Abstract
The second exon of lymphocyte antigen receptor genes is assembled in developing lymphocytes from component V, J and, in some cases, D gene segments through the process of V(D)J recombination. This process is initiated by an endonuclease comprised of the Rag-1 and Rag-2 proteins, collectively referred to as Rag. Rag binds to recombination signals (RSs) and catalyzes the pair-wise introduction of DNA double strand breaks (DSBs) at recombining gene segments. DNA cleavage by Rag is restricted both by intrinsic features of RSs, as well as the activity of other cis-acting elements, such as promoters and enhancers that regulate the accessibility of gene segments to Rag. In the TCRβ locus, accessibility of the Dβ1-Jβ1 gene segment cluster relies on the function of an enhancer, Eβ, and a promoter, PDβ1. Here we demonstrate that deletion of a small genomic region containing 5 of the 6 Jβ1 gene segments, but no known transcriptional regulatory elements, leads to a marked decrease in transcription and rearrangements involving the Dβ1 and Jβ1.1 gene segments. Surprisingly, point mutations in the RS of the Jβ1.1 gene segment not only impact Rag cleavage, but also lead to diminished transcription through the Dβ1-Jβ1 gene segment cluster. Our findings demonstrate that cis-acting elements that regulate transcription and accessibility of the TCRβ locus may functionally overlap with RS sequences, which are known primarily to direct Rag-mediated cleavage.
Keywords: T cell receptor, recombination signal, accessibility, transcription, VDJ rearrangement
1. Introduction
Developing B and T lymphocytes must express heterodimeric B cell receptors (BCRs) and T cell receptors (TCRs), respectively, in order to fully traverse their different developmental checkpoints. The genes that encode these receptor chains are assembled during development from component variable (V), joining (J) and, in some cases, diversity (D) gene segments by the process of V(D)J recombination (Tonegawa, 1983). This process is initiated by the recombinase activating gene (Rag) -1 and -2 proteins, that together form an endonuclease, hereafter referred to as Rag, which introduces DNA double strand breaks (DSBs) at the border of two recombining gene segments and their flanking recombination signals (RSs) (Fugmann et al., 2000; Gellert, 2002; Oettinger, 1999). This DNA cleavage generates a pair of coding ends and a pair of signal ends that are processed into a coding joint and a signal joint, respectively, by proteins of the non-homologous end-joining pathway of DNA DSB repair (Bassing et al., 2002; Rooney et al., 2004).
The V(D)J recombination reaction is regulated at several important levels. Firstly, it is lineage specific with complete assembly of immunoglobulin (Ig) heavy (H) and light (L) chain genes occurring only in B cells, and complete assembly of TCR α and β chain genes occurring only in T cells (Bassing et al., 2002; Cobb et al., 2006). Secondly, recombination is developmental stage specific, with IgH chain genes being assembled prior to IgL chain genes during B cell development and with TCRβ chain genes being assembled prior to TCRα chain genes during T cell development (Bassing et al., 2002; Cobb et al., 2006). Thirdly, intra-allelic constraints are imposed upon the assembly of some loci with D gene segments, such as the IgH and TCRβ chain genes, with D to J rearrangement preceding V to DJ rearrangement (Alt et al., 1984; Bassing et al., 2002; Cobb et al., 2006; Khor and Sleckman, 2005). Finally, inter-allelic regulation allows rearrangement of the TCRβ, IgH and IgL chain genes to be regulated in the context of allelic exclusion, which is enforced at the V to DJ step of rearrangement, ensuring that mature B and T cells each express a single antigen receptor (Bassing et al., 2002; Bergman, 1999; Cobb et al., 2006; Khor and Sleckman, 2002).
The appropriate assembly of antigen receptor chain genes relies on the coordinated activities of several cis-acting DNA elements. These include promoters and enhancers that drive the transcription and modulate the chromatin structure of unrearranged V, D and J gene segments in a way that permits their accessibility to the Rag proteins (Bassing et al., 2002; Cobb et al., 2006). Assembly is also regulated by RSs, the sequences recognized by the Rag proteins, which restrict the gene segments that can undergo recombination. RSs are composed of conserved heptamer and nonamer sequences that flank either a non-conserved 12 or 23 base pair spacer sequence (Tonegawa, 1983). Synapsis and Rag cleavage only occurs between pairs of RSs with dissimilar spacer lengths, a restriction known as the 12/23 rule (Fugmann et al., 2000; Gellert, 2002; Oettinger, 1999; Tonegawa, 1983). However, not all 12/23 RS combinations mediate efficient V(D)J recombination, demonstrating that RSs impose additional restrictions on this reaction, termed B12/23 (Bassing et al., 2000; Bassing et al., 2002).
The murine TCRβ locus spans approximately 250 kb and includes 23 Vβ gene segments distributed in a 150kb region in the 5′ portion of the locus, followed by two Dβ-Jβ gene segment clusters (Dβ1-Jβ1 and Dβ2-Jβ2) each with one Dβ and six Jβ gene segments and an associated constant region gene (Cβ1 and Cβ2) (Fig. 1A) (Glusman et al., 2001). A single Vβ gene segment, Vβ14, lies in the most 3′ region of the locus (Fig. 1A). The Vβ and Jβ gene segments are flanked by 23-RSs and 12-RSs, respectively, while the Dβ gene segments have 5′ 12-RSs and 3′ 23-RSs. Assembly of TCRβ chain genes is regulated intra- and inter- allelically as well as within the context of allelic exclusion (Khor and Sleckman, 2002; Khor and Sleckman, 2005).
Figure 1. Generation of the Jβ1σ allele.
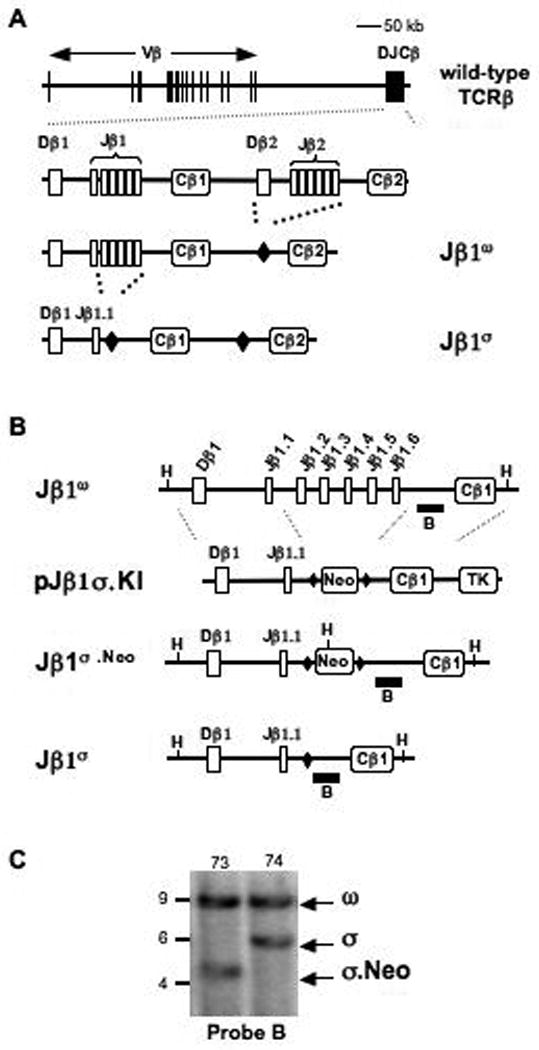
A-B. Schematics of the wild-type TCRβ locus drawn to scale and enlarged DJβ1-Cβ1-DJβ2-Cβ2 regions on the Jβ1ω allele where the DJβ2 gene cluster has been deleted and replaced by a single loxP site (filled diamond) and the Jβ1σ allele where Jβ1.2 to Jβ1.6 on the Jβ1ω allele has been deleted and replaced with a single loxP site. A schematic of the pJβ1σ.KI targeting construct is shown including the loxP flanked neomycin resistance gene (Neo) and the thymidine kinase (TK) gene. Relevant HindIII sites (H), and the location of probe B (solid bar) are indicated. C. Southern analysis of HindIII-digested genomic DNA from Jβ1ω/σ.Neo (73) and Jβ1ω/σ (74) ES cells that was probed with probe B. The bands generated by the Jβ1ω (ω), Jβ1σ.Neo (σ.Neo), and Jβ1σ (σ) alleles are indicated as are the molecular weight standards (kb).
The TCRβ locus has a single defined enhancer, Eβ, which lies in the most 3′ region of the locus with deletion of this element leading to a near complete block in TCRβ chain gene assembly (Bories et al., 1996; Bouvier et al., 1996; McDougall et al., 1988). Eβ functions with a promoter, PDβ1, which lies just upstream of Dβ1, to promote transcription and accessibility of the Dβ1-Jβ1 gene segment cluster (Cobb et al., 2006; Doty et al., 1999; Oestreich et al., 2006; Sikes et al., 1998; Sikes et al., 1999; Whitehurst et al., 1999; Whitehurst et al., 2000). There is also a promoter, PDβ2, associated with the Dβ2-Jβ2 gene segment cluster that would be expected to function with Eβ to promote accessibility of the Dβ2-Jβ2 gene segments (McMillan and Sikes, 2008). The RSs associated with the Vβ, Dβ and Jβ gene segments regulate TCRβ chain gene assembly through both 12/23 restrictions and B12/23 restrictions. At the TCRβ locus the B12/23 ruleprevents the direct joining of Vβ and Jβ gene segments despite the 12/23 compatibility of their flanking RSs (Bassing et al., 2000).
Here, through the generation and analysis of several modified TCRβ alleles, we show that deleting a small region of the TCRβ locus harboring 5 of the 6 Jβ1 gene segments and their RSs leads to a dramatic reduction in germline transcription through this region and a concomitant reduction in Dβ1 to Jβ1 rearrangement. Furthermore, point mutations of the Jβ 12-RS, but not of the 3′Dβ 23-RS, that diminish Rag activity also lead to an unexpected reduction in germline transcription and accessibility of this region. These findings demonstrate a novel overlap between the functions of cis-acting elements that regulate transcription and accessibility and those which control Rag binding and cleavage.
2. Materials and Methods
2.1 Generating the targeting constructs
The pJβ1σ.KI targeting vector was generated from pLNTK using a 5′ homology arm that extends 4.5 kb 3′ of the KpnI site upstream of Dβ1, and a 3′ homology arm that extends 6.4 kb 3′ of the SacI site just downstream of Jβ1.6. Standard PCR based site-directed mutagenesis techniques were used to modify the Jβ1.1 and Dβ1 3′ RS sequences as shown in Fig. 4 to generate pJβ1σ.1.KI and pJβ1σ.2.KI, which were identical to the pJβ1σ.KI targeting vector except for the indicated point mutations (Khor et al., 2006).
Figure 4. Transcription on the Jβ1σ allele during development.
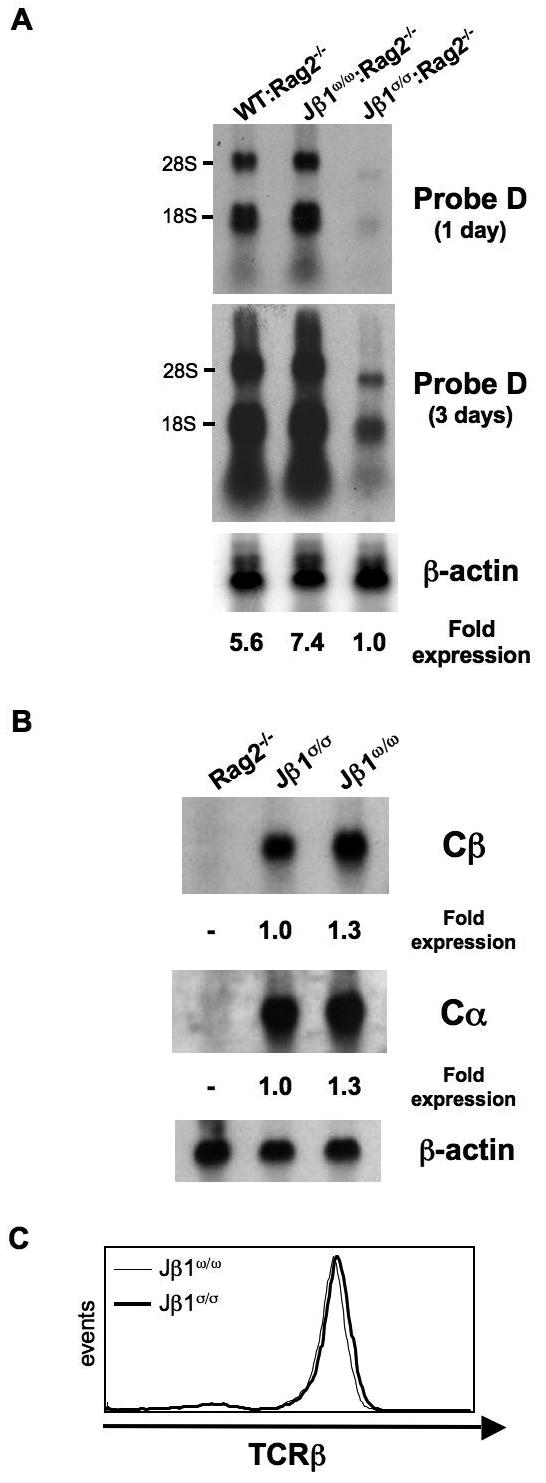
A. Northern blot analyses of thymocytes from Rag-2-/- mice with a wild type TCRβ locus (WT:Rag-2-/-) and from Jβ1ω/ω:Rag-2-/- and Jβ1σ/σ:Rag-2-/- mice using probe D (see Fig. 3D). The relative level of probe D hybridizing germline Dβ1-Jβ1 transcripts in each lane is indicated, normalized to β-actin expression. B. Northern blot analyses of splenocytes from Jβ1ω/ω and Jβ1σ/σ mice using probes to the Cβ and Cα constant region genes. The relative level of Cβ and Cα hybridizing mature transcripts is indicated, normalized to β-actin expression. C. Flow cytometric analyses showing TCRβ expression on Thy1 expressing (T cells) Jβ1ω/ω and Jβ1σ/σ splenocytes.
2.2 Embryonic stem cells
The generation, culture and gene targeting of embryonic stem cells was carried out as previously described (Khor et al., 2006; Kim et al., 2005).
2.3 Flow cytometric analyses
Flow cytometric analyses were carried out on thymocytes, splenocytes and peripheral lymph node cells as previously described using FITC-conjugated anti-CD8 and anti-TCRβ (Pharmingen), PE-conjugated anti-CD4 (Pharmingen) and CyC-conjugated anti-CD8 (Pharmingen)(Huang et al., 2005). Flow cytometric analyses were performed on a FACSVantage (Becton-Dickenson).
2.4 Southern blot, Northern blot and PCR analyses
Genomic DNA and RNA were isolated and analyzed by Southern and Northern blot, respectively, as previously described (Khor et al., 2006; Khor and Sleckman, 2005). TCRβ locus probes B and D and the probes to the TCRβ and TCRα constant region genes (Cβ and Cα), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), β actin and RAG-2 have been previously described (Khor and Sleckman, 2005; Sleckman et al., 1997). Quantification was carried out using a Molecular Dynamics phosphoimager and Imagequant software. PCR amplification was performed using 500 ng of genomic DNA, 20 pmol of each primer and Taq polymerase in 50 μl reaction volume with 1 mM MgCl2. Amplification conditions were 92°C for 60 seconds, 65°C for 90 seconds and 72°C for 90 seconds cycled 30 times. Vβ-Dβ1 rearrangements were amplified using Vβ-specific primers and a primer downstream of Dβ1 as previously described (Sleckman et al., 2000).
2.5 Generation of αβ T cell hybridomas
αβ T cell hybridomas were generated using ConA stimulated lymph node T cells from Jβ1ω/ω, Jβ1σ/σ and Jβ1σ/ω mice as previously described (Khor and Sleckman, 2005).
3. Results
3.1 A TCRβ allele (Jβ1σ) with a single Dβ and Jβ gene segment
To generate a TCRβ allele (Jβ1σ) with a single Dβ (Dβ1) and Jβ (Jβ1.1) gene segment, we initially made embryonic stem cells from Jβ1ω/ω mice (Fig. 1A) (Bassing et al., 2000). The Jβ1ω allele is identical to the TCRβ allele except that a 2kb region containing the Dβ2 gene segment and the 6 Jβ2 gene segments has been replaced with a single loxP site (Fig. 1A) (Bassing et al., 2000). Therefore, TCRβ rearrangements on the Jβ1ω allele are restricted to the Dβ1-Jβ1 gene segment cluster, which contains one Dβ (Dβ1) and six Jβ (Jβ1.1 to Jβ1.6) gene segments. In order to generate a TCRβ allele with only a single Dβ and Jβ gene segment, we used the pJβ1σ.KI targeting vector to replace a 1.5 kb region of the Jβ1ω allele, which contains 5 of the 6 Jβ1 gene segments (Jβ1.2 thru Jβ1.6), with the loxP-flanked neomycin resistance gene, creating the Jβ1σ.Neo allele (Fig. 1B and C). The Jβ1σ allele was derived from the Jβ1σ.Neo allele through Cre-mediated deletion of the neomycin resistance gene (Fig. 1B and C). Thus, the Jβ1σ allele is identical to the wild-type TCRβ locus except for the deletion of a 2 kb region, containing the Dβ2-Jβ2 gene segment cluster, and a 1.5 kb region, containing 5 Jβ1 gene segments. As a result, all TCRβ chain genes assembled on the Jβ1σ allele use the Dβ1 and Jβ1.1 gene segments.
3.2 Reduced efficiency of TCRβ chain gene assembly on the Jβ1σ allele
Analyses of Jβ1σ/σ mice revealed that they have similar numbers of total thymocytes, as well as normal distributions of CD4-/CD8- (double negative, DN), CD4+/CD8+ (double positive, DP), CD4+ and CD8+ (single positive, SP) thymocytes as compared to wild-type mice (Fig. 2). The Jβ1ω allele rearranges efficiently, as evidenced by analyses of T cell hybridomas generated from mature Jβ1ω/ω αβ T cells. These studies showed that none of the cells had an un-rearranged Jβ1ω allele, and that an expectedly large fraction of these cells had completely rearranged both Jβ1ω alleles in the VDJβ/VDJβ configuration (Fig. 3A) (Bassing et al., 2000; Khor and Sleckman, 2002; Khor and Sleckman, 2005). In surprising contrast, analyses of T cell hybridomas generated from mature Jβ1σ/σ αβ T cells revealed that 22% of those cells had a un-rearranged Jβ1σ allele (Fig. 3A).
Figure 2. Thymocyte development in Jβ1σ/σ mice.
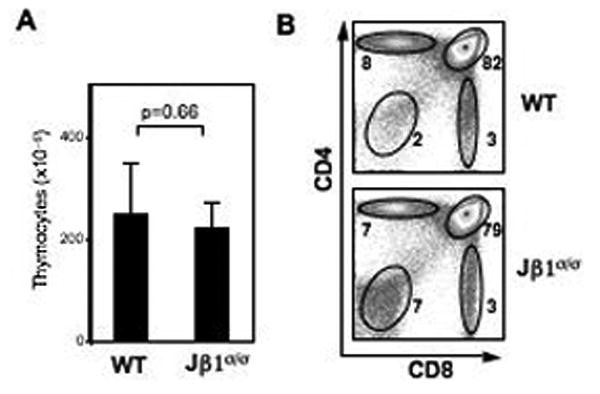
A. Numbers of total thymocytes in Jβ1σ/σ (n=4) and wild-type (WT) (n=4) mice. B. Flow cytometric analysis of thymocyte development, using anti-CD4 and anti-CD8, in WT and Jβ1σ/σ mice. Percentages are indicated beside each gate. Results are representative of at least 3 independent experiments.
Figure 3. Inefficient rearrangement of the Jβ1σ allele.
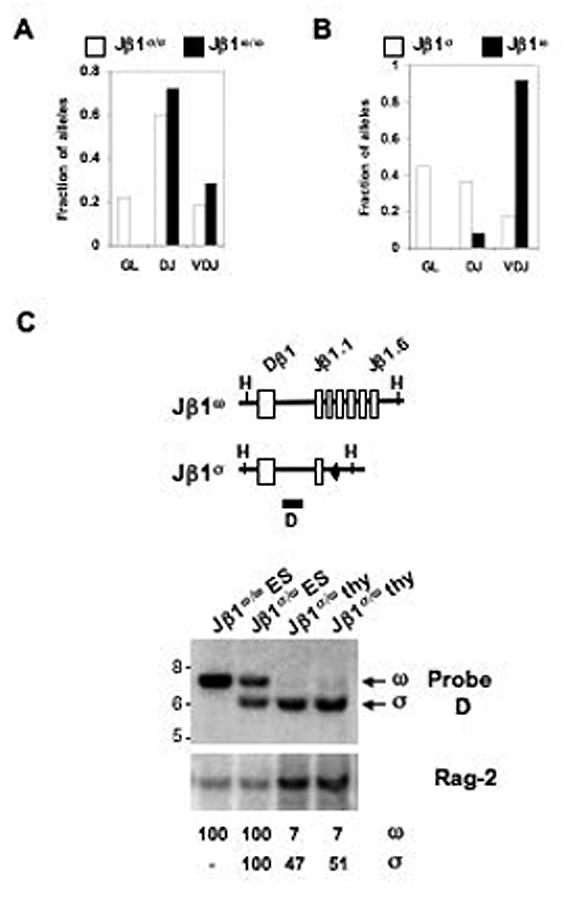
A. Analyses of Jβ1σ/σ (open) and Jβ1ω/ω (solid) αβ T cell hybridomas. All hybridomas have one complete in-frame TCRβ allele. The fraction in which the other allele was un-rearranged (GL), in the DJβ (DJ) and in the VDJβ (VDJ) configurations are indicated. B. Analyses of Jβ1σ/ω αβ T cell hybridomas. Shown is the fraction of Jβ1σ (open) and Jβ1ω (solid) alleles in the GL, DJβ and VDJβ configurations. C. Southern blot analysis of rearrangement of the Jβ1σ and Jβ1ω alleles in thymocytes (thy) from two Jβ1σ/ω mice and Jβ1ω/ω and Jβ1σ/ω ES cells as a reference for maximal retention of the germline bands. Genomic DNA was digested with HindIII and probed with probe D or a Rag-2 probe as a DNA loading control. The bands resulting from the germline Jβ1σ (σ) and Jβ1ω (ω) alleles are indicated. The molecular weight markers are indicated. The percent retention of the germline Jβ1σ and Jβ1ω alleles in the Jβ1σ/ω thymocytes was calculated based on the intensity of the specific band in Jβ1σ/ω thymocytes relative to the same band in Jβ1σ/ω ES cells corrected for DNA loading using the intensity of the Rag-2 band.
To directly compare the efficiency of rearrangement on the Jβ1ω and Jβ1σ alleles, we generated αβ T cell hybridomas from hemizygous Jβ1ω/σ mice. Whereas the Jβ1ω allele was rearranged in all Jβ1ω/σ αβ T cell hybridomas, the Jβ1σ allele was un-rearranged in 45% of these cells (Fig. 3B). Similarly, Southern blot analysis of genomic DNA from Jβ1σ/ω thymocytes revealed that 7% of the Jβ1ω alleles were in the un-rearranged configuration, as compared to 49% of the Jβ1σ alleles (Fig. 3C). Taken together, these analyses demonstrate that deletion of a 1.5 kb region containing 5 of the 6 Jβ1 gene segments on the Jβ1ω allele leads to a significant reduction in V(D)J recombination on this allele.
3.3 Germline transcription is significantly diminished on the Jβ1σ allele
The decreased rearrangement observed on the Jβ1σ allele could reflect a requirement for a minimal number of Jβ gene segments to achieve optimal levels of Dβ-Jβ rearrangement (see discussion). Alternatively, we reasoned that deletion of the Jβ1 gene segments on the Jβ1σ allele could have disrupted cis-acting elements that impact transcription and accessibility of the region. To test this notion, we generated Jβ1ω/ω and Jβ1σ/σ mice that were also deficient in Rag-2 (Jβ1ω/ω:Rag-2-/- and Jβ1σ/σ:Rag-2-/- mice, respectively) in order to assay for germline TCRβ transcripts on the Jβ1ω and Jβ1σ alleles. Northern blot analyses performed on DN thymocytes, using a probe (probe D) between the Dβ1 and Jβ1.1 segments, revealed similar levels of germline TCRβ transcripts in Jβ1ω/ω:Rag-2-/- and Rag-2-/- thymocytes, which have a wild-type TCRβ locus (WT:Rag-2-/-) (Figs. 3C and 4). In striking contrast, the level of transcripts through the Dβ1-Jβ1 region was decreased 7-fold in Jβ1σ/σ:Rag-2-/- thymocytes as compared to Jβ1ω/ω:RAG-2-/- thymocytes (Fig. 4A). These data demonstrate that deleting a 1.5 kb region containing 5 of the 6 Jβ1 gene segments leads to a diminished level of germline Dβ1-Jβ1 transcripts. Notably, whereas germline transcripts are decreased in developing thymocytes, Northern blot analyses revealed splenic Jβ1σ/σ and Jβ1ω/ω αβ T cells have similar levels of Cβ hybridizing transcripts (Fig. 4B). Moreover, mature splenic Jβ1σ/σ and Jβ1ω/ω αβ T cells express similar levels of αβ TCR on their cell surfaces (Fig. 4C). Thus, transcription of the Jβ1σ allele is not universally compromised throughout ontogeny.
3.4 RS sequence-specific effects on TCRβ germline transcripts and locus accessibility
Our results suggest that the Jβ1 gene segment cluster may contain sequences that regulate germline transcription and accessibility. Since this region is primarily composed of Jβ gene segments and their RSs, we considered the possibility that these sequences could overlap with the RSs themselves. In this regard, the Dβ1 and Dβ2 23-RSs were recently shown to contain functional c-fos binding motifs (Wang et al., 2008). To test this notion, we used gene targeting approaches to generate two modified versions of the Jβ1σ allele, Jβ1σ.1 and Jβ1σ.2 (Fig. 5). The Jβ1σ.1 and Jβ1σ.2 alleles are identical to the Jβ1σ allele except for similar point mutations in the heptamer and nonamer sequences of the Jβ1.1 RS and 3′Dβ1 RS, respectively (Fig. 5). These mutations are expected to abrogate Rag binding and cleavage at the mutated RS. As expected, mutating either the Jβ1.1 RS or the 3′Dβ1 RS prevents Dβ1 to Jβ1 rearrangement (data not shown). Northern blot analyses of Jβ1σ.2/σ.2:Rag-2-/- thymocytes revealed similar levels of germline Dβ1-Jβ1 transcripts, as compared to Jβ1σ/σ:Rag-2-/- thymocytes (Fig. 6a). In contrast, these transcripts were not detected in Jβ1σ.1/σ.1:Rag-2-/- thymocytes (Fig. 6a). Thus, mutation of the Jβ1.1 RS heptamer and nonamer leads to a significant reduction in the level of germline Dβ1-Jβ1 transcripts, whereas similar mutation of the 3′Dβ1 RS does not.
Figure 5. The Jβ1σ.1 and Jβ1σ.2 alleles.
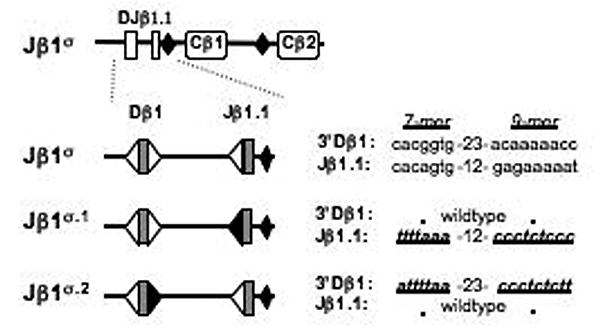
Schematic of the Jβ1.1 12-RS and 3′ Dβ1 23-RS heptamer (7-mer) and nonamer (9-mer) sequences on the Jβ1σ, Jβ1σ.1 and Jβ1σ.2 alleles. RSs are indicated as triangles with the mutated RSs shown as filled triangles.
Figure 6. Jβ1.1 RS sequences influence transcription and accessibility.
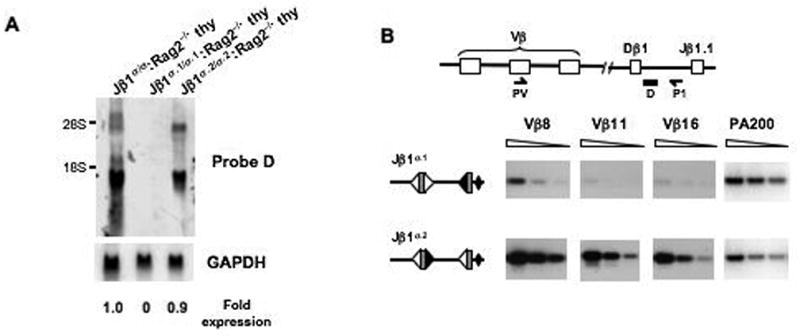
A. Northern blot analysis of germline probe D (see Fig. 3C) hybridizing germline transcripts from Jβ1σ/σ:Rag-2-/-, Jβ1σ.1/σ.1:Rag-2-/- and Jβ1σ.2/σ.2:Rag-2-/- thymocytes. Expression is normalized to GAPDH. B. PCR analysis of Vβ-Dβ rearrangement on the Jβ1σ.1 and Jβ1σ.2 alleles in Jβ1ω/σ.1 and Jβ1ω/σ.2 thymocytes. The schematic illustrates the PCR strategy using a Vβ-specific primer (PV) and a primer between Dβ1 and Jβ1.1 (P1). PCR analyses were carried out on serial 3-fold dilutions of thymocyte DNA. The PA200 gene is amplified for a DNA loading control. Shown is a representative experiment of three Jβ1ω/σ.1 and three Jβ1ω/σ.2 mice analyzed. That the VDβ PCR products were from the Jβ1σ.1 and Jβ1σ.2 alleles was confirmed by restriction digestion using unique restriction sites introduced during the targeting.
To determine whether the mutation of the Jβ1.1 RS on the Jβ1σ.1 allele also leads to diminished accessibility, we assayed for rearrangement on the Jβ1σ.1 and Jβ1σ.2 alleles in Jβ1ω/σ.1 and Jβ1ω/σ.2 thymocytes, respectively. Because the Jβ1σ.1 and Jβ1σ.2 allele RS mutations both prohibit Dβ to Jβ1.1 rearrangements, we analyzed Vβ to Dβ1 rearrangements on these alleles. Whereas Vβ to Dβ rearrangements were readily detected on the Jβ1σ.2 allele in Jβ1ω/σ.2 thymocytes, they were detected at much lower levels on the Jβ1σ.1 allele in Jβ1ω/σ.1 thymocytes (Fig. 6b). Together, these findings demonstrate that in addition to their critical role in Rag binding and cleavage, the heptamer and nonamer sequences of the Jβ1.1 RS also contribute to the regulation of germline transcription and accessibility of the Dβ-Jβ1 gene segment cluster.
4. Discussion
Two cis-acting elements in the TCRβ locus, Eβ and PDβ1, have been shown to regulate germline transcription and accessibility of the Dβ1-Jβ1 gene segment cluster. Here we make the rather surprising finding that deletion of a small (1.5kb) region of the Dβ1-Jβ1 gene segment cluster, which consists primarily of Jβ1.2 through Jβ1.6 and their associated RSs, leads to a dramatic reduction in Dβ1-Jβ1.1 rearrangement on the Jβ1σ allele in developing thymocytes. This reduction is most likely due to alterations in accessibility of the Jβ1σ allele as evidenced by the diminished levels of germline TCRβ transcripts from this allele. However, it is conceivable that the reduction in germline transcripts could also be due, in part, to alterations in the stability of germline transcripts templated by the Jβ1σ allele.
The Jβ1.1 gene segment is frequently used in complete VDJβ rearrangements on the wild type TCRβ allele, making it unlikely that our findings are due primarily to intrinsic features of the Jβ1.1 gene segment or its RS that make it a poor substrate for V(D)J recombination. It is possible that reducing the number of Jβ1 gene segments on the Jβ1σ allele to a single gene segment, Jβ1.1, leads to a reduced efficiency of Dβ1 to Jβ1 rearrangement. This could occur, for example, if multiple Jβ 12-RSs are important for increasing the local concentration of the Rag proteins. In this regard, it is notable that in vivo studies have suggested that the Rag proteins first bind a 12-RS and then capture a 23-RS to form a synaptic complex (Curry et al., 2005). However, while such a requirement could contribute to the decreased rearrangement, it would not be expected to lead to the decrease in germline transcripts observed on the Jβ1σ allele.
The small genomic deletion on the Jβ1σ allele, that contains primarily Jβ1 gene segments, could perturb the functional association of two cis-acting elements, such as Eβ and PDβ1, disrupting optimal transcription and accessibility of this region (Oestreich et al., 2006). However, this would not explain the additional reduction in germline transcription and accessibility on the Jβ1σ.1 allele, which differs from the Jβ1σ allele only by point mutations in the Jβ1.1 heptamer and nonamer sequences and not in the deletion of any sequences. Notably, modifying the 3′ Dβ1 RS heptamer and nonamer (Jβ1σ.2 allele) did not similarly reduce levels of germline Dβ1-Jβ1 transcripts as on the Jβ1σ.1 allele. Given the RS mutations introduced, neither the Jβ1σ.1 nor the Jβ1σ.2 allele undergoes Dβ1 to Jβ1.1 rearrangement, as expected. The link between the integrity of heptamer and nonamer sequences in the Jβ1.1 RS and accessibility of the region is evidenced by the decreased Vβ to Dβ rearrangement on the Jβ1σ.1 allele (Jβ1.1 RS mutation) as compared to the Jβ1σ.2 allele (3′Dβ1 RS mutation). Thus, a very precise mutation of the 16 nucleotides that comprise the heptamer and nonamer of the Jβ1.1 RS, but not similar mutation of the heptamer and nonamer of the 3′ Dβ1 RS, leads to reduced levels of Dβ1-Jβ1 germline transcripts and decreased accessibility of the Dβ1 gene segment.
Several factors could contribute to the reduced transcription and accessibility of the Dβ1-Jβ1 region on the Jβ1σ allele. Our data suggest that this is due, at least in part, to a functional overlap between RSs that direct Rag binding and synaptic complex formation, and cis-acting sequences that regulate germline transcription and accessibility. It is possible that the Jβ1.1 RS, and other regions in the Jβ1 gene segment cluster, are part of or even synonymous with a novel cis-acting element that regulates transcription through the Dβ1-Jβ1 gene segment cluster. Alternatively, these sequences could augment and optimize the function of PDβ1 in promoting Dβ1 to Jβ1 rearrangement. Finally, mutation of the Jβ1.1 RSs could affect nucleosome phasing in this region in a way that impacts both transcription and accessibility (Baumann et al., 2003; Golding et al., 1999; Kwon et al., 1998; Nightingale et al., 2007). By whatever mechanism, our findings demonstrate that there is a functional overlap between RSs and cis-acting sequences that regulate transcription. As such they mandate that future analyses of RS function in regulating V(D)J recombination in vivo consider the possibility that introduced mutations effect not only on interactions with the Rag endonuclease, but also elements that regulate transcription and accessibility.
Acknowledgments
This work is supported in part by the National Institutes of Health grants AI47829 and AI49934 (B.P.S.). B.P.S. is a recipient of a Research Scholar Award from the American Cancer Society. Mice were produced by a transgenic core facility supported by the Rheumatic Diseases Core Center at Washington University (NIH P30-AR48335) and housed in a facility supported by NCRR grant RR012466.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alt FW, Yancopoulos GD, Blackwell TK, Wood C, Thomas E, Boss M, Coffman R, Rosenberg N, Tonegawa S, Baltimore D. Ordered rearrangement of immunoglobulin heavy chain variable region segments. EMBO J. 1984;3:1209–19. doi: 10.1002/j.1460-2075.1984.tb01955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassing CH, Alt FW, Hughes MM, D'Auteuil M, Wehrly TD, Woodman BB, Gartner F, White JM, Davidson L, Sleckman BP. Recombination signal sequences restrict chromosomal V(D)J recombination beyond the 12/23 rule. Nature. 2000;405:583–6. doi: 10.1038/35014635. [DOI] [PubMed] [Google Scholar]
- Bassing CH, Swat W, Alt FW. The mechanism and regulation of chromosomal V(D)J recombination. Cell. 2002;109(Suppl):S45–55. doi: 10.1016/s0092-8674(02)00675-x. [DOI] [PubMed] [Google Scholar]
- Baumann M, Mamais A, McBlane F, Xiao H, Boyes J. Regulation of V(D)J recombination by nucleosome positioning at recombination signal sequences. EMBO J. 2003;22:5197–207. doi: 10.1093/emboj/cdg487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman Y. Allelic exclusion in B and T lymphopoiesis. Semin Immunol. 1999;11:319–28. doi: 10.1006/smim.1999.0188. [DOI] [PubMed] [Google Scholar]
- Bories JC, Demengeot J, Davidson L, Alt FW. Gene-targeted deletion and replacement mutations of the T-cell receptor β-chain enhancer: the role of enhancer elements in controlling V(D)J recombination accessibility. Proc Natl Acad Sci U S A. 1996;93:7871–6. doi: 10.1073/pnas.93.15.7871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvier G, Watrin F, Naspetti M, Verthuy C, Naquet P, Ferrier P. Deletion of the mouse T-cell receptor β gene enhancer blocks αβ T-cell development. Proc Natl Acad Sci USA. 1996;93:7877–81. doi: 10.1073/pnas.93.15.7877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb RM, Oestreich KJ, Osipovich OA, Oltz EM. Accessibility control of V(D)J recombination. Adv Immunol. 2006;91:45–109. doi: 10.1016/S0065-2776(06)91002-5. [DOI] [PubMed] [Google Scholar]
- Curry JD, Geier JK, Schlissel MS. Single-strand recombination signal sequence nicks in vivo: evidence for a capture model of synapsis. Nat Immunol. 2005;6:1272–9. doi: 10.1038/ni1270. [DOI] [PubMed] [Google Scholar]
- Doty RT, Xia D, Nguyen SP, Hathaway TR, Willerford DM. Promoter element for transcription of unrearranged T-cell receptor β-chain gene in pro-T cells. Blood. 1999;93:3017–25. [PubMed] [Google Scholar]
- Fugmann SD, Lee AI, Shockett PE, Villey IJ, Schatz DG. The RAG proteins and V(D)J recombination: complexes, ends, and transposition. Annu Rev Immunol. 2000;18:495–527. doi: 10.1146/annurev.immunol.18.1.495. [DOI] [PubMed] [Google Scholar]
- Gellert M. V(D)J recombination: rag proteins, repair factors, and regulation. Annu Rev Biochem. 2002;71:101–32. doi: 10.1146/annurev.biochem.71.090501.150203. [DOI] [PubMed] [Google Scholar]
- Glusman G, Rowen L, Lee I, Boysen C, Roach JC, Smit AF, Wang K, Koop BF, Hood L. Comparative genomics of the human and mouse T cell receptor loci. Immunity. 2001;15:337–49. doi: 10.1016/s1074-7613(01)00200-x. [DOI] [PubMed] [Google Scholar]
- Golding A, Chandler S, Ballestar E, Wolffe AP, Schlissel MS. Nucleosome structure completely inhibits in vitro cleavage by the V(D)J recombinase. EMBO J. 1999;18:3712–23. doi: 10.1093/emboj/18.13.3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CY, Sleckman BP, Kanagawa O. Revision of T cell receptor α chain genes is required for normal T lymphocyte development. Proc Natl Acad Sci U S A. 2005;102:14356–61. doi: 10.1073/pnas.0505564102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khor B, Bredemeyer AL, Huang CY, Turnbull IR, Evans R, Maggi LB, Jr, White JM, Walker LM, Carnes K, Hess RA, Sleckman BP. Proteasome activator PA200 is required for normal spermatogenesis. Mol Cell Biol. 2006;26:2999–3007. doi: 10.1128/MCB.26.8.2999-3007.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khor B, Sleckman BP. Allelic exclusion at the TCRβ locus. Curr Opin Immunol. 2002;14:230–4. doi: 10.1016/s0952-7915(02)00326-6. [DOI] [PubMed] [Google Scholar]
- Khor B, Sleckman BP. Intra- and inter-allelic ordering of T cell receptor β chain gene assembly. Eur J Immunol. 2005;35:964–70. doi: 10.1002/eji.200425806. [DOI] [PubMed] [Google Scholar]
- Kim JM, White JM, Shaw AS, Sleckman BP. MAPK p38 α is dispensable for lymphocyte development and proliferation. J Immunol. 2005;174:1239–44. doi: 10.4049/jimmunol.174.3.1239. [DOI] [PubMed] [Google Scholar]
- Kwon J, Imbalzano AN, Matthews A, Oettinger MA. Accessibility of nucleosomal DNA to V(D)J cleavage is modulated by RSS positioning and HMG1. Mol Cell. 1998;2:829–39. doi: 10.1016/s1097-2765(00)80297-x. [DOI] [PubMed] [Google Scholar]
- McDougall S, Peterson CL, Calame K. A transcriptional enhancer 3′ of the Cβ2 in the T cell receptor β locus. Science. 1988;241:205–208. doi: 10.1126/science.2968651. [DOI] [PubMed] [Google Scholar]
- McMillan RE, Sikes ML. Differential activation of dual promoters alters Dβ2 germline transcription during thymocyte development. J Immunol. 2008;180:3218–28. doi: 10.4049/jimmunol.180.5.3218. [DOI] [PubMed] [Google Scholar]
- Nightingale KP, Baumann M, Eberharter A, Mamais A, Becker PB, Boyes J. Acetylation increases access of remodelling complexes to their nucleosome targets to enhance initiation of V(D)J recombination. Nucleic Acids Res. 2007;35:6311–21. doi: 10.1093/nar/gkm650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oestreich KJ, Cobb RM, Pierce S, Chen J, Ferrier P, Oltz EM. Regulation of TCRβ gene assembly by a promoter/enhancer holocomplex. Immunity. 2006;24:381–91. doi: 10.1016/j.immuni.2006.02.009. [DOI] [PubMed] [Google Scholar]
- Oettinger MA. V(D)J recombination: on the cutting edge. Curr Opin Cell Biol. 1999;11:325–9. doi: 10.1016/S0955-0674(99)80044-1. [DOI] [PubMed] [Google Scholar]
- Rooney S, Chaudhuri J, Alt FW. The role of the non-homologous end-joining pathway in lymphocyte development. Immunol Rev. 2004;200:115–31. doi: 10.1111/j.0105-2896.2004.00165.x. [DOI] [PubMed] [Google Scholar]
- Sikes ML, Gomez RJ, Song J, Oltz EM. A developmental stage-specific promoter directs germline transcription of Dβ Jβ gene segments in precursor T lymphocytes. J Immunol. 1998;161:1399–405. [PubMed] [Google Scholar]
- Sikes ML, Suarez CC, Oltz EM. Regulation of V(D)J recombination by transcriptional promoters. Mol Cell Biol. 1999;19:2773–81. doi: 10.1128/mcb.19.4.2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleckman BP, Bardon CG, Ferrini R, Davidson L, Alt FW. Function of the TCR α enhancer in αβ and γδ T cells. Immunity. 1997;7:505–15. doi: 10.1016/s1074-7613(00)80372-6. [DOI] [PubMed] [Google Scholar]
- Sleckman BP, Bassing CH, Hughes MM, Okada A, D'Auteuil M, Wehrly TD, Woodman BB, Davidson L, Chen J, Alt FW. Mechanisms that direct ordered assembly of T cell receptor β locus V, D, and J gene segments. Proc Natl Acad Sci U S A. 2000;97:7975–80. doi: 10.1073/pnas.130190597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonegawa S. Somatic generation of antibody diversity. Nature. 1983;302:575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- Wang X, Xiao G, Zhang Y, Wen X, Gao X, Okada S, Liu X. Regulation of Tcrβ recombination ordering by c-Fos-dependent RAG deposition. Nat Immunol. 2008;9:794–801. doi: 10.1038/ni.1614. [DOI] [PubMed] [Google Scholar]
- Whitehurst CE, Chattopadhyay S, Chen J. Control of V(D)J recombinational accessibility of the Dβ 1 gene segment at the TCRβ locus by a germline promoter. Immunity. 1999;10:313–22. doi: 10.1016/s1074-7613(00)80031-x. [DOI] [PubMed] [Google Scholar]
- Whitehurst CE, Schlissel MS, Chen J. Deletion of germline promoter PDβ1 from the TCRβ locus causes hypermethylation that impairs Dβ1 recombination by multiple mechanisms. Immunity. 2000;13:703–14. doi: 10.1016/s1074-7613(00)00069-8. [DOI] [PubMed] [Google Scholar]


