Abstract
MyD88 is a key downstream adapter for most Toll-like receptors (TLRs) and interleukin-1 receptors (IL-1Rs). MyD88 deficiency in mice leads to susceptibility to a broad range of pathogens in experimental settings of infection. We describe a distinct situation in a natural setting of human infection. Nine children with autosomal recessive MyD88 deficiency suffered from life-threatening, often recurrent pyogenic bacterial infections, including invasive pneumococcal disease. However, these patients were otherwise healthy, with normal resistance to other microbes. Their clinical status improved with age, but not due to any cellular leakiness in MyD88 deficiency. The MyD88-dependent TLRs and IL-1Rs are therefore essential for protective immunity to a small number of pyogenic bacteria, but redundant for host defense to most natural infections.
The search for human genetic etiologies of pediatric infectious diseases aims to decipher the molecular mechanism of disease and to reveal the function of immune genes in natura (1). The immunological investigation of children with invasive pneumococcal disease (IPD) led to the discovery of children lacking interleukin-1 receptor–associated kinase 4 (IRAK-4), which is selectively recruited to Toll-like receptors (TLRs) and interleukin-1 receptors (IL-1Rs) by MyD88 (2). The patients present with a life-threatening but narrow and transient predisposition to infection, apparently restricted to pyogenic bacterial diseases, particularly IPD, during the first 10 years of life (3). This clinical phenotype is surprising given the central role commonly attributed to both TLRs and IL-1Rs, and the high susceptibility of MyD88-deficient mice to experimental infections with at least 35 pathogens—19 bacteria, seven viruses, five parasites, and four fungi (4) (tables S1 to S3). However, IRAK-4–deficient mice have thus far been challenged with only a few pathogens (5). Fibroblasts and individual leukocyte subsets from IRAK-4–deficient patients fail to respond to the TLR agonists tested, at least for the orthologs of the mouse MyD88-dependent target genes tested (3, 6). The resistance of IRAK-4–deficient patients might thus be explained by IRAK-4–independent but MyD88-dependent TLR or IL-1R responses in other cell types and/or for other target genes.
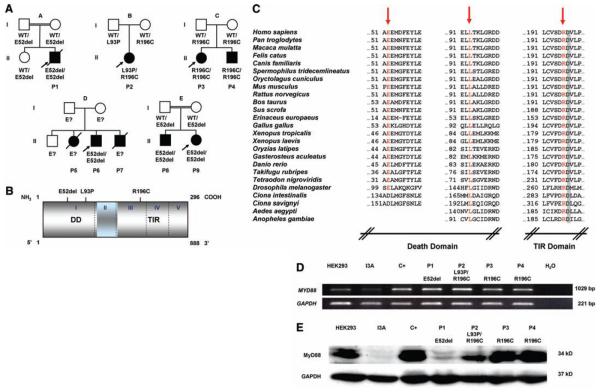
We investigated nine children (P1 to P9) with invasive pyogenic bacterial diseases and without IRAK-4 deficiency, from five unrelated kindreds (supplementary note 1). Three children died between 1 and 11 months of age and six are now between 3 and 16 years old. A homozygous in-frame MYD88 deletion was found in P1, P6, P8, and P9 (160del3, designated E52del), compound heterozygous missense mutations in P2 (278 T→C, L93P; 586 C→T, R196C), and a homozygous missense mutation in P3 and P4 (586 C→T, R196C) (Fig. 1, A and B, and fig. S1). The deletion and missense mutations were not found in 100 and 1728 healthy individuals, respectively. These mutations are nonconservative and affect residues conserved across species (Fig. 1C). Residues 195 to 197 are crucial for Toll/IL-1 receptor (TIR)/TIR interaction (7). The segregation of the MYD88 genotype and of the clinical phenotype is consistent with an autosomal recessive trait (supplementary note 1, Fig. 1A, and figs. S2 to S5). The MYD88 mRNA in fibroblasts from patients P1 to P4 (representing the three combinations of alleles) was of normal molecular weight and abundance (Fig. 1D and fig. S6). MyD88 protein was detected in SV40-transformed fibroblasts in trace amounts for P1, small amounts for P2, and normal amounts for P3 and P4, as shown by Western blotting (Fig. 1E). These data suggest that our patients have functional MyD88 deficiency, with low (P1 and P2, P5 to P9) or normal (P3 and P4) MyD88 protein levels.
Fig. 1.
(A) Kindreds and patients with MYD88 mutations. (B) Positions of the MyD88 mutations in the death domain (DD) or the TIR domain of the protein. (C) Parts of the DD and TIR domain of MyD88 in humans and the corresponding regions in 24 other species. The residues mutated are indicated in red. Amino acid D197 (gray) is conserved in all species. (D) Full-length MYD88 transcripts in SV40-transformed fibroblasts from a healthy control donor (C+), four MyD88-deficient patients (P1 to P4), the MyD88-deficient HEK293 cell line (I3A), and the parental MyD88-positive HEK293 cell line. (E) MyD88 protein expression in SV40-transformed fibroblasts from a healthy control (C+), four patients (P1 to P4), the I3A line, and the parental HEK293 cell line. The MyD88-specific antibody recognizes residues 279 to 296. Abbreviations for the amino acid residues are as follows: A, Ala; C, Cys; D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; W, Trp; and Y, Tyr.
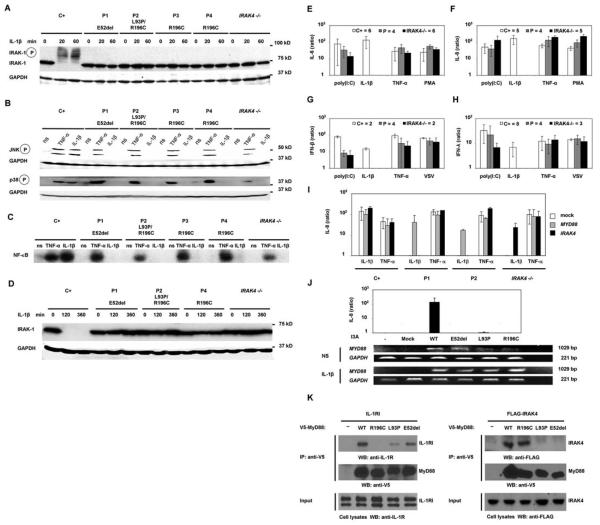
The functional impact of the MyD88 mutations was then tested in cell lines derived from the patients. As in IRAK-4–deficient cells (2, 3, 6), IRAK-1 was not degraded in the patients' SV40-transformed fibroblasts up to 6 hours after stimulation with IL-1β (Fig. 2, A and D). Similarly, IRAK-1 was not degraded in the patients' Epstein-Barr virus–transformed B (EBV-B) cells up to 2 hours after stimulation with the TLR7/TLR8 agonist R-848 (fig. S7, A and B). Phosphorylation of the mitogen-activated protein kinases (MAPKs) p38 and c-Jun N-terminal kinase (JNK) and the DNA binding activity of nuclear factor κB (NF-κB) were impaired in the patients' SV40-transformed fibroblasts after stimulation with IL-1β (Fig. 2, B and C). Furthermore, the patients' EBV-B cells showed a complete lack of response to stimulation with R-848 for 24 hours, in terms of tumor necrosis factor-α (TNF-α) induction (fig. S7C). Similarly, the production of IL-6, IL-8, interferon-β (IFN-β), and IFN-λ was abolished after 24 hours of incubation of the patients' SV40-transformed fibroblasts with IL-1β, although these cells responded normally to TNF-α and poly(I:C), a TLR3-dependent agonist in human fibroblasts (8) (Fig. 2, E to H). Thus, two cell types derived from our patients carrying two mutant MYD88 alleles, with normal (P3 and P4) or impaired (P1 and P2) MyD88 protein production, showed a selective failure to respond to the stimulation of two key IRAK-4–dependent signaling pathways (TLR7/8 and IL-1R). This uniform cellular phenotype suggests that each of the patients presents an autosomal recessive, functionally complete deficiency in MyD88.
Fig. 2.
(A) IRAK-1 phosphorylation and degradation upon activation by IL-1β in SV40-transformed fibroblasts from a healthy control (C+), MyD88-deficient patients (P1 to P4), and an IRAK-4–deficient patient (IRAK4 −/−). (B) Phosphorylation of MAP kinases and (C) electrophoretic mobility shift assay for NF-κB upon 20 min of activation by TNF-α and IL-1β of SV40-fibroblasts from a healthy control (C+), MyD88-deficient patients (P1 to P4), and an IRAK-4–deficient patient (IRAK4 −/−). (D) IRAK-1 degradation upon activation by IL-1β in SV40-transformed fibroblasts between 2 and 6 hours after stimulation. (E) IL-6 production, (F) IL-8 production, (G)IFN-β production, and (H) IFN-λ production by SV40-transformed fibroblasts upon 18 hours of activation by poly(I:C), IL-1β, TNF-α, and phorbol 12-myristate 13-acetate (PMA)/ionomycin (E and F) or poly(I:C), IL-1β, TNF-α, and vesicular stomatitis virus (VSV) (G and H). Results for healthy controls (C+) are shown in white, for MyD88-deficient patients (P1 to P4) in gray, and for IRAK-4–deficient patients (IRAK4 −/−) in black (numbers of individuals tested are indicated next to these boxes). Cytokine production is presented as the ratio of cytokine production by stimulated cells to that by nonstimulated cells. (I) IL-8 production upon activation of SV40-transformed fibroblasts from healthy donors (C+), MyD88-deficient patients (P1 and P2), and an IRAK-4–deficient patient (IRAK4 −/−)after transfection with the pUNO empty plasmid (white), wild-type pUNO-MyD88 (gray), and wild-type pcDNA3–IRAK-4 (black). Cells were also transfected with the pcDNA3 empty plasmid, giving results similar to those of the pUNO empty plasmid (not shown). (J) IL-8 production upon activation of I3A cells (MyD88-deficient HEK293 cells) without transfection and after transfection with pcDNA3.2 empty plasmid (mock), or pcDNA3.2 encoding V5-tagged wild-type MYD88, V5-tagged MYD88-E52del, V5-tagged MYD88-L93P, and V5-tagged MYD88-R196C. RT-PCR in nonactivated (NS) and activated I3A cells (IL-1β)servedto check that the cells were correctly transfected with MYD88. Western blotting for the V5 tag was also done (fig. S8). (K) Coimmunoprecipitation in I3A cells, transfected with expression vectors encoding V5-tagged MyD88 and FLAG-tagged IRAK-4. Immunoprecipitation was carried out with V5- or FLAG-specific antibodies and Western blotting with V5-, FLAG-, or IL-1R–specific antibodies. Coimmunoprecipitation of IL-1R1 with the MyD88 mutants (left panel). MyD88-L93P and MyD88-E52del (in the DD) interact weakly with IL-1R1, whereas MyD88-R196C (in the TIR domain) cannot associate with IL-1R. Coimmunoprecipitation of IRAK-4 with MyD88 mutants (right panel). MyD88-L93P and MyD88-E52del cannot interact with IRAK-4, whereas MyD88-R196C still associates with IRAK-4.
Fibroblasts from a healthy control and from P1 and P2 (representing the three mutant alleles) and from an IRAK-4–deficient child were transiently transfected with expression vectors encoding wild-type (WT) MyD88 or IRAK-4. Cells from P1 and P2 regained IL-1β responsiveness after transfection with the WT MYD88 gene only, as shown by the levels of IL-8 production (Fig. 2I). IRAK-4–deficient cells, used as a control, were complemented with WT IRAK4 only (Fig. 2I). The human embryonic kidney (HEK) 293– derived MyD88-deficient I3A cell line (9) was then transiently transfected with WT, E52del, L93P, and R196C MYD88 alleles. All of the MYD88 alleles used for transfection were expressed, as shown by reverse transcription–polymerase chain reaction (RT-PCR) and Western blotting (Fig. 2J and fig. S8). I3A cells responded to IL-1β after transfection with the WT MYD88 allele, but not after transfection with any of the three mutant MYD88 alleles, as shown by IL-8 production levels (Fig. 2J). These analyses confirmed that all three MYD88 mutant alleles were loss-of-function. Finally, immunoprecipitation and Western blotting experiments showed that the R196C mutation in the TIR domain prevents interaction with IL-1R, whereas the E52del and L93P mutations in the death domain (DD) prevent interaction with IRAK-4 (Fig. 2K). These results demonstrate that all nine patients had complete MyD88 deficiency, resulting from the inheritance of two loss-of-function MYD88 alleles.
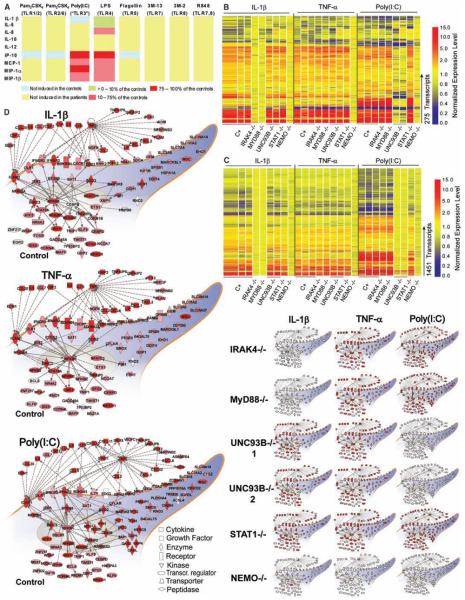
We then investigated the potential effect of MyD88 deficiency on blood TLR and IL-1R responses. IL-6 production in peripheral blood mononuclear cells (PBMCs) after stimulation with agonists of TLR4, TLR7 and TLR8 (P1), and in whole blood after stimulation with agonists of IL-1R and TLR4 (P2, P3, and P4) was abolished (figs. S2 and S3). The shedding of CD62L by granulocytes normally seen in response to TLR1/2, TLR4, TLR7, and TLR8 stimulation was also impaired (figs. S4 and S5). Whole blood from P2, P3, and P4 was then stimulated with a broad range of TLR agonists, and cytokine secretion was measured by means of a multiplex cytometry-based system. Whole blood from MyD88-deficient patients showed no cytokine response to six of the eight TLR agonists tested, for any of the nine cytokines induced in controls by at least one TLR (Fig. 3A). After activation with poly(I:C), whole-blood cells from the patients displayed the induction of several cytokines to levels similar to those in healthy control cells, but no production of IL-6 and IL-8 was observed. After activation with lipopolysaccharide (LPS), whole-blood cells from the patients displayed normal induction of a smaller number of cytokines, including IP10. These data are similar to those previously reported for IRAK-4–deficient PBMCs (3, 6). Thus, MyD88 deficiency generally abolishes cytokine responses to the TLR stimulation of blood cells, although most cytokines were produced after poly(I:C) treatment [possibly via receptors other than TLR3 (8)] and some were produced in response to LPS.
Fig. 3.
(A) Multiple cytokine secretion by whole-blood cells from three healthy donors and three MyD88-deficient patients (P2, P3, and P4), activated by incubation with various TLR agonists for 24 hours. Cytokine levels are presented as ratios of secretion by cells from MyD88-deficient patients to secretion by cells from the healthy control. (B) Transcriptional profiles of fibroblasts from healthy controls and patients stimulated with IL-1β, TNF-α, and poly(I:C) for 2 hours. Transcriptional signature of 275 genes differentially regulated upon stimulation with IL-1β,TNF-α, or poly(I:C) in at least two of three control fibroblast lines. Genes were arranged by hierarchical clustering and, for each donor, changes in expression with respect to the corresponding untreated conditions are represented on a heat map. Red indicates a relative increase in expression levels, blue indicates a relative decrease, and yellow indicates no change in expression level. Samples are grouped by stimulus and ordered by donor: controls (1 to 3), IRAK-4–deficient patient (3), MyD88-deficient patient, UNC-93B–deficient patients (1 and 2) (11), STAT1-deficient patient (12), and NEMO-deficient patient (10). (C) Transcriptional profiles of fibroblasts stimulated with IL-1β, TNF-α, and poly(IC) for 8 hours. Transcriptional signature of 1451 genes differentially regulated upon stimulation with IL-1β,TNF-α, or poly(IC)in at least two of three control fibroblast lines. (D) Functional pathways regulated in fibroblasts treated with IL-1β, TNF-α, or poly(IC) for 2 hours. Genes or gene products regulated by these factors are represented as nodes, and the biological relation between two nodes is represented as an edge (line). Solid and dashed lines indicate direct and indirect relations, respectively. All edges are supported by at least one reference from the literature. Nodes are arranged according to the cellular distribution of the corresponding gene products. Expression levels of individual genes are represented on a color scale on the main network: white denotes <1.5-fold difference from nonstimulated conditions; solid red denotes >5-fold difference from nonstimulated conditions. The main network indicates the average expression level for the three control cell lines. The scaled-down networks indicate the levels of expression obtained for cells derived from patients.
We investigated whether residual, MyD88-independent signaling occurred by evaluating the IL-1R pathway of MyD88-deficient patients in more detail, through the analysis of genome-wide transcriptional profiles (48,701 probes) in fibroblasts. In control fibroblasts, 275 transcripts were regulated by IL-1β, TNF-α, or poly(I:C) after 2 hours of culture and 1451 transcripts were regulated after 8 hours (tables S4 to S8). The prototypic signatures in cell lines derived from patients were as follows: NEMO-deficient cells (10) were unresponsive to all three stimuli, UNC-93B–deficient cells (11) did not respond to poly(I:C), and signal transducers and activators of transcription 1 (STAT1)–deficient cells (12) responded only weakly to poly(I:C) after 8 hours (Fig. 3, B and C). MyD88- and IRAK-4–deficient cells had indistinguishable phenotypes, as both were unresponsive to IL-1β at both time points. The signatures obtained in response to TNF-α and poly(I:C) [via TLR3 (8)] in these cells were similar to that of control fibroblasts (Fig. 3, B and C, and supplementary note 2). We then focused on the functional pathways regulated by IL-1β,TNF-α, and poly(I:C) in control fibro-blasts and in fibroblasts derived from patients. Control fibroblasts treated with IL-1β responded by a rapid increase in production of inflammatory cytokines and chemokines (including TNF-α, IL-1β, IL-6, and IL-8) and cell surface receptors (including ICAM1, OLR1, IFN-γR2, and IFN-αR2) (Fig. 3D). Differences in the activation status of the IL-1β, TNF-α, and poly(I:C) functional networks among fibroblasts derived from patients clearly identified a complete, specific lack of response to IL-1β as a defining characteristic of IRAK-4 and MyD88 deficiencies (Fig. 3D).
We thus report nine children with inherited MyD88 deficiency. The immunological pheno-type is similar to that of MyD88-deficient mice (tables S9 and S10) (13-15), but the infectious phenotype is different. Like MyD88-deficient mice, the patients are vulnerable to Streptococcus pneumoniae (16, 17), Staphylococcus aureus (18, 19), and Pseudomonas aeruginosa (20-23). However, the nine MyD88-deficient patients were normally resistant to most common bacteria, viruses, fungi, and parasites (supplementary note 1 and tables S10 and S11). By contrast, MyD88-deficient mice are vulnerable to almost all pathogens tested (at least 35 microbes, tables S1 to S3). Because MyD88- and IRAK-4–deficient patients have indistinguishable cellular phenotypes, the clinical phenotype of the 32 known IRAK-4–deficient patients may be taken into account when comparing mice and humans. The broad vulnerability of MyD88-deficient mice to experimental infections suggested that TLRs were the principal pathogen-associated molecular pattern receptors (24) or the principal microbial sensors (25) of innate immunity in vivo. The natural history of 41 patients with MyD88 or IRAK-4 deficiency suggests that the MyD88-and IRAK-4–dependent TLRs (and IL-1Rs) play a narrow, nonredundant role in protective immunity in natura (1, 26). Although narrow, this role is vital early in life, because all affected children would probably have died before the advent of antibiotics. TIR signaling seems to be less important for survival later in life (supplementary note 1) (3). This may be due to the compensatory effect of adaptive immunity (27, 28) and/or the maturation of TIR-independent innate immunity (29, 30).
Supplementary Material
Acknowledgments
We thank the patients and their families, the members of the laboratory, and our collaborators A. Alcais, C. Bidalled, M. Courat, M. N'Guyen, T. Leclerc, K. von Bernuth, E. von Bernuth, A. von Bernuth, J. von Bernuth, M. Gahr, F. de la Rocque, C. Levy, A. Lecuyer, M. Albert, F. Barrat, and R. Miller. This work was supported by grants from the INSERM, Agence Nationale de la Recherche, Institut des Maladies Rares, Programme Hospitalier de Recherche Clinique, Banque nationale de Paris Paribas Foundation, and the Dana Foundation (to J.-L.C.); grants from the NIH (U19 AIO57234-02) and the Dana Foundation (to D.C.); grants from the Hungarian Research Fund (to L.M.); and grants FIS/PI060241 (to J.Y.) and FIS/PI070329 (to M.J.) from Spain's Ministry of Health. H.v.B. was supported by the Deutsche Forschungsgemeinschaft, Legs Poix, and University San Rafaele Salute. J.-L.C. is an International Scholar of the Howard Hughes Medical Institute. The authors declare that they have no financial conflict of interest. The microarray data used in this study have been deposited in NCBI's Gene Expression Omnibus (GEO) with the accession number GSE 12124.
Footnotes
References and Notes
- 1.Casanova JL, Abel L. Science. 2007;317:617. doi: 10.1126/science.1142963. [DOI] [PubMed] [Google Scholar]
- 2.Picard C, et al. Science. 2003;299:2076. doi: 10.1126/science.1081902. [DOI] [PubMed] [Google Scholar]
- 3.Ku CL, et al. J. Exp. Med. 2007;204:2407. doi: 10.1084/jem.20070628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Supporting material is available on Science Online.
- 5.Suzuki N, et al. Nature. 2002;416:750. doi: 10.1038/nature736. [DOI] [PubMed] [Google Scholar]
- 6.Yang K, et al. Immunity. 2005;23:465. [Google Scholar]
- 7.Li C, Zienkiewicz J, Hawiger J. J. Biol. Chem. 2005;280:26152. doi: 10.1074/jbc.M503262200. [DOI] [PubMed] [Google Scholar]
- 8.Zhang SY, et al. Science. 2007;317:1522. doi: 10.1126/science.1139522. [DOI] [PubMed] [Google Scholar]
- 9.Jiang Z, Mak TW, Sen G, Li X. Proc. Natl. Acad. Sci. U.S.A. 2004;101:3533. doi: 10.1073/pnas.0308496101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smahi A, et al. Nature. 2000;405:466. doi: 10.1038/35013114. [DOI] [PubMed] [Google Scholar]
- 11.Casrouge A, et al. Science. 2006;314:308. doi: 10.1126/science.1128346. [DOI] [PubMed] [Google Scholar]
- 12.Chapgier A, et al. J. Immunol. 2006;176:5078. doi: 10.4049/jimmunol.176.8.5078. [DOI] [PubMed] [Google Scholar]
- 13.Adachi O, et al. Immunity. 1998;9:143. doi: 10.1016/s1074-7613(00)80596-8. [DOI] [PubMed] [Google Scholar]
- 14.Kawai T, Adachi O, Ogawa T, Takeda K, Akira S. Immunity. 1999;11:115. doi: 10.1016/s1074-7613(00)80086-2. [DOI] [PubMed] [Google Scholar]
- 15.Hoebe K, et al. Nature. 2003;424:743. doi: 10.1038/nature01889. [DOI] [PubMed] [Google Scholar]
- 16.Albiger B, et al. Cell. Microbiol. 2005;7:1603. doi: 10.1111/j.1462-5822.2005.00578.x. [DOI] [PubMed] [Google Scholar]
- 17.Khan AQ, Chen Q, Wu ZQ, Paton JC, Snapper CM. Infect. Immun. 2005;73:298. doi: 10.1128/IAI.73.1.298-307.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller LS, et al. Immunity. 2006;24:79. [Google Scholar]
- 19.Takeuchi O, Hoshino K, Akira S. J. Immunol. 2000;165:5392. doi: 10.4049/jimmunol.165.10.5392. [DOI] [PubMed] [Google Scholar]
- 20.Power MR, Peng Y, Maydanski E, Marshall JS, Lin TJ. J. Biol. Chem. 2004;279:49315. doi: 10.1074/jbc.M402111200. [DOI] [PubMed] [Google Scholar]
- 21.Power MR, Marshall JS, Yamamoto M, Akira S, Lin TJ. Clin. Exp. Immunol. 2006;146:323. doi: 10.1111/j.1365-2249.2006.03210.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Skerrett SJ, Liggitt HD, Hajjar AM, Wilson CB. J. Immunol. 2004;172:3377. doi: 10.4049/jimmunol.172.6.3377. [DOI] [PubMed] [Google Scholar]
- 23.Skerrett SJ, Wilson CB, Liggitt HD, Hajjar AM. Am. J. Physiol. Lung Cell. Mol. Physiol. 2007;292:L312. doi: 10.1152/ajplung.00250.2006. [DOI] [PubMed] [Google Scholar]
- 24.Janeway CA, Jr., Medzhitov R. Annu. Rev. Immunol. 2002;20:197. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 25.Beutler B, et al. Annu. Rev. Immunol. 2006;24:353. doi: 10.1146/annurev.immunol.24.021605.090552. [DOI] [PubMed] [Google Scholar]
- 26.Quintana-Murci L, Alcais A, Abel L, Casanova JL. Nat. Immunol. 2007;8:1165. doi: 10.1038/ni1535. [DOI] [PubMed] [Google Scholar]
- 27.Gavin AL, et al. Science. 2006;314:1936. doi: 10.1126/science.1135299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meyer-Bahlburg A, Khim S, Rawlings DJ. J. Exp. Med. 2007;204:3095. doi: 10.1084/jem.20071250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hirsch MS, Zisman B, Allison AC. J. Immunol. 1970;104:1160. [PubMed] [Google Scholar]
- 30.Pham LN, Dionne MS, Shirasu-Hiza M, Schneider DS. PLoS Pathog. 2007;3:e26. doi: 10.1371/journal.ppat.0030026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.