthes J, Ludwig A, et al. Mechanism of Cav1.2 channel modulation by the amino terminus of cardiac β2-subunits. FASEB J. 2007;21:1527–1538. doi: 10.1096/fj.06-7377com. [DOI] [PubMed] [Google Scholar]
- 30.Castellano A, Wei X, Birnbaumer L, Perez-Reyes E. Cloning and expression of a third calcium channel β subunit. J BiolChem. 1993;268:3450–3455. [PubMed] [Google Scholar]
- 31.Collin T, Lory P, Taviaux S, Courtieu C, Guilbault P, et al. Cloning, chromosomal location and functional expression of the human voltage-dependent calcium-channel β3 subunit. Eur J Biochem. 1994;220:257–262. doi: 10.1111/j.1432-1033.1994.tb18621.x. [DOI] [PubMed] [Google Scholar]
- 32.Hullin R, Singer-Lahat D, Freichel M, Biel M, Dascal N, et al. Calcium channel β subunit heterogeneity: Functional expression of cloned cDNA from heart, aorta and brain. EMBO J. 1992;11:885–890. doi: 10.1002/j.1460-2075.1992.tb05126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murakami M, Yamamura H, Suzuki T, Kang M-G, Ohya S, et al. Modified cardiovascular L-type channels in mice lacking the voltage-dependent Ca2+ channel β3 subunit. J Biol Chem. 2003;278:43261–43267. doi: 10.1074/jbc.M211380200. [DOI] [PubMed] [Google Scholar]
- 34.Papadopoulos S, Leuranguer V, Bannister RA, Beam KG. Mapping sites of potential proximity between the dihydropyridine receptor and RyR1 in muscle using a cyan fluorescent protein-yellow fluorescent protein tandem as a fluorescence resonance energy transfer probe. J Biol Chem. 2004;279:44046–44056. doi: 10.1074/jbc.M405317200. [DOI] [PubMed] [Google Scholar]
- 35.Malo GD, Pouwels LJ, Wang M, Weichsel A, Montfort WR, et al. X-ray structure of cerulean GFP: A tryptophan-based chromophore useful for fluorescence lifetime imaging. Biochemistry. 2007;46:9865–9873. doi: 10.1021/bi602664c. [DOI] [PubMed] [Google Scholar]
- 36.Merzlyak EM, Goedhart J, Shcherbo D, Bulina ME, Shcheglov AS, et al. Bright monomeric red fluorescent protein with an extended fluorescence lifetime. Nat Meth. 2007;4:555–557. doi: 10.1038/nmeth1062. [DOI] [PubMed] [Google Scholar]
- 37.Wiedenmann J, Vallone B, Renzi F, Nienhaus K, Ivanchenko S, et al. Red fluorescent protein eqFP611 and its genetically engineered dimeric variants. J Biomed Optics. 2005;10:014003-014001-014007. doi: 10.1117/1.1854680. [DOI] [PubMed] [Google Scholar]
- 38.Zacharias DA, Violin JD, Newton AC, Tsien RY. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 2002;296:913–916. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]
- 39.Galperin E, Verkhusha VV, Sorkin A. Three-chromophore FRET microscopy to analyze multiprotein interactions in living cells. Nat Meth. 2004;1:209–217. doi: 10.1038/nmeth720. [DOI] [PubMed] [Google Scholar]
- 40.Kobrinsky E, Kepplinger KJF, Yu A, Harry JB, Kahr H, et al. Voltage-gated rearrangements associated with differential β-subunit modulation of the L-type Ca2+ channel inactivation. Biophys J. 2004;87:844–857. doi: 10.1529/biophysj.104.041152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Berney C, Danuser G. FRET or no FRET: A quantitative comparison. Biophys J. 2003;84:3992–4010. doi: 10.1016/S0006-3495(03)75126-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen H, Puhl HL, 3rd, Koushik SV, Vogel SS, Ikeda SR. Measurement of FRET efficiency and ratio of donor to acceptor concentration in living cells. Biophys J. 2006;91:L39–41. doi: 10.1529/biophysj.106.088773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lakowicz JR. Principles of Fluorescence Spectroscopy. New York: Springer; 2006. [Google Scholar]
- 44.Lao QZ, Kobrinsky E, Harry JB, Ravindran A, Soldatov NM. New determinant for the Cavβ2 subunit modulation of the Cav1.2 calcium channel. J Biol Chem. 2008;283:15577–15588. doi: 10.1074/jbc.M802035200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Protasi F, Sun XH, Franzini-Armstrong C. Formation and maturation of the calcium release apparatus in developing and adult avian myocardium. Dev Biol. 1996;173:265–278. doi: 10.1006/dbio.1996.0022. [DOI] [PubMed] [Google Scholar]
- 46.Mager DE, Kobrinsky E, Masoudieh A, Maltsev A, Abernethy DR, et al. Analysis of functional signaling domains from fluorescence imaging and the two-dimensional continuous wavelet transform. Biophys J. 2007;93:2900–2910. doi: 10.1529/biophysj.106.102582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wheeler DG, Barrett CF, Groth RD, Safa P, Tsien RW. CaMKII locally encodes L-type channel activity to signal to nuclear CREB in excitation-transcription coupling. J Cell Biol. 2008;183:849–863. doi: 10.1083/jcb.200805048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen-Izu Y, McCulle SL, Ward CW, Soeller C, Allen BM, et al. Three-dimensional distribution of ryanodine receptor clusters in cardiac myocytes. Biophys J. 2006;91:1–13. doi: 10.1529/biophysj.105.077180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun XH, Protasi F, Takahashi M, Takeshima H, Ferguson DG, et al. Molecular architecture of membranes involved in excitation-contraction coupling of cardiac muscle. J Cell Biol. 1995;129:659–671. doi: 10.1083/jcb.129.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scriven DRL, Klimek A, Lee KL, Moore EDW. The molecular architecture of calcium microdomains in rat cardiomyocytes. Ann NY Acad Sci. 2002;976:488–499. doi: 10.1111/j.1749-6632.2002.tb04783.x. [DOI] [PubMed] [Google Scholar]
- 51.Wang S-Q, Song L-S, Lakatta EG, Cheng H. Ca2+ signalling between single L-type Ca2+ channels and ryanodine receptors in heart cells. Nature. 2001;410:592–596. doi: 10.1038/35069083. [DOI] [PubMed] [Google Scholar]
- 52.Vacher H, Mohapatra DP, Trimmer JS. Localization and targeting of voltage-dependent ion channels in mammalian central neurons. Physiol Rev. 2008;88:1407–1447. doi: 10.1152/physrev.00002.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Di Biase V, Obermair GJ, Szabo Z, Altier C, Sanguesa J, et al. Stable membrane expression of postsynaptic Cav1.2 calcium channel clusters is independent of interactions with AKAP79/150 and PDZ proteins. J Neurosci. 2008;28:13845–13855. doi: 10.1523/JNEUROSCI.3213-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bers DM. Calcium cycling and signaling in cardiac myocytes. Annu Rev Physiol. 2008;70:23–49. doi: 10.1146/annurev.physiol.70.113006.100455. [DOI] [PubMed] [Google Scholar]
- 55.Calin-Jageman I, Lee A. Cav1 L-type Ca2+ channel signaling complexes in neurons. J Neurochem. 2008;105:573–583. doi: 10.1111/j.1471-4159.2008.05286.x. [DOI] [PubMed] [Google Scholar]
- 56.Floyd R, Wray S. Calcium transporters and signalling in smooth muscles. Cell Calcium. 2007;42:467–476. doi: 10.1016/j.ceca.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 57.Rizzo MA, Piston DW. High-contrast imaging of fluorescent protein FRET by fluorescence polarization microscopy. Biophys J. 2005;88:L14–16. doi: 10.1529/biophysj.104.055442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kammermeier P. Surface clustering of metabotropic glutamate receptor 1 induced by long Homer proteins. BMC Neurosci. 2006;7:1. doi: 10.1186/1471-2202-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee KL, Orr M, Lithgow B. :7664–7667. doi: 10.1109/IEMBS.2005.1616287. A Novel Wavelet-Statistics Based Feature Detection System for Detecting Microcalcifications; 2005. [DOI] [PubMed] [Google Scholar]
- 60.Papadopoulos A, Fotiadis DI, Costaridou L. Improvement of microcalcification cluster detection in mammography utilizing image enhancement techniques. Comput Biol Med. 2008;38:1045–1055. doi: 10.1016/j.compbiomed.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 61.Kremers GJ, Goedhart J, vanMunster EB, Gadella TWJ. Cyan and yellow super fluorescent proteins with improved brightness, protein folding, and FRET Förster radius. Biochemistry. 2006;45:6570–6580. doi: 10.1021/bi0516273. [DOI] [PubMed] [Google Scholar]
- 62.Van Petegem F, Clark KA, Chatelain FC, Minor DL., Jr Structure of a complex between a voltage-gated calcium channel β-subunit and an α-subunit domain. Nature. 2004;429:671–675. doi: 10.1038/nature02588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kobrinsky E, Tiwari S, Maltsev VA, Harry JB, Lakatta E, et al. Differential role of the α1C subunit tails in regulation of the Cav1.2 channel by membrane potential, β subunits, and Ca2+ ions. J Biol Chem. 2005;280:12474–12485. doi: 10.1074/jbc.M412140200. [DOI] [PubMed] [Google Scholar]
- 64.Chen Y-h, Li M-h, Zhang Y, He L-l, Yamada Y, et al. Structural basis of the α1-β subunit interaction of voltage-gated Ca2+ channels. Nature. 2004;429:675–680. doi: 10.1038/nature02641. [DOI] [PubMed] [Google Scholar]
- 65.Opatowsky Y, Chen C-C, Campbell KP, Hirsch JA. Structural analysis of the voltage-dependent calcium channel β subunit functional core and its complex with the α1 interaction domain. Neuron. 2004;42:387–399. doi: 10.1016/s0896-6273(04)00250-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FRET efficiency and donor/acceptor ratio of FRET standards. Shown are bar graphs summarizing the mean FRET efficiency (A) and the D/A ratio (B) for the indicated FRET calibration constructs. Data are presented as mean±SEM. (A) FRET efficiency values were: C4R (0.110±0.005), R39C (0.081±0.003), CTV (0.023±0.004), V4R (0.433±0.011), RTV (0.191±0.009), C5V (0.474±0.014), C39V (0.266±0.014) and CTV (0.179±0.006). (B) D/A ratios were: C4R (0.99±0.009), R39C (1.00±0.10), RTC (1.00±0.06), CRC (1.96±0.10), RCR (0.48±0.03), V4R (1.00±0.07), RTV (1.00±0.10), RVR (0.53±0.09), C5V (1.00±0.04), C39V (0.95±0.09), CTV (1.00±0.06), CVC (2.00±0.10) and VCV (0.54±0.02). The number of tested cells is shown in the bars. As one can see, increasing the length of the linker between the fluorophores significantly reduced FRET efficiency consistent with an increased distance between donor and acceptor. The measured mean D/A ratio corresponds well to the expected values of 1.0 (1∶1), 2.0 (2∶1) and 0.5 (1∶2). D/A ratios were also determined for the three-color construct CRV. D/A ratio and FRET efficiency were calculated in CRV independently for each pair of fluorophores. D/A ratio (EFRET) are: for CV, 1.00±0.07 (0.45±0.02); for VR, 1.10±0.10 (0.41±0.02), and for CR, 0.99±0.07 (0.20±0.01), n = 11. Thus, the D/A ratio of three-color standards well corresponds to the expected 1∶1 ratio.
(0.29 MB EPS)