The isolation, crystallization and preliminary X-ray crystallographic data of the transamidosome from T. thermophilus, a ribonucleoprotein particle that provides the translational machinery with Asn-tRNAAsn, are reported.
Keywords: transamidosome, ribonucleoproteins, asparagine formation, Thermus thermophilus
Abstract
Thermus thermophilus deprived of asparagine synthetase synthesizes Asn on tRNAAsn via a tRNA-dependent pathway involving a nondiscriminating aspartyl-tRNA synthetase that charges Asp onto tRNAAsn prior to conversion of the Asp to Asn by GatCAB, a tRNA-dependent amidotransferase. This pathway also constitutes the route of Asn-tRNAAsn formation by bacteria and archaea deprived of asparaginyl-tRNA synthetase. The partners involved in tRNA-dependent Asn formation in T. thermophilus assemble into a ternary complex called the transamidosome. This particule produces Asn-tRNAAsn in the presence of free Asp, ATP and an amido-group donor. Crystals of the transamidosome from T. thermophilus were obtained in the presence of PEG 4000 in MES–NaOH buffer pH 6.5. They belonged to the primitive monoclinic space group P21, with unit-cell parameters a = 115.9, b = 214.0, c = 127.8 Å, β = 93.3°. A complete data set was collected to 3 Å resolution. Here, the isolation and crystallization of the transamidosome from T. thermophilus and preliminary crystallographic data are reported.
1. Introduction
Aminoacylation of tRNAs catalyzed by aminoacyl-tRNA synthetases (aaRS) is a key step of the translational machinery since the products of the reaction, the aminoacyl-tRNAs (aa-tRNAs), constitute adaptators that are capable of translating the genetic information contained in messenger RNA into the amino-acid sequence of proteins (reviewed by Schimmel & Söll, 1979 ▶; Lapointe & Giegé, 1991 ▶). The fidelity of the translation of the genetic information and the synthesis of functional proteins depend on the accurate aminoacylation of tRNAs by aaRSs (Chapeville et al., 1962 ▶). Until the beginning of the 1990s, it was believed that the formation of homologous aa-tRNAs was dependent on the existence of 20 distinct aaRSs: one to charge each of the 20 canonical amino acids onto the family of homologous isoaccepting tRNAs (Kern et al., 1977 ▶; Kern & Lapointe, 1979 ▶). However, biochemical analysis and in silico comparisons of genome sequences showed that most prokaryotes do not contain the complete set of 20 aaRSs (Ibba & Söll, 2004 ▶), although their proteins contain the 20 canonical amino acids. This raised the question of how the orphan tRNA is aminoacylated. Many prokaryotes, both bacteria and archaea, are deprived of asparaginyl-tRNA synthetase (AsnRS) and/or glutaminyl-tRNA synthetase (GlnRS) (Kern et al., 2005 ▶; Feng et al., 2005 ▶). In the absence of a particular aaRS, the homologous aa-tRNA is formed by mischarging of the orphan tRNA by one of the remaining aaRSs followed by conversion of the amino acid to form the cognate aa-tRNA (Kern et al., 2005 ▶; Feng et al., 2005 ▶). A similar pathway is used for selenocysteinylation of tRNASec in organisms of the three phylae that contain selenoproteins (reviewed by Böck et al., 2005 ▶) and for cysteinylation of tRNACys in methanoarchaea deprived of CysRS (Sauerwald et al., 2005 ▶).
Organisms deprived of AsnRS (e.g. Helicobacter pylori and Methanothermobacter thermautotrophicus) or more exceptionally of Asn synthetase (e.g. Thermus thermophilus and Deinococcus radiodurans) cannot directly form Asn-tRNAAsn by charging preformed Asn onto tRNAAsn. These organisms contain a nondiscriminating aspartyl-tRNA synthetase of relaxed specificity (ND-AspRS) that is able to charge Asp onto tRNAAsn as efficiently as onto the cognate tRNAAsp and a tRNA-dependent amidotransferase called GatCAB that subsequently amidates the mischarged Asp to Asn (Becker & Kern, 1998 ▶). Likewise, organisms deprived of GlnRS synthesize Gln-tRNAGln via amidation of Glu mischarged on tRNAGln by a nondiscriminating glutamyl-tRNA synthetase (ND-GluRS; Curnow et al., 1997 ▶). In bacteria tRNA-dependent conversion of Glu to Gln is promoted by the trimeric GatCAB, which exhibits a relaxed amidation specificity for Asp and Glu bound on tRNAAsn and tRNAGln, respectively, and in archaea it is promoted by the dimeric GatDE, which has a strict amidation specificity for tRNAGln-bound Glu (Kern et al., 2005 ▶; Feng et al., 2005 ▶; Schmitt et al., 2005 ▶; Oshikane et al., 2006 ▶). It has been proposed that the indirect pathways of aa-tRNA formation are the remnants of ancestral routes that formed the aa-tRNA before the metabolic pathways of free amino-acid formation and the modern aaRSs capable of charging the free amino acid onto the cognate tRNA emerged. Therefore, investigation of the nonconventional aminoacylation systems contributes to our understanding of the evolution of the translational machinery. The indirect pathways of aa-tRNA formation raise two questions: (i) how does the transfer of the mischarged aa-tRNA from aaRS to transamidase occur in a way that protects its labile ester bond from hydrolysis and (ii) what mechanism prevents the use of mischarged aa-tRNA by the translation machinery and the misincorporation of the wrong amino acid into proteins?
Analysis of the properties of the partners of the indirect pathway of Asn-tRNAAsn formation from T. thermophilus revealed that they associate in a ribonucleoprotein particle, called the transamidosome, of molecular weight 380 kDa that contains a dimeric ND-AspRS, two tRNAAsn molecules and two trimeric GatCAB molecules (Bailly et al., 2007 ▶). A model of this complex based on the three-dimensional structures of the AspRS–tRNAAsp complex (Cavarelli et al., 1993 ▶; Charron et al., 2003 ▶) and of Staphylococcus aureus GatCAB (Nakamura et al., 2006 ▶) suggested that the tRNA is sandwiched between the AspRS subunits and the GatCAB (Bailly et al., 2007 ▶). Kinetic investigations demonstrated that the particle synthesizes Asn-tRNAAsn in the presence of ATP, MgCl2, l-Asp and l-Gln as an amido-group donor. Comparison of the uncoupled and the transamidosome-coupled reactions revealed the functional advantages conferred by the assembly of the partners: (i) the affinity of GatCAB for tRNAAsn increases by two orders of magnitude when bound on AspRS, (ii) the steady-state rate of Asn-tRNAAsn formation increases fourfold when the partners are bound in a complex and (iii) the half-life of the ester bond of the mischarged Asp-tRNAAsn increases twofold inside the transamidosome (Bailly et al., 2007 ▶). Most complexes involved in tRNA aminoacylation are formed exclusively of proteins, whereas nucleoproteins such as the transamidosome are exceptional. It is noteworthy that the transamidosome dedicated to tRNA asparaginylation is an atypical ribonucleoprotein since beside its contribution to the stability of the ternary complex, the RNA is also the substrate of both enzymatic protein partners. This property confers a nonconventional dynamic character to this ribonucleoprotein particle.
To obtain a better insight into the structural properties of the transamidosome, we crystallized it in order to establish its three-dimensional structure. Here, we describe the isolation of the complex from T. thermophilus and its crystallization. Preliminary crystallographic data and structural analysis are reported.
2. Materials and methods
2.1. Preparation of the transamidosome from T. thermophilus
The ND-AspRS, the GatCAB amidotransferase and tRNAAsn from T. thermophilus overexpressed in Escherichia coli were purified as described previously (Bailly et al., 2008 ▶). The complex was isolated by preparative gel filtration on a 145 ml Superdex G200 column equilibrated with 50 mM HEPES–NaOH buffer pH 7.2, 30 mM KCl, 6 mM MgCl2, 5 mM β-mercaptoethanol and 0.1 mM Na2EDTA in a 1.5 ml reaction mixture containing a 17 µM concentration of each of the ND-AspRS subunits and GatCAB and a 25 µM concentration of tRNAAsn in the equilibration buffer. Under these experimental conditions, the equilibrium is strongly displaced toward the ternary complex and the complex can be separated from excess tRNAAsn by chromatography. The concentration of the complex was estimated using the molar extinction coefficient (∊260nm = 1.48 × 106 M −1 cm−1) of a solution containing each partner at a concentration of 10 µM.
2.2. Activity measurements of the transamidosome
A 100 µl reaction mixture containing 10 µM of the transamidosome in gel-filtration buffer was incubated for 5 min at 310 K in the presence of 40 µM [14C]Asp and 1 mM ATP to aspartylate the tRNAAsn present in the particle. The reaction mixture was then divided into two 50 µl samples. One of the samples was supplemented with 2 mM Gln as an amido-group donor and incubated for 5 min at 310 K to convert the tRNAAsn-bound Asp to Asn. The two samples were then submitted to phenol extraction and the [14C]aa-tRNAAsn isolated by ethanol precipitation was hydrolyzed by 30 min incubation in 0.1 N NaOH at 310 K. The neutralized hydrolyzates were dried and then dissolved in 3 µl water before analysis of the released [14C]-amino acid by TLC (Bailly et al., 2008 ▶).
2.3. Static and dynamic light-scattering measurements
Dynamic light-scattering (DLS) measurements were performed with a Zetasizer Nano S instrument (Malvern Instruments, Malvern, UK). Samples consisting of 40 µl transamidosome at 5–20 µM in gel-filtration buffer were transferred into quartz cells. The laser light (λ = 633 nm) backscattered at an angle of 173° was measured at 293 K. Data were processed using the DTS software (Dispersion Technology Software) provided by the manufacturer.
2.4. Crystallization
Crystallization conditions were searched for using the vapour-diffusion technique in sitting drops with sparse matrices from Nextal, Hampton Research, Jena Bioscience and Qiagen. Drops were prepared with a Mosquito robot (TTP LabTech, Melbourn, UK). Transamidosome at 3.5 mg ml−1 was mixed with reservoir solution in ratios of 0.5:1, 1:1 and 1.5:1 at 293 K. Only the condition containing 0.1 M MES–NaOH buffer pH 6.5, 0.2 M MgCl2 and 10%(v/v) PEG 4000 gave tiny crystals. Streak-seeding was used to optimize this condition. A sitting drop was first prepared by mixing 5 µl transamidosome solution with 2 µl reservoir solution and equilibrated against 500 µl reservoir solution at 293 K. After 2 d, streak-seeding was performed with a horse hair soaked in a reservoir solution containing crystal fragments obtained by strongly stirring larger crystals in a vortex apparatus. Streak-seeding produced slightly larger crystals measuring 100–200 µm; these crystals were then used for a new round of seeding. Using the same approach as before, several crystals were crushed in the reservoir solution and 48 drops were then seeded with the same hair soaked in this mixture only once. Crystal growth took about three weeks.
3. Results and discussion
3.1. Purification and characterization of the transamidosome
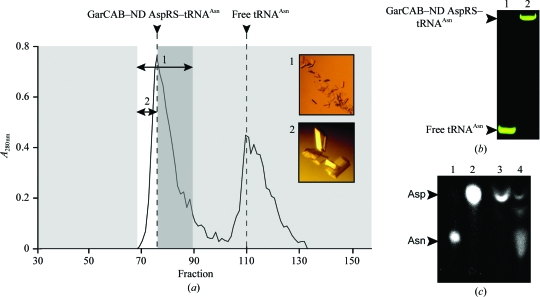
The three partners of the transamidosome from T. thermophilus, ND-AspRS, GatCAB and tRNAAsn, were isolated in high yields after overexpression in E. coli strains. 50 mg pure ND-AspRS (specific activity 8 nmol Asp-tRNAAsn formed per minute per milligram of protein at 310 K) was obtained from 50 g of cells and 30 mg GatCAB (specific activity 1700 nmol Asn-tRNAAsn formed per minute per milligram of protein at 310 K) was obtained from 60 g of cells. 40 mg pure tRNAAsn (accepting capacity 38 nmol mg−1) and 70 mg tRNAAsn of 80% purity were isolated from 200 g of cells. Preparative gel filtration led to the isolation of 9.7 mg (25.5 nmol) homogeneous transamidosome (Fig. 1 ▶ a), which was stored at 277 K. The dissociation constants of ∼2 µM for the binary AspRS–tRNAAsn complex and ∼0.6 µM for the GatCAB–AspRS–tRNAAsn complex (Bailly et al., 2007 ▶) indicate that the equilibrium of association is strongly displaced toward the ternary complex under the experimental conditions of gel filtration when AspRS subunits, GatCAB and tRNAAsn are mixed at concentrations of 10, 10 and 20 µM, respectively (Bailly et al., 2007 ▶). Consequently, the strong affinity between the partners combined with the use of high concentrations is favourable for the isolation of the complex under non-equilibrium conditions without significant dissociation. The purity of the transamidosome, analyzed by native PAGE, shows that the complex migrates as a single population of particles (Fig. 1 ▶ b). Its activity was checked by measurement of aminoacylation of the tRNAAsn bound to the complex in the presence of Asp and ATP (Fig. 1 ▶ c, lane 3) and subsequently by conversion of the tRNA-bound Asp into Asn in the presence of ATP and Gln as an amido-group donor (Fig. 1 ▶ c, lane 4).
Figure 1.
Isolation and structural and functional characterization of the transamidosome from T. thermophilus. (a) Isolation of the ribonucleoprotein particle by gel filtration. The elution profile A 280nm for fractions of the mixture containing 10 µM of each of the ND-AspRS subunits and GatCAB and 20 µM tRNAAsn is represented. The inserts show the crystals obtained from mixed fractions from elution peak (1) or from a mixture of the three first fractions from the beginning of peak (2). (b) Native PAGE analysis of the transamidosome eluted in (a). Lane 1, tRNAAsn eluted in the second peak of the elution profile in (a); lane 2, transamidosome eluted in the first peak of the elution profile in (a). (c) Functional analysis of the transamidosome eluted in (a). The transamidation reaction was conducted and the reaction mix was analyzed as described in §2. Lane 1, free [14C]Asn; lane 2, free [14C]Asp; lane 3, [14C]-amino acid released from [14C]aa-tRNA formed by incubation of the transamidosome in the presence of [14C]Asp and ATP; lane 4, [14C]-amino acid released from [14C]aa-tRNA formed by incubation of the transamidosome in the presence of [14C]Asp, ATP and Gln as the amido-group donor.
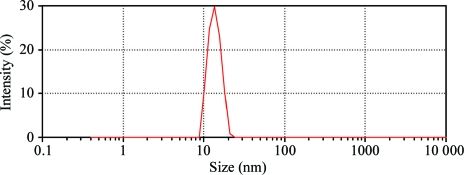
The complex was stable and reproducibly crystallized after storage for several weeks at 277 K in 0.1 M HEPES–NaOH buffer pH 7.2 containing 30 mM KCl, 6 mM MgCl2, 5 mM β-mercaptoethanol and 0.1 mM Na2EDTA. The unchanged retention time of the pure complex on the gel-filtration column after 3–4 weeks storage confirmed the absence of either denaturation or degradation of the partners. In DLS the complex exhibited a narrow intensity peak corresponding to a single population of particles with a mean diffusion coefficient of 3.0 ± 0.1 × 10−7 cm2 s−1 (Fig. 2 ▶). Assuming that the latter have a globular shape, their hydrodynamic diameter and their molecular weight would be 13.6 ± 0.4 nm and 300 kDa, respectively. Additionally, static light scattering (SLS) allowed the determination of a molecular weight of 400 kDa. These values agree with that determined by gel filtration (380 kDa) and with assembly of the three partners, i.e. the dimeric AspRS, the trimeric GatCAB and the tRNAAsn, in a 1:2:2 stoichiometry. The transmidosome particles were characterized by a very low polydispersity, which was reproducibly ≤8% at 277, 293, 310 and 323 K. This property was very encouraging for attempting crystallization assays.
Figure 2.
Dynamic light-scattering analysis of pure transamidosome particle. The measurements were performed as described in §2. The graph represents the distribution of the intensity of light scattered at 633 nm as a function of particle size (in nm).
3.2. Data collection and X-ray diffraction analysis
Prior to flash-freezing in liquid nitrogen, crystals were dehydrated overnight at 281 K by replacing the reservoir solution with 0.1 M MES–NaOH buffer pH 6.5, 0.2 M MgCl2, 30%(v/v) PEG 400 and 10%(v/v) PEG 4000. Several other dehydration methods using either lower PEG 400 concentrations or glycerol were unsuccessful, as was soaking crystals in different solutions containing glycerol or PEG. Crystals of the best quality were obtained using the transamidosome particles in the three first fractions of the gel-filtration elution peak (Fig. 1 ▶ a).
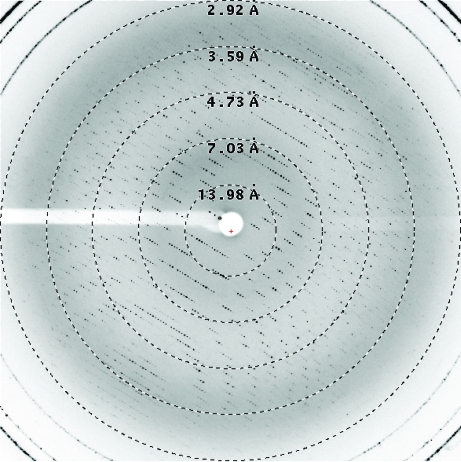
Diffraction data were collected to a resolution limit of 3 Å (Fig. 3 ▶). A total of 400 oscillation images were collected with an interval of 0.5° at a wavelength of 0.9807 Å using a crystal-to-detector distance of 320 nm on the X10SA beamline of the Swiss Light Source (Paul Scherer Institute, Villigen, Switzerland). Reflections were indexed and scaled with XDS (Kabsch, 1993 ▶). The data statistics are displayed in Table 1 ▶. Dehydration improved the diffraction resolution from 4 to 3 Å and changed the space group from P212121 to P21. Indeed, data collected from crystals diffracting to between 3.5 and 4 Å were only processable in the orthorhombic space group P212121 despite a high overall R merge (>20%). However, molecular replacement was possible on these data sets (data not shown).
Figure 3.
X-ray diffraction pattern of the transamidosome. Dashed circles indicate the diffraction resolution limits.
Table 1. Data-collection statistics.
Values in parentheses are for the last resolution shell (3.1–3.0 Å).
| Beamline | X10SA, Swiss Light Source |
| Space group | P21 |
| Wavelength (Å) | 0.9807 |
| Resolution range (Å) | 50–3.0 |
| Unit-cell parameters (Å, °) | a = 115.9, b = 214.0, c = 127.8, β = 93.3 |
| Total No. of reflections | 527469 |
| No. of unique reflections | 123945 |
| Completeness | 99.9 (99.9) |
| Rmeas† (%) | 15.4 (80.5) |
| Rmrgd-F† (%) | 14 (63.9) |
| 〈I/σ(I)〉 | 10.3 (2.4) |
| Matthews coefficient (Å3 Da−1) | 3.17 |
| Solvent content (%) | 65.3 |
R meas and R mrgd-F, defined according to Diederichs & Karplus (1997 ▶), are quality measures of the individual intensity observations and of the reduced structure-factor amplitudes, respectively.
After dehydration, crystals belonged to the monoclinic space group, with unit-cell parameters a = 115.9, b = 214.0, c = 127.8 Å, α = γ = 90.00, β = 93.3°.
3.3. Preliminary structure analysis
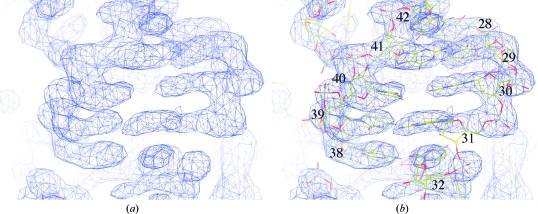
The structure was solved by molecular replacement performed using the program Phaser from the PHENIX package (Storoni et al., 2004 ▶; Adams et al., 2002 ▶). The structures of ND-AspRS from T. thermophilus (PDB code 1n9w; Charron et al., 2003 ▶) and of S. aureus GatCAB (PDB code 2f2a; Nakamura et al., 2006 ▶) were used as search models. Two molecules of dimeric AspRS and two molecules of trimeric GatCAB were found per asymmetric unit, but no tRNA could be located in the transamidosome particle when this procedure was applied using two search models, i.e. tRNAAsp from E. coli (PDB code 1c0a; Eiler et al., 1999 ▶) and S. cerevisiae (PDB 1tra; Westhof & Sundaralingam, 1986 ▶). After density modification using the program RESOLVE (Terwilliger, 2000 ▶), clear electron density appeared in the map indicating the presence of four copies of tRNAAsn (Figs. 4 ▶ a and 4 ▶ b). The structure model of the ribonucleoprotein particle was then built with Coot (Emsley & Cowtan, 2004 ▶) and refined with PHENIX. The composition of the asymmetric unit shows the addition of one dimeric AspRS and two tRNAAsn molecules compared with the complex characterized in solution. Structure determination is in progress.
Figure 4.
(a) Electron-density map after density modification, (b) nucleotides of the tRNAAsn anticodon arm fitted into the density; the nucleotides are numbered according to tRNA-numbering nomenclature.
Acknowledgments
The work was supported by the Centre National de la Recherche Scientifique (CNRS), the Université Louis Pasteur (Strasbourg) and the Association pour la Recherche sur le Cancer (ARC). We thank the team at beamline X10SA and in particular Dr Takashi Tomizaki from the Swiss Light Source and the members of ESRF beamlines ID29, ID23 and ID14. We are also grateful to Jens Preben Morth, Laure Yatime and Thomas Boesen for helpful discussions and advice, Eric Westhof (IBMC Strasbourg) and Poul Nissen (Aarhus University, Denmark) for constant support, E. Ennifar and G. Bec (IBMC, Strasbourg) for facilitating some steps of this work and M. Brayé for technical assistance. M. Blaise was the recipient of funding from the Association Française contre les Myopathies (AFM) and Danscatt, and M. Bailly was the recipient of a fellowship from ARC.
References
- Adams, P. D., Grosse-Kunstleve, R. W., Hung, L.-W., Ioerger, T. R., McCoy, A. J., Moriarty, N. W., Read, R. J., Sacchettini, J. C., Sauter, N. K. & Terwilliger, T. C. (2002). Acta Cryst. D58, 1948–1954. [DOI] [PubMed]
- Bailly, M., Blaise, M., Lorber, B., Becker, H. D. & Kern, D. (2007). Mol. Cell, 28, 228–239. [DOI] [PubMed]
- Bailly, M., Blaise, M., Roy, H., Deniziak, M., Lorber, B., Birck, C., Becker, H. D. & Kern, D. (2008). Methods, 44, 146–163. [DOI] [PubMed]
- Becker, H. D. & Kern, D. (1998). Proc. Natl Acad. Sci. USA, 95, 12832–12837. [DOI] [PMC free article] [PubMed]
- Böck, A., Thanbichler, M., Rother, M. & Resch, A. (2005). The Aminoacyl-tRNA Synthetases, edited by M. Ibba, C. Francklyn & S. Cusack, pp. 320–327. Georgetown, USA: Landes Bioscience.
- Cavarelli, J., Rees, B., Ruff, M., Thierry, J.-C. & Moras, D. (1993). Nature (London), 362, 181–184. [DOI] [PubMed]
- Chapeville, F., Lipman, F., Ehrenstein, G. V., Weisblum, B., Ray, W. H. & Benzer, S. (1962). Proc. Natl Acad. Sci. USA, 48, 1086–1092. [DOI] [PMC free article] [PubMed]
- Charron, C., Roy, H., Blaise, M., Giegé, R. & Kern, D. (2003). EMBO J.22, 1632–1643. [DOI] [PMC free article] [PubMed]
- Curnow, A. W., Hong, R., Yuan, R., Kim, S. I., Martins, O., Winkler, W., Henkin, T. M. & Söll, D. (1997). Proc. Natl Acad. Sci USA, 94, 11819–11826. [DOI] [PMC free article] [PubMed]
- Diederichs, K. & Karplus, P. A. (1997). Nature Struct. Biol.4, 269–275. [DOI] [PubMed]
- Eiler, S., Dock-Bregeon, A. C., Moulinier, L., Thierry, J. C. & Moras, D. (1999). EMBO J.18, 6532–6541. [DOI] [PMC free article] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Feng, L., Tumbula-Hansen, D., Min, S., Namgoong, S., Salazar, O., Orellana, O. & Söll, D. (2005). The Aminoacyl-tRNA Synthetases, edited by M. Ibba, C. Francklyn & S. Cusack, pp. 314–318. Georgetown, USA: Landes Bioscience.
- Ibba, M. & Söll, D. (2004). Genes Dev.18, 731–738. [DOI] [PubMed]
- Kabsch, W. (1993). J. Appl. Cryst.26, 795–800.
- Kern, D., Dietrich, A., Fasiolo, F., Renaud, M., Giegé, R. & Ebel, J. P. (1977). Biochimie, 59, 453–462. [DOI] [PubMed]
- Kern, D. & Lapointe, J. (1979). Biochimie, 61, 1257–1272. [DOI] [PubMed]
- Kern, D., Roy, H. & Becker, H. (2005). The Aminoacyl-tRNA Synthetases, edited by M. Ibba, C. Francklyn & S. Cusack, pp. 193–205. Georgetown, USA: Landes Bioscience.
- Lapointe, J. & Giegé, R. (1991). Translation in Eukaryotes, edited by H. Trachsel, pp. 35–69. Boca Raton: CRC Press.
- Nakamura, A., Yao, M., Chimnaronk, S., Sakai, N. & Tanaka, I. (2006). Science, 312, 1954–1958. [DOI] [PubMed]
- Oshikane, H., Sheppard, K., Fukai, S., Nakamura, Y., Ishitani, R., Numata, T., Sherrer, R. L., Feng, L., Schmitt, E., Panvert, M., Blanquet, S., Mechulam, Y., Söll, D. & Nureki, O. (2006). Science, 312, 1950–1954. [DOI] [PubMed]
- Sauerwald, A., Zhu, W., Major, T. A., Roy, H., Palioura, S., Jahn, D., Whitman, W. B., Jates, J. R. III, Ibba, M. & Söll, D. (2005). Science, 307, 1969–1972. [DOI] [PubMed]
- Schimmel, P. & Söll, D. (1979). Annu. Rev. Biochem.48, 601–648. [DOI] [PubMed]
- Schmitt, E., Panvert, M., Blanquet, S. & Mechulam, Y. (2005). Structure, 42, 932–939. [DOI] [PubMed]
- Storoni, L. C., McCoy, A. J. & Read, R. J. (2004). Acta Cryst. D60, 432–438. [DOI] [PubMed]
- Terwilliger, T. C. (2000). Acta Cryst. D56, 965–972. [DOI] [PMC free article] [PubMed]
- Westhof, E. & Sundaralingam, M. (1986). Biochemistry, 25, 4868–4878. [DOI] [PubMed]