Abstract
Lithium is an anti-depressant drug that also possesses immunomodulatory functions. The anti-inflammatory effect of lithium is thought to involve activation of the transcription factor CREB, although the underlying mechanism is incompletely understood. We show here that in macrophages lithium stimulates Tpl2, a MAP kinase kinase kinase (MAP3K) known to mediate activation of extracellular signal regulated kinase (ERK) and the downstream target CREB. Lithium activates Tpl2 by inducing degradation of p105, an NF-κB precursor protein that functions as a physiological inhibitor of Tpl2. This novel function of lithium does not involve inhibition of a well-characterized lithium target, GSK3β, since other known GSK3β inhibitors do not induce p105 degradation or Tpl2 activation. Lithium also promotes the activation of Tpl2 and ERK by the TLR4 ligand LPS. On the other hand, prolonged incubation of macrophages with lithium results in dramatic loss of p105 and inhibition of LPS-stimulated NF-κB activation. Consequently, lithium both attenuates LPS-mediated pro-inflammatory gene induction and induces apoptosis in macrophages. These results provide novel insight into the anti-inflammatory function of lithium.
Keywords: Lithium, Tpl2, ERK, NF-κB, apoptosis, macrophages
1. Introduction
Inflammation serves as an important innate immune mechanism against infections [1]. A typical inflammatory response is initiated through the exposure of innate immune cells, such as macrophages and neutrophils, to microbial products [2]. The surface of these host cells have pattern recognition receptors, most importantly the toll-like receptors (TLRs), which recognize various microbial products and trigger the production of a plethora of cytokines and other pro-inflammatory mediators. These soluble immune factors in turn mediate induction of inflammation at the site of an infection, thereby preventing the spread of pathogens and facilitating the recruitment of additional immune cells/factors to the infected tissue [1]. However, deregulated inflammatory responses are associated with various human diseases, including chronic inflammatory disorders and systemic acute inflammatory diseases such as septic shock [3–5].
A well-studied TLR member, TLR4, responds to lipopolysaccharide (LPS), a bacterial cell-wall component that is responsible for the induction of septic shock by gram-negative bacteria [6]. In response to LPS stimulation, TLR4 recruits signaling adaptors and elicits activation of receptor-proximal signaling molecules, such as the interleukin-1 receptor-associated kinases (IRAKs) and the ubiquitin ligase TRAF6 [7]. The TLR signals can then be amplified to initiate several downstream signaling pathways, including those that lead to the activation of IκB kinase (IKK) and three families of MAP kinases (MAPKs), the extracellular signal-regulated kinases (ERK), the c-Jun NH2-terminal kinases (JNK), and p38. The primary function of IKK is to mediate activation of NF-κB, a transcription factor regulating genes involved in immune response, inflammation, and cell survival [8, 9]. Like IKK, the MAPKs are important for TLR-mediated induction of various target genes. Activation of MAPKs is mediated by specific MAPK kinases (MAP2K), which in turn are activated by MAPK kinase kinases (MAP3K) [10]. In particular, MEK1 and MEK2 are the MAP2Ks of two major ERK members, ERK1 and ERK2. Although different MAP3Ks are involved in the activation of MEK1/2, Tpl2 is the specific MAP3K that mediates MEK1/2 activation by a subset of inflammatory stimuli, including TLR ligands and TNF-α [11, 12].
Recent studies from us and others demonstrate that the stability and activity of Tpl2 are tightly regulated by the NF-κB1 precursor protein, p105 [13, 14]. Tpl2 is expressed as a longer and shorter isoforms, Tpl2L and Tpl2S, both being stably associated with p105 [15]. Activation of Tpl2 by LPS is mediated through degradation of p105, which in turn involves p105 phosphorylation by IKKβ [16, 17]. Upon liberation from p105, Tpl2 accesses and phosphorylates MEK1/2, leading to activation of the downstream ERK signaling pathway. The activated Tpl2, particularly Tpl2L, is rapidly degraded, which may serve as a feedback mechanism that prevents persistent ERK activation. Genetic evidence suggests that the Tpl2/ERK signaling pathway may have both pro- and anti-inflammatory functions. Tpl2 positively regulates the posttranscriptional expression and secretion of the pro-inflammatory cytokine TNF-α [11, 18]. On the other hand, Tpl2 negatively regulates the transcriptional induction of IL-12 induced by both LPS and the TLR9 ligand, CpG [19–21]. Additionally, Tpl2 mediates the production of prostaglandin E2 (PGE2) [22], an important anti-inflammatory lipid mediator that is generated during the resolving phase of an inflammation [23, 24]. Thus, understanding the mechanism of Tpl2 activation is important for developing more effective and specific approaches to modulate inflammatory responses.
The anti-depressant drug lithium is known to have immunomodulatory functions. Accumulating evidence suggests that lithium and other anti-depressant drugs have both immuno-stimulatory and immuno-suppressive functions [25–28]. Regarding the latter function, both in vitro and in vivo studies have revealed strong anti-inflammatory effect of lithium [28–30]. The anti-inflammatory function of lithium may be important for its anti-depressant efficacy, since inflammation contributes to the pathogenesis of major depression [31, 32]. It is known that depressed patients have increased levels of pro-inflammatory cytokines and acute phase proteins in the plasma. Furthermore, lithium treatment significantly reduces the number of monocytes, which may contribute to the reduction of pro-inflammatory mediators [28].
Although how lithium mediates immunomodulation is poorly understood, it has been suggested that the anti-inflammatory action of lithium involves activation of the transcription factor CREB [29, 33, 34]. It is thought that CREB inhibits the transactivation function of NF-κB by competing for nuclear coactivators [29]. Additionally, CREB may also suppress inflammation through induction of the anti-inflammatory mediator PGE2. Since the Tpl2 signaling pathway plays a critical role in CREB activation [22], we examined whether lithium functions as an inducer of Tpl2. We show that lithium induces Tpl2 activation and also potentiates LPS-stimulated Tpl2 activation. Although a well-known molecular target of lithium is glycogen synthase kinase 3 beta (GSK3β) [35], the Tpl2-activating function of lithium is unlikely mediated through GSK3β inhibition, since other GSK3β inhibitors do not display this novel activity. We further show that lithium is a potent inducer of p105 degradation, a central step in Tpl2 activation. However, prolonged incubation of macrophages with lithium causes depletion of p105 and inhibition of LPS-stimulated NF-κB activation. Consequently, lithium attenuates LPS-mediated induction of several pro-inflammatory genes and induces apoptosis in primary macrophages. Together, these findings provide molecular insight into the anti-inflammatory effect of lithium.
2. Materials and methods
2.1. Mice
C57BL6/129 mice were purchased from Jackson Laboratories and housed under specific pathogen-free conditions. Animal experiments were performed in accordance with protocols approved by the Institutional Animal Care and Use Committee of the University of Texas MD Anderson Cancer Center.
2.2. Antibodies and agents
The C-terminal specific anti-p105 and anti-p100 antibodies were provided by Dr. Nancy Rice. Antibodies for Tpl2 (anti-Cot M20), HRP-conjugated anti-Tpl2, anti-p50 (D17), c-Rel (sc-70), anti-RelB (N-17), anti-Actin (C2), iNOS (M-19), IRAK1 (H-273), Tubulin (TU-02), ERK (K-23), and phopho-ERK (P-ERK, E-4) were from Santa Cruz. Antibody for cleaved caspase-3 was from Cell Signaling. LiCl was from Fisher Scientific. NaCl, propidium iodide (PI), and LPS derived from E. coli 0127:B8 were obtained from Sigma. FITC-conjugated annexin V (annexin-FITC) was purchased from BD Biosciences, and myelin basic protein (MBP) was from Millipore.
2.3. Macrophage preparation and stimulation
Bone marrow derived macrophages were prepared from the femours and tibious of wildtype mice under the C57/BL6 and 129 mixed background as we previously described [13]. The cells were plated in growth medium and starved overnight before stimulation. Stimulated cells were washed once in PBS and used for lysate preparation.
2.4. Immunoblotting (IB) and in vitro kinase assays
Cell lysates were prepared by lysing the cells in a kinase cell lysis buffer supplemented with protease inhibitors and phosphatase inhibitors [36]. Tpl2 and IRAK1 were isolated from cell lysates by immunoprecipitation (IP) and subjected to kinase assays [36] using GST-MEK1 [13] and MBP substrates, respectively. For IB assays, cell lysates were fractionated by SDS-PAGE, and the proteins were transferred onto nitrocellulose and detected by the indicated antibodies using Enhanced Chemiluminescence Reagent (PerkinElmer Life Sciences, Inc.). Protein bands were quantified using NIH Image J software.
2.5. Electrophoresis mobility shift assays (EMSA)
Nuclear extracts were prepared from macrophages, quantitated using a protein assay reagent (Bio-Rad), and subjected to EMSA using a 32P-radiolabeled κB probe (CAA CGG CAG GGG AAT TCC CCT CTC CTT), as previously described [37].
2.6. Apoptosis assay
Apoptosis was analyzed by staining the cells with FITC-conjugated annexin V (annexin-FITC) and PI as previously described [38]. Statistical analysis was performed using the Prism software.
2.7. Real-time quantitative RT-PCR
Total cellular RNA was isolated from the stimulated macrophages and subjected to real-time quantitative RT-PCR as described [39]. The gene-specific primer sets (all for murine genes) included: IL-6, 5′-CACAGAGGATACCACTCCCAACA-3′ and 5′-TCCACGATTTCCCAGAGAACA-3′; IL-12 p40, 5′-CCA TTG AAC TGG CGT TGG AAG-3′ and 5′-ACT TGA GGG AGA AGT AGG AAT GG-3′; TNF-α, 5′-CATCTTCTCAAAATTCGAGTGACAA-3′ and 5′-CCAGCTGCTCCTCCACTTG-3′; Actin, 5′-CGTGAAAAGATGACCCAGATCA-3′ and 5′-CACAGCCTGGATGGCTACGT-3′.
3. Results
3.1. Lithium is a stimulator of Tpl2 and ERK in macrophages
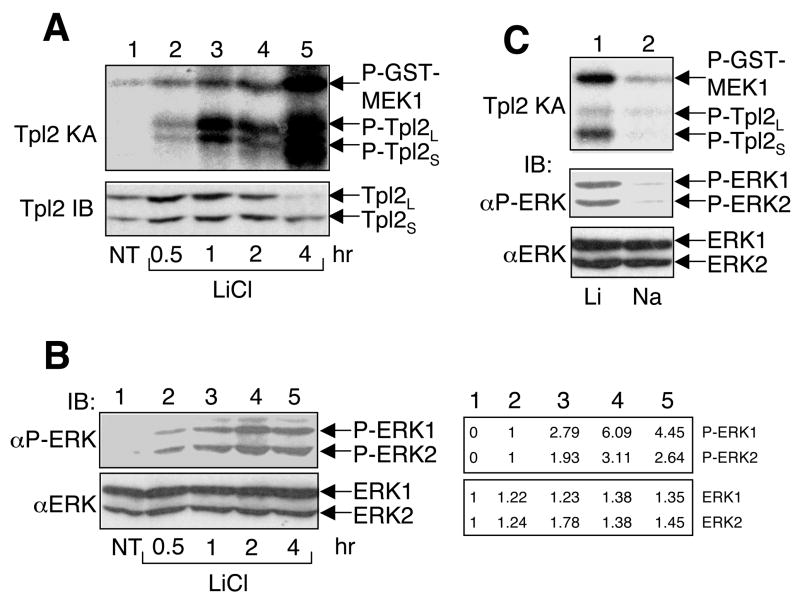
To examine the effect of lithium on Tpl2 signaling pathway, we incubated bone marrow derived macrophages (BMDM) with lithium for different times. Interestingly, lithium stimulated the catalytic activity of Tpl2, as demonstrated by phosphorylation of its substrate MEK1 and auto-phosphorylation of the longer and shorter Tpl2 isoforms (Tpl2L and Tpl2S) in an in vitro kinase assay (Fig. 1A, upper panel). The kinetics of Tpl2 activation was slow and persistent, with the maximal level of Tpl2 activation being detected at 4 hours after lithium stimulation. Prolonged incubation of macrophages with lithium also resulted in the induction of Tpl2L degradation (Fig. 1A, lower panel), an event coupled with Tpl2 activation [13].
Fig. 1.
Stimulation of Tpl2 activation and ERK phosphorylation by lithium. (A) Bone marrow derived macrophages were either not treated (NT) or incubated with LiCl (30 mM) for the indicated times. The cells were subjected to in vitro kinase assays to determine the Tpl2 kinase activity using GST-MEK1 as substrate (upper panel). The phosphorylated substrate (P-GST-MEK1) and the auto-phosphorylated Tpl2 isoforms were detected based on their 32P incorporation. Expression of the short (Tpl2S) and long (TPL2L) Tpl2 isoforms [13] was analyzed by IB using a C-terminal specific anti-Tpl2 (lower panel). (B) Cell lysates from A were subjected to IB to detect the phosphorylated ERKs (upper panel) and total ERKs (lower panel). Bands were quantified using NIH image J, and results are presented on the right of the panels. (C) Bone marrow macrophages were incubated with either LiCl (30 mM) or NaCl (30 mM) for 4 hr. Cell lysates were subjected to Tpl2 kinase assay (top panel) and phospho-ERK or total ERK IB assays (bottom two panels). Data in A-C are representative of three independent experiments.
Since a major downstream signaling event of Tpl2 is activation of ERK, we analyzed the effect of lithium on ERK activation based on its site-specific phoshorylation. As expected, lithium stimulated the phosphorylation of ERK1 and ERK2 (Fig. 1B, upper panel). This effect was not due to alteration in ERK expression (Fig. 1B, lower panel). Moreover, in contrast to the transient activation of ERK by TLR signals, lithium stimulated persistent activation of ERK, lasting for at least 4 hours (Fig. 1B). This signaling kinetics is consistent with that of Tpl2 activation. To assure that the Tpl2/ERK-stimulatory function of lithium was not due to alteration in salt concentration of the medium, we incubated the cells with sodium. In contrast to lithium, sodium did not induce the activation of Tpl2 or phosphorylation of ERK (Fig. 1C). Thus, lithium is a potent and persistent inducer of Tpl2/ERK signaling pathway in macrophages.
3.2. Lithium-stimulated Tpl2 activation is not mediated by GSK3β inhibition
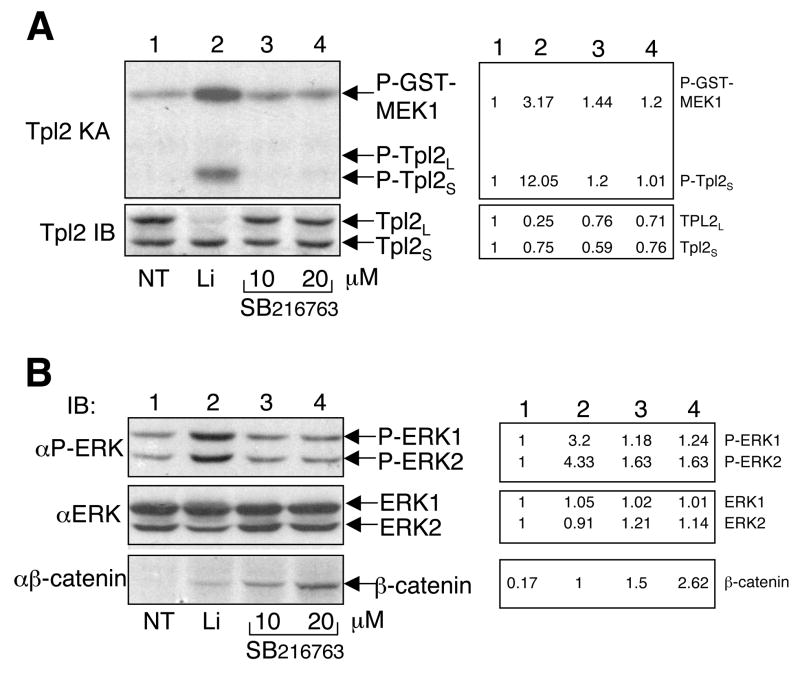
Lithium is known as an inhibitor of GSK3β, a kinase with diverse biological functions [40, 41]. A well-characterized function of GSK3β is to phosphorylate b-catenin, thereby triggering b-catenin degradation and activation of genes involved in Wnt signaling [41, 42]. By inhibiting GSK3β, lithium and other GSK3β inhibitors are capable of stabilizing b-catenin, which is often used as an indicator of GSK3β inhibition. To examine whether lithium activates Tpl2 via inhibition of GSK3β, we incubated macrophages with either lithium or a more specific GSK3β inhibitor, SB216763 [40]. As expected, lithium potently activated Tpl2 (Fig. 2A, upper panel), which was associated with degradation of the Tpl2L (Fig. 2A, lower panel). Surprisingly, SB216763 did not activate Tpl2 or induces Tpl2L degradation (Fig. 2A). Similarly, lithium, but not SB216763, induced the phosphorylation of ERK (Fig. 2B, panel 1). These results were unlikely due to a weaker GSK3β-inhibitory activity of SB216763, since this agent even more potently inhibited GSK3β, as indicated by the stabilization of b-catenin (Fig. 2B, panel 3).
Fig. 2.
Inhibition of GSK3β does not leads to Tpl2 activation or ERK phosphorylation. (A) Bone marrow derived macrophages were either not treated (NT) or incubated with LiCl (30 mM) and the indicated doses of a selective GSK3β inhibitor, SB216763. After 4 hr, Tpl2 kinase assays and IB assays were performed as in Fig. 1A. (B) Cell lysates from A were subjected to IB assays to detect phopshorylated ERK (P-ERK), total ERK, and β-catenin. For both A and B, bands were quantified by NIH Image J, and results are shown on the right side. Data are representative of three independent experiments.
3.3. Lithium induces degradation of p105 and liberation of Tpl2
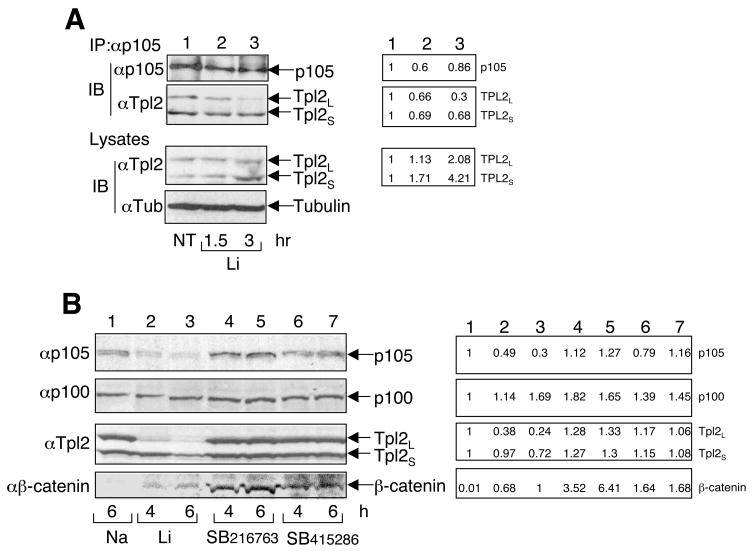
Activation of Tpl2 by LPS involves degradation of the NF-κB1 precursor protein p105 and the liberation of Tpl2 [13, 14]. Since p105 functions as a stabilizer and an inhibitor of Tpl2, p105 degradation contributes to both the activation and the subsequent degradation of Tpl2. Consistent with the kinetics of Tpl2 activation (Fig. 1A), lithium induced release of Tpl2L from p105 at 1.5 hr, which became more profound at 3 hr (Fig. 3A, panel 2). At these time points, the level of p105 was also reduced (Fig. 3A, panel 1), thus supporting the idea that the release of Tpl2 is due to the degradation of its associated p105 [13, 14]. Interestingly, prolonged incubation of macrophages with lithium led to drastic loss of p105 and Tpl2L (Fig. 3B, panels 1 and 3). This prominent action of lithium was specific, since the NF-κB2 precursor protein, p100, was not degraded under the same conditions (Fig. 3B, panel 2). Furthermore, consistent with its inability to activate Tpl2, the more selective GSK3β inhibitor SB216763 failed to stimulate the degradation of p105 and Tpl2L (Fig. 3B, lanes 4 and 5). Similar results were obtained with another GSK3β selective inhibitor, SB415286 (Fig. 3B, lanes 6 and 7). On the other hand, both SB216763 and SB415286 efficiently inhibited GSK3β activity, as evidenced from the accumulation of b-catenin (Fig. 3B, panel 4). Thus, induction of p105 degradation likely serves as a mechanism by which lithium stimulates Tpl2 activation.
Fig. 3.
Induction of p105 degradation and Tpl2 liberation by lithium but not other GSK3β inhibitors. (A) Macrophages were treated with LiCl (30 mM) for the indicated times. The p105 immune complexes were isolated by IP, and the precipitated p105 and its associated Tpl2 isoforms were detected by IB (top two panels). The expression levels of Tpl2 and the loading control tubulin were analyzed by IB (bottom two panels). (B) Macrophages were treated with NaCl (30 mM), LiCl (30 mM), SB216763 (10 μM), or SB415286 (50 μM) for the indicated times. Cell lysates were subjected to IB to detect the indicated proteins. For both A and B, bands were quantified NIH Image J, and results are presented on the right side. Data are representative of 2 (A) and 3 (B) independent experiments.
3.4. Lithium promotes LPS-stimulated activation of Tpl2 but impairs LPS-stimulated activation of NF-κB
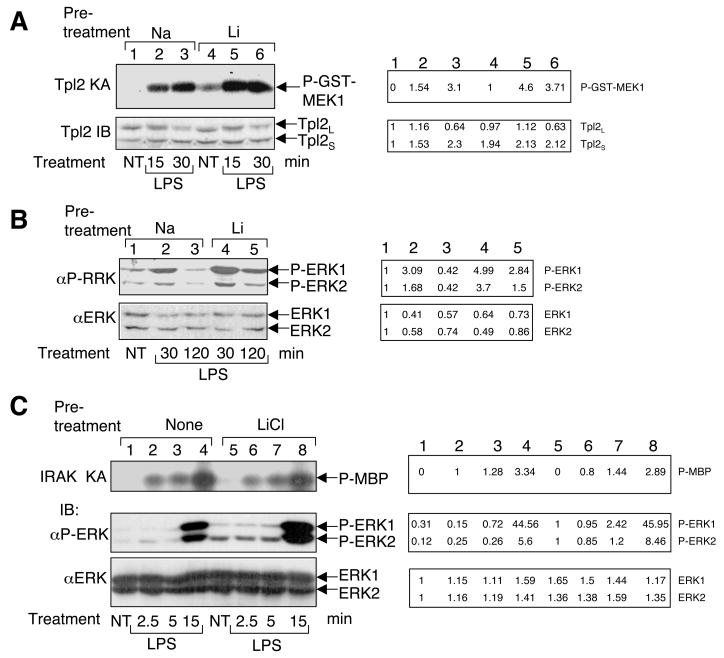
The finding that lithium induces persistent activation of Tpl2/ERK pathway prompted us to examine the effect of lithium on LPS-stimulated signaling in macrophages. For these studies we preincubated macrophages with either sodium or lithium and then stimulated the cells with LPS. As expected, lithium alone weakly stimulated Tpl2 activity within the time period of preincubation (1.5 h, Fig. 4A, lane 4). Moreover, lithium drastically enhanced LPS-stimulated Tpl2 activation (Fig. 4A, compare lanes 2 and 3 with lanes 5 and 6). Similarly, lithium also potentiated and prolonged LPS-stimulated ERK phosphorylation (Fig. 4B). On the other hand, lithium does not seem to stimulate the overall TLR signaling activity, since it did not induce the activation or promote LPS-stimulated activation of an upstream signaling molecule, IRAK1 (Fig. 4C, top panel).
Fig. 4.
Synergistic activation of Tpl2 and ERK but not IRAK by lithium and LPS. (A) Macrophages were preincubated for 1.5 hr with either NaCl (30 mM) or LiCl (30 mM) and then stimulated with LPS (2.5 mg/ml) for the indicated times or not treated (NT). Cell lysates were subjected to Tpl2 kinase assay and IB as described in Fig. 1A. (B) Macrophages were preincubated for 1.5 hr with either NaCl or LiCl and then stimulated with LPS for the indicated times. Cell lysates were analyzed by IB to detect phospho-ERK and total ERK. (C) Macrophages were preincubated with either NaCl or LiCl and then stimulated with LPS for the indicated times. The cell lysates were subjected to IRAK1 kinase assay using MBP as substrate (top panel) and IB assays to detect phospho-ERK and ERK (bottom two panels). Data in A, B, and C are representative of 5, 4, and 2 independent experiments, respectively. Bands were quantified by NIH Image J (numbers on right side).
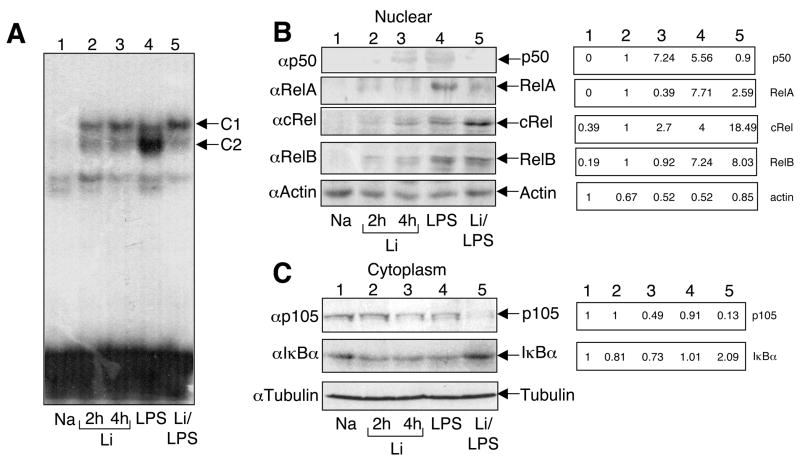
We next examined the effect of lithium on LPS-stimulated activation of NF-κB, a key transcription factor mediating the induction of pro-inflammatory genes. As expected, LPS potently stimulated the nuclear DNA-binding activity of NF-κB (Fig. 5A, lane 4). Lithium alone also induced some weak DNA-binding activity of NF-κB (Fig. 5A, lanes 2 and 3). However, the lithium-induced NF-κB species appeared to be different from those induced by LPS, as indicated by the differential migration of the complexes. More importantly, preincubation of macrophages with lithium largely blocked the LPS-stimulated NF-κB complex (Fig. 5A, lane 5). Parallel IB assays revealed that lithium inhibited LPS-stimulated nuclear translocation of two major NF-κB members, p50 and RelA (Fig. 5B). On the other hand, lithium had no effect on the activation of RelB and even enhanced the activation of c-Rel. To assess the potential mechanism of the differential effect of lithium on the induction of different NF-κB subunits, we examined the cytoplasmic level of the NF-κB inhibitors. Remarkably, in macrophages treated with lithium and LPS, the intracellular p105 was almost depleted (Fig. 5C, panel 1, lane 5). In sharp contrast, we repeatedly detected increased levels of IκBα under the same stimulation conditions (Fig. 5C, panel 2, lane 5), which might contribute to the reduced nuclear expression of RelA. The loss of p105 did not generate the mature NF-κB1 product, p50, which explains the defect in p50 activation. Furthermore, since p105 is known as a major inhibitor of c-Rel [43], the loss of p105 may contribute to the enhanced nuclear translocation of c-Rel in the lithium/LPS-treated cells. Together, these data suggest that lithium inhibits the nuclear translocation and DNA binding function of canonical NF-κB members.
Fig. 5.
Inhibition of LPS-stimulated NF-κB nuclear expression and DNA binding activity. (A) Macrophages were incubated with the indicated agents: NaCl (4 hr), LiCl (2 and 4 hr), LPS (2 hr) or LiCl plus LPS. In the case of LiCl/LPS costimulation, LPS was added after 2 hr of LiCl pretreatment and then continued for another 2 hr. Nuclear extracts were subjected to EMSA to examine the DNA binding activity of NF-κB. (B) Nuclear extracts from A were subjected to IB to detect the nuclear expression of the indicated proteins. Bands were quantified by NIH Image J (right side). (C) Cytoplasmic extracts from A were subjected to IB to examine the expression of the indicated proteins. Bands were quantified by NIH Image J (numbers on right side). Data in A-C are representative of three independent experiments.
3.5. Lithium inhibits pro-inflammatory gene induction and induces apoptosis
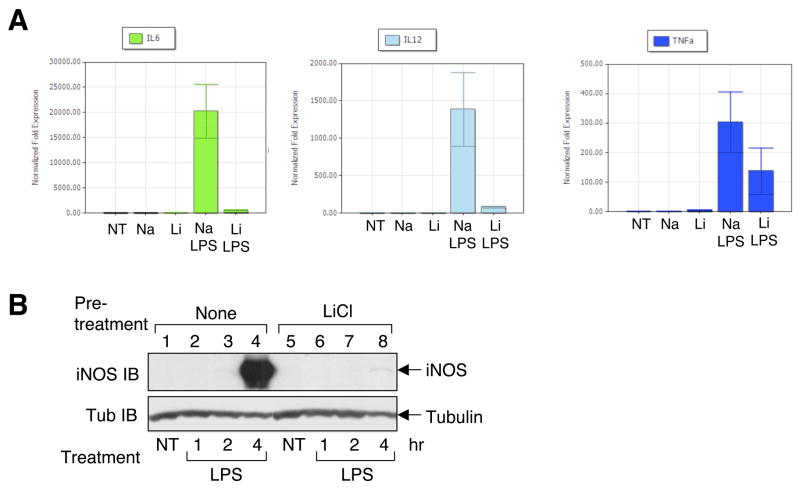
The strong effect of lithium on TLR signaling pathways prompted to examine whether lithium modulates LPS-stimulated gene expression in primary macrophages. Real-time RT-PCR analyses revealed that lithium alone was unable to induce significant expression of TNF-α, IL-6, or IL-12 (Fig. 6A). Importantly, lithium potently inhibited LPS-stimulated expression of both IL-6 and IL-12. The induction of TNF-α by LPS was also moderately inhibited by lithium. Immunoblotting assays further revealed a potent inhibitory effect of lithium on LPS-stimulated expression of iNOS (Fig. 6B).
Fig. 6.
Inhibition of LPS-stimulated gene expression by LiCl. (A) Macrophages were either not treated (NT) or treated with the indicated agents. NaCl and LiCl were both incubated for 4 hr. In the case of NaCl/LPS and LiCl/LPS double treatments, LPS was added after 1 hr of NaCl or LiCl treatment. Total cellular RNA was subjected to real-time quantitative RT-PCR. The data are presented as relative expression of the indicated genes to their corresponding untreated levels (means of three independent experiments ±SEM). (B) Macrophages were pretreated with either medium (none) or LiCl for 1 hr and then stimulated with LPS for the indicated times. Total cell lysates were subjected to IB to examine expression of iNOS and tubulin. Data are representative of 4 independent experiments.
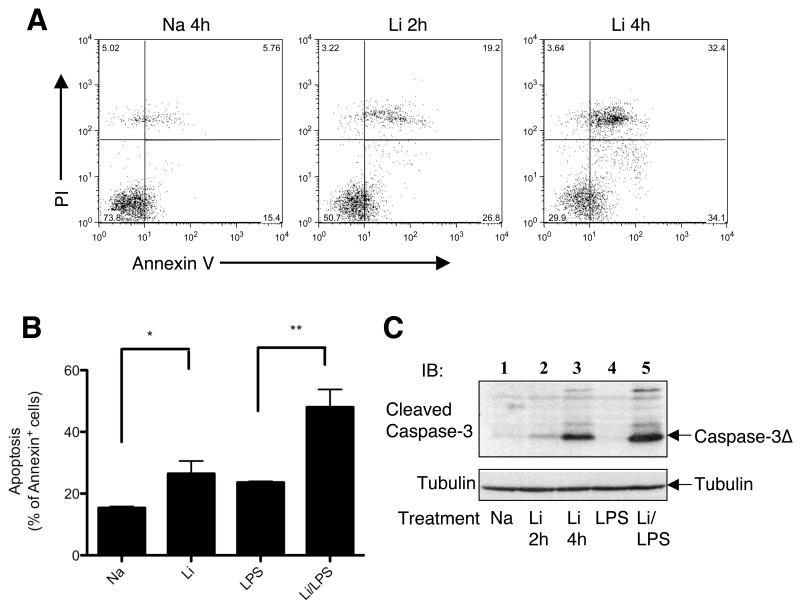
Since NF-κB is a critical survival factor in many cell types [44] and is potently inhibited by lithium, we examined whether lithium promotes macrophage apoptosis based on staining with annexin V and propidium iodide (PI). Early apoptotic cells are annexin V positive, and late apoptotic cells also become sensitive to PI staining. Remarkably, incubation of macrophages with lithium caused massive apoptosis (Fig. 7A). When the cells were incubated with LPS together with lithium, the apoptotic cells were further increased (Fig. 7B). Parallel immunoblotting assays revealed that induction of apoptosis by lithium was associated with generation of cleaved caspase-3 that is known to be the active form of this executioner caspase (Fig. 7C). Lithium together with LPS induced more robust production of the cleaved caspase-3, a result that is consistent with that of annexin staining (Fig. 7B). Thus, lithium not only inhibits pro-inflammatory gene expression but also induces apoptosis in macrophages, which provide mechanistic insight into the potent anti-inflammatory effect of this agent.
Fig. 7.
Induction of apoptosis in macrophages by lithium. (A) Macrophages were incubated with either NaCl (4 hr) or LiCl (2 and 4 hr). Apoptosis was analyzed by flow cytometry based on staining with annexin V and PI. Early apoptotic cells are annexin positive, whereas late apoptotic cells are annexin/PI double positive. (B) Macrohages were treated with NaCl (4 hr), LiCl (4 hr), LPS (4 hr), or LiCl plus LPS (4 hr). The cells were subjected to apoptosis assays as in A and presented as percentage of annexin positive cells. Data are presented as means ± s.d. *P<0.05; **P<0.01. (C) Macrophages were treated with NaCl (4 hr), LiCl (2 and 4 hr), LPS (2 hr), or LiCl (2 hr) followed by LPS (2 hr). Activation of caspase-3 was detected based on its cleavage using an antibody that specifically detects the cleaved caspase-3. Tubulin was used as loading control. Data are representative of three independent experiments.
4. Discussion
The anti-depressant drug lithium has potent anti-inflammatory functions, but the underlying signaling mechanisms are incompletely understood. In this study, we have investigated the molecular mechanism by which lithium modulates macrophage signaling and survival. We have shown that lithium not only inhibits LPS-stimulated gene expression but also stimulates apoptosis in macrophages. A major signaling function of lithium is to stimulate degradation of the NF-κB1 precursor protein p105. This action of lithium is associated with persistent activation of the Tpl2/ERK and attenuated activation of NF-κB.
Lithium is known as an inhibitor of GSK3β, although it also has other targets [35]. Interestingly, GSK3β is reported to interact with p105 and modulate the stability of p105 [45]. We thus initially thought that the induction of p105 degradation and Tpl2 activation by lithium might be due to inhibition of GSK3β. However, this idea was challenged by the finding that two other GSK3β inhibitors, SB216763 and SB415286, did not display this signaling activity. LPS-stimulated p105 degradation is mediated through phosphorylation of p105 by IKKβ [16, 17]. The fact that lithium potently potentiated and greatly prolonged LPS-stimulated p105 degradation suggests the existence of additional mechanisms. Such a synergistic effect was not seen in the degradation of IκBα. However, it is still possible that activation of IKKβ may contribute to the induction of p105 degradation by lithium. Studies are in progress to test this possibility.
Prior studies suggest that lithium suppresses the transactivation function of NF-κB through activating CREB [29]. Our finding that lithium stimulates the Tpl2/ERK signaling pathway, which is known to activate CREB [22], supports this model. However, our present study also reveals an additional mechanism of NF-κB modulation by lithium. Incubation of macrophages with lithium potently inhibits LPS-stimulated NF-κB DNA binding activity. This function of lithium involves inhibition of nuclear translocation of two major NF-κB members, p50 and RelA. It is remarkable that the major RelA inhibitor, IκBα, is accumulated when macrophages are treated with LPS in the presence of lithium. This molecular event is in sharp contrast to the drastic loss of the p50 precursor, p105. Since the loss of p105 does not lead to the generation of p50, this action of lithium may contribute to the attenuation of LPS-stimulated p50 activation. The degradation of p105 also explains how lithium induces Tpl2 activation and its subsequent degradation, since p105 serves as an inhibitor and stabilizer of Tpl2 [13, 14]. Moreover, the persistent degradation of p105 is also correlated with the induction of c-Rel nuclear translocation, which supports the previous finding that p105 is an inhibitor of c-Rel [43].
The finding that lithium induces macrophage apoptosis shed new light on the potent anti-inflammatory function of this anti-depressant drug. This function of lithium is consistent with its inhibition of LPS-stimulated NF-κB activation, since NF-κB is a major survival factor. However, it is important to note that lithium alone also induces potent apoptosis in macrophages, which cannot be explained solely through inhibition of NF-κB activation. Our preliminary data suggest that lithium inhibits the ubiquitination of RIP1 (data not shown). Recent studies suggest that ubiquitination of RIP1 is an important mechanism that regulates apoptosis induction via the caspase pathway [46–48]. When ubiquitinated, RIP1 cannot bind caspase-8 or induce apoptosis. However, upon deubiquitination, RIP1 associates with caspase-8 and triggers the apoptotic pathway. Whether deubiquitination of RIP1 is involved in lithium-induced apoptosis requires further studies.
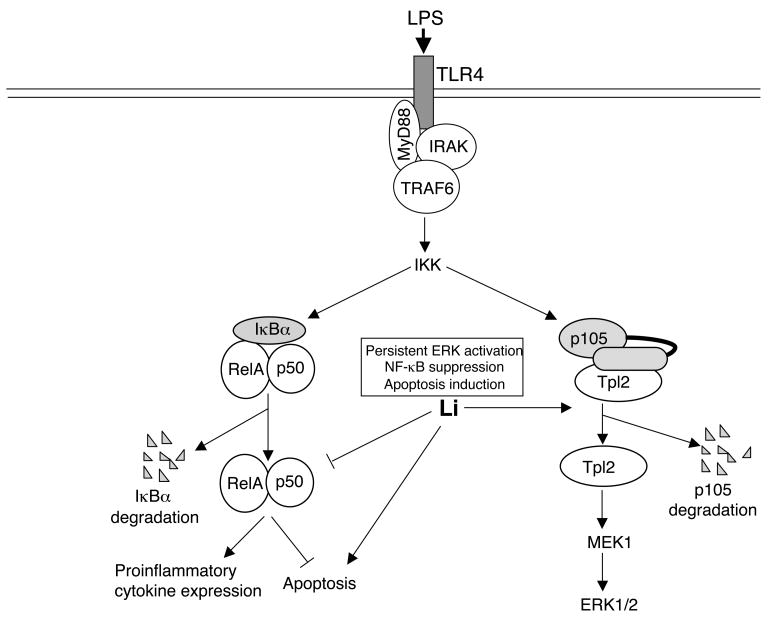
In conclusion, we demonstrate that the anti-depressant drug lithium is a potent stimulator of p105 degradation (Fig. 8). This action of lithium causes persistent activation of Tpl2 and its downstream kinase ERK. The depletion of p105 in lithium-treated cells also causes the deficiency in NF-κB1 p50, which together with the accumulation of the RelA inhibitor IκBα, contributes to the inhibition of NF-κB activation TLR4 signals. Consequently, lithium inhibits TLR4-mediated induction of pro-inflammatory genes and promotes apoptosis in macrophages. Lithium also induces macrophage apoptosis in the absence of TLR ligands, although the underlying mechanism remains to be further investigated. These findings provide molecular insight into the potent anti-inflammatory function of lithium.
Fig. 8.
Model of lithium-mediated regulation of macrophage activation and apoptosis. Lithium induces degradation of the NF-κB1 precursor protein, p105, leading to persistent activation of Tpl2 and its downstream target kinase ERK. The depletion of p105 by lithium also reduces the intracellular level of its processing product p50, which may contribute to the inhibition of LPS-stimulated canonical NF-κB activation. Through inhibition of NF-κB, lithium inhibits LPS-stimulated expression of proinflammatory cytokines and promotes LPS-stimulated macrophage apoptosis. Lithium also induces macrophage apoptosis in the absence of LPS, although the mechanism is unclear.
Acknowledgments
We thank Dr. Nancy Rice for the anti-p105 and anti-p100 antibodies. This study was supported by grants from National Institutes of Health (AI064639, GM084459, and AI057555) and start-up fund from the University of Texas MD Anderson Cancer Center.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Janeway CA, Jr, Medzhitov R. Annu Rev Immuno. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 2.Medzhitov R. Nature. 2008;454:428–435. doi: 10.1038/nature07201. [DOI] [PubMed] [Google Scholar]
- 3.Cook DN, Pisetsky DS, Schwartz DA. Nat Immunol. 2004;5:975–979. doi: 10.1038/ni1116. [DOI] [PubMed] [Google Scholar]
- 4.Liew FY, Xu D, Brint EK, O'Neill LA. Nat Rev Immunol. 2005;5:446–458. doi: 10.1038/nri1630. [DOI] [PubMed] [Google Scholar]
- 5.Andreasen AS, Krabbe KS, Krogh-Madsen R, Taudorf S, Pedersen BK, Møller K. Curr Med/Chem. 2008;15:1697–1705. doi: 10.2174/092986708784872393. [DOI] [PubMed] [Google Scholar]
- 6.Downey JS, Han J. Front Biosci. 1998;3:d468–476. doi: 10.2741/a293. [DOI] [PubMed] [Google Scholar]
- 7.Akira S, Takeda K. Nat Rev Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 8.Luo JL, Kamata H, Karin M. J Clinic Invest. 2005;115:2625–2632. doi: 10.1172/JCI26322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hayden MS, West AP, Ghosh S. Oncogene. 2006;25:6758–6780. doi: 10.1038/sj.onc.1209943. [DOI] [PubMed] [Google Scholar]
- 10.Symons A, Beinke S, Ley SC. Trends Immunol. 2006;27:40–48. doi: 10.1016/j.it.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 11.Dumitru CD, Ceci JD, Tsatsanis C, Kontoyiannis D, Stamatakis K, Lin JH, Patriotis C, Jenkins NA, Copeland NG, Kollias G, Tsichlis PN. Cell. 2000;103:1071–1083. doi: 10.1016/s0092-8674(00)00210-5. [DOI] [PubMed] [Google Scholar]
- 12.Eliopoulos AG, Wang C-C, Dumitru CD, Tsichlis PN. EMBO J. 2003;15:3855–3864. doi: 10.1093/emboj/cdg386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Waterfield M, Zhang M, Norman LP, Sun SC. Mol Cell. 2003;11:685–694. doi: 10.1016/s1097-2765(03)00070-4. [DOI] [PubMed] [Google Scholar]
- 14.Beinke S, Deka J, Lang V, Belich MP, Walker PA, Howell S, Smerdon SJ, Gamblin SJ, Ley SC. Mol Cell Biol. 2003;23(14):4739–4752. doi: 10.1128/MCB.23.14.4739-4752.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Belich MP, Salmeron A, Johnston LH, Ley SC. Nature. 1999;397:363–368. doi: 10.1038/16946. [DOI] [PubMed] [Google Scholar]
- 16.Waterfield M, Wei J, Reiley W, Zhang MY, Sun S-C. Mol Cell Biol. 2004;24:6040–6048. doi: 10.1128/MCB.24.13.6040-6048.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beinke S, Robinson MJ, Hugunin M, Ley SC. Mol Cell Biol. 2004;24:9658–9667. doi: 10.1128/MCB.24.21.9658-9667.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rousseau S, Papoutsopoulou M, Symons A, Cook D, Lucocq JM, Prescott AR, O'Garra A, Ley SC, Cohen P. J Cell Sci. 2008;121(Pt2):149–154. doi: 10.1242/jcs.018671. [DOI] [PubMed] [Google Scholar]
- 19.Feng GJ, Goodridge HS, Harnett MM, Wei XQ, Nikolaev AV, Higson AP, Liew FY. J Immunol. 1999;163:6403–6412. [PubMed] [Google Scholar]
- 20.Sugimoto K, Ohata M, Miyoshi J, Ishizaki H, Tsuboi N, Masuda A, Yoshikai Y, Takamoto M, Sugane K, Matsuo S, Shimada Y, Matsuguchi T. J Clin Invest. 2004;114:857–866. doi: 10.1172/JCI20014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tomczak MF, Gadjeva M, Wang YY, Brown K, Maroulakou I, Tsichlis PN, Erdman SE, Fox JG, Horwitz BH. J Immunol. 2006;176:1244–1251. doi: 10.4049/jimmunol.176.2.1244. [DOI] [PubMed] [Google Scholar]
- 22.Eliopoulos AG, Dumitru CD, Wang C-C, Cho J, Tsichlis PN. EMBO J. 2002;21:4831–4840. doi: 10.1093/emboj/cdf478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grbic JT, Mannick JA, Gough DB, Rodrick ML. Ann Surg. 1991;214:253–262. doi: 10.1097/00000658-199109000-00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Serhan CN, Chiang N, Van Dyke TE. Nat Rev Immunol. 2008;8:349–361. doi: 10.1038/nri2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fernandez LA, Fox RA. Clin Exp Immunol. 1980;41:527–532. [PMC free article] [PubMed] [Google Scholar]
- 26.Rybakowski JK. Pharmacopsychiatry. 2000;33:159–164. [PubMed] [Google Scholar]
- 27.Szuster-Ciesielska A, Tustanowska-Stachura A, Slotwinska M, Marmurowska-Michalowska H, Kandefer-Szerszen M. Pol J Pharmacol. 2003;55:353–362. [PubMed] [Google Scholar]
- 28.Maes M, Song C, Lin AH, Pioli R, Kenis G, Kubera M, Bosmans E. Psychopharmacology. 1999;143:401–407. doi: 10.1007/s002130050965. [DOI] [PubMed] [Google Scholar]
- 29.Martin M, Rehani K, Jope RS, Michalek SM. Nat Immunol. 2005;6:777–784. doi: 10.1038/ni1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.De Sarno P, Axtell RC, Raman C, Roth KA, Alessi DR, Jope RS. J Immunol. 2008;181:338–345. doi: 10.4049/jimmunol.181.1.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leonard BE. Neurochem Res. 2007;32:1749–1756. doi: 10.1007/s11064-007-9385-y. [DOI] [PubMed] [Google Scholar]
- 32.Dantzer R, O'Connor JC, Freund GG, Johnson RW, Kelley KW. Nat Rev Neurosci. 2008;9:46–56. doi: 10.1038/nrn2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Böer U, Eglins J, Krause D, Schnell S, Schöfl C, Knepel W. Biochem J. 2007;408:69–77. doi: 10.1042/BJ20070796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liang MH, Wendland JR, Chuang DM. Mol Cell Neurosci. 2008;37:440–453. doi: 10.1016/j.mcn.2007.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Phiel CJ, Klein PS. Annu Rev Pharmacol Toxicol. 2001;41:789–813. doi: 10.1146/annurev.pharmtox.41.1.789. [DOI] [PubMed] [Google Scholar]
- 36.Babu GR, Jin W, Norman L, Waterfield M, Chang M, Wu X, Zhang M, Sun SC. Biochim Biophys Acta. 2006;1763:174–181. doi: 10.1016/j.bbamcr.2005.12.010. [DOI] [PubMed] [Google Scholar]
- 37.Sun S-C, Ganchi PA, Ballard DW, Greene WC. Science. 1993;259:1912–1915. doi: 10.1126/science.8096091. [DOI] [PubMed] [Google Scholar]
- 38.Rivera-Walsh I, Cvijic ME, Xiao G, Sun SC. J Biol Chem. 2000;275:25222–25230. doi: 10.1074/jbc.M000444200. [DOI] [PubMed] [Google Scholar]
- 39.Zhang M, Wu X, Lee AJ, Jin W, Chang M, Wright A, Imaizumi T, Sun SC. J Biol Chem. 2008;283:18621–18626. doi: 10.1074/jbc.M801451200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meijer L, Flajolet M, Greengard P. Trends Pharmacol Sci. 2004;25:471–480. doi: 10.1016/j.tips.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 41.Cohen P, Frame S. Nat Rev Mol Cell Biol. 2001;2:769–776. doi: 10.1038/35096075. [DOI] [PubMed] [Google Scholar]
- 42.Gurvich N, Klein PS. Pharmacol Ther. 2002;96:45–66. doi: 10.1016/s0163-7258(02)00299-1. [DOI] [PubMed] [Google Scholar]
- 43.Rice NR, MacKichan ML, Israel A. Cell. 1992;71:243–253. doi: 10.1016/0092-8674(92)90353-e. [DOI] [PubMed] [Google Scholar]
- 44.Dutta J, Fan Y, Gupta N, Fan G, Gélinas C. Oncogene. 2006;25:6800–6816. doi: 10.1038/sj.onc.1209938. [DOI] [PubMed] [Google Scholar]
- 45.Demarchi F, Bertoli C, Sandy P, Schneider C. J Biol Chem. 2003;278:39583–39590. doi: 10.1074/jbc.M305676200. [DOI] [PubMed] [Google Scholar]
- 46.O'Donnell MA, Legarda-Addison D, Skountzos P, Yeh WC, Ting AT. Curr Biol. 2007;17:418–424. doi: 10.1016/j.cub.2007.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bertrand MJ, Milutinovic S, Dickson KM, Ho WC, Boudreault A, Durkin J, Gillard JW, Jaquith JB, Morris SJ, Barker PA. Mol Cell. 2008;30:689–700. doi: 10.1016/j.molcel.2008.05.014. [DOI] [PubMed] [Google Scholar]
- 48.Wang L, Du F, Wang X. Cell. 2008;133:693–703. doi: 10.1016/j.cell.2008.03.036. [DOI] [PubMed] [Google Scholar]