Oih-1, a β-lactamase enzyme isolated from the deep-sea bacterium O. iheyensis, has been crystallized and a complete X-ray diffraction data set has been collected to 1.65 Å resolution.
Keywords: Oih-1, β-lactamases, Oceanobacillus iheyensis
Abstract
Bacterial resistance to the β-lactam family of antibiotics is primarily the result of the deactivation of the drugs by β-lactamase enzymes. The gene encoding the proficient β-lactamase Oih-1 from the alkaliphilic and halotolerant Gram-positive bacterium Oceanobacillus iheyensis has been cloned and the mature wild-type protein (comprising 274 amino-acid residues) has been expressed in Escherichia coli and subsequently purified to homogeneity. Oih-1 crystallized in two crystal forms both belonging to the trigonal space group P3121 but with distinctly different unit-cell parameters. Synchrotron diffraction data were collected to high resolution (1.65–1.75 Å) from both crystal forms on beamlines BL7-1 and BL11-1 at SSRL (Stanford, California, USA).
1. Introduction
β-Lactams are the most common antibiotics in clinical use and represent more than 60% of the total world consumption of antimicrobial drugs (Livermore & Woodford, 2006 ▶). A major mechanism of bacterial resistance to β-lactam antibiotics is the production of β-lactamases, enzymes which hydrolyze the conserved four-membered ring of β-lactams, rendering them inactive (Poole, 2004 ▶). Four classes of β-lactamases (A–D) have been identified (Ambler et al., 1991 ▶), with class A enzymes being the most abundant in both Gram-negative and Gram-positive pathogens. Originating as relatively narrow-spectrum enzymes capable of hydrolyzing mostly penicillins and early (first- and second-generation) cephalosporins, some class A β-lactamase enzymes such as TEM and SHV have evolved into large superfamilies of extended-spectrum β-lactamases (ESBLs) that are capable of hydrolyzing modern cephalosporins, monobactams (Jacoby & Bush, 2007 ▶) and, more recently in the case of the GES family, carbapenems (Smith et al., 2007 ▶). The mechanism of action of the class A β-lactamases involves the deprotonation of a serine residue (Minasov et al., 2002 ▶) followed by nucleophilic attack on the β-lactam ring by the deprotonated serine to produce an acyl-enzyme intermediate. This intermediate is subsequently deacylated in a step involving an activated water molecule (Strynadka et al., 1992 ▶).
Bacteria in the environment are continuously trying to gain an advantage over other organisms by secreting specific chemicals and it is thought that the first β-lactamase enzymes may have originated from the penicillin-binding proteins (PBPs), a group of cell-wall biosynthetic enzymes (Fisher et al., 2005 ▶), as a defense against β-lactam molecules produced by other microorganisms. The clinical use of these antibiotics has markedly accelerated the evolution and spread of the genes for these enzymes, leading to the current resistance problem. Analysis of the genome sequence of the novel deep-sea bacterium Oceanobacillus iheyensis (Takami et al., 2002 ▶) allowed us to identify the gene encoding a putative class A β-lactamase that we called Oih-1. O. iheyensis is an alkaliphilic and halotolerant bacterium collected from a depth of 1050 m off the coast of Japan (Lu et al., 2001 ▶) and, given its extreme habitat, this β-lactamase is a prime example of an antibiotic resistance enzyme which has evolved in the complete absence of clinical selection.
The Oih-1 β-lactamase shows about 55% sequence identity to class A β-lactamases from a number of Bacillus species and has the fingerprint sequence motifs SxxK, SD(G)N and KTG. The enzyme has been expressed in Escherichia coli, purified to homogeneity and crystallized; here, we report the preliminary X-ray analysis of the Oih-1 crystals.
2. Materials and methods
2.1. Cloning, expression and purification of Oih-1
The gene for wild-type Oih-1 β-lactamase (GenBank accession No. FJ905107) including its own predicted leader sequence was optimized for expression in E. coli and custom-synthesized such that it had a hexahistidine tag fused to the C-terminus. Recognition sequences for NdeI and HindIII endonucleases were introduced at the 5′- and 3′-ends of the Oih-1 β-lactamase gene, respectively, and the gene was cloned into the unique NdeI and HindIII sites of the pET24a(+) expression vector (Novagen). E. coli BL21 (DE3) harboring the pET24a(+) vector with the gene encoding mature Oih-1 β-lactamase was grown overnight in LB medium supplemented with 30 µg ml−1 kanamycin A. The bacterial suspension was diluted 100-fold in 300 ml of the same medium and incubated at 310 K with shaking until the OD600 was 1.0. Production of β-lactamase was induced with 0.8 mM IPTG and the culture was incubated overnight at 295 K. Cells were pelleted by centrifugation (3000g for 30 min) and disrupted by sonication in buffer A (25 mM HEPES pH 7.5, 1 mM EDTA, 0.2 mM DTT). After two cycles of centrifugation (20 000g for 30 min and 150 000g for 1 h) the resulting supernatant was applied onto a DEAE anion-exchange column equilibrated with buffer A. Oih-1 was eluted with a linear gradient of buffer A (without EDTA) and buffer B (25 mM HEPES pH 7.5, 1 M NaCl, 0.2 mM DTT). Fractions were analyzed by their ability to hydrolyze the chromogenic substrate nitrocefin (O’Callaghan et al., 1972 ▶) and by SDS–PAGE. Fractions containing β-lactamase were pooled together and further purified on a HiTrap Chelating Affinity column (GE Healthcare) equilibrated with 25 mM HEPES pH 7.5, 0.5 M NaCl. The column was washed with buffer C (20 mM sodium phosphate pH 7.4, 0.5 M NaCl, 20 mM imidazole) and Oih-1 was eluted with buffer C containing increasing concentrations of imidazole. Fractions were analyzed by SDS–PAGE and those containing pure β-lactamase were combined, dialyzed against 25 mM HEPES pH 7.5, concentrated to 53 mg ml−1 and stored at 193 K. The purity of the protein was confirmed by the presence of a single band on an overloaded 12% SDS–PAGE run under reducing conditions (Fig. 1 ▶). The mature enzyme comprises 274 amino acids with an estimated molecular weight of 30.4 kDa.
Figure 1.
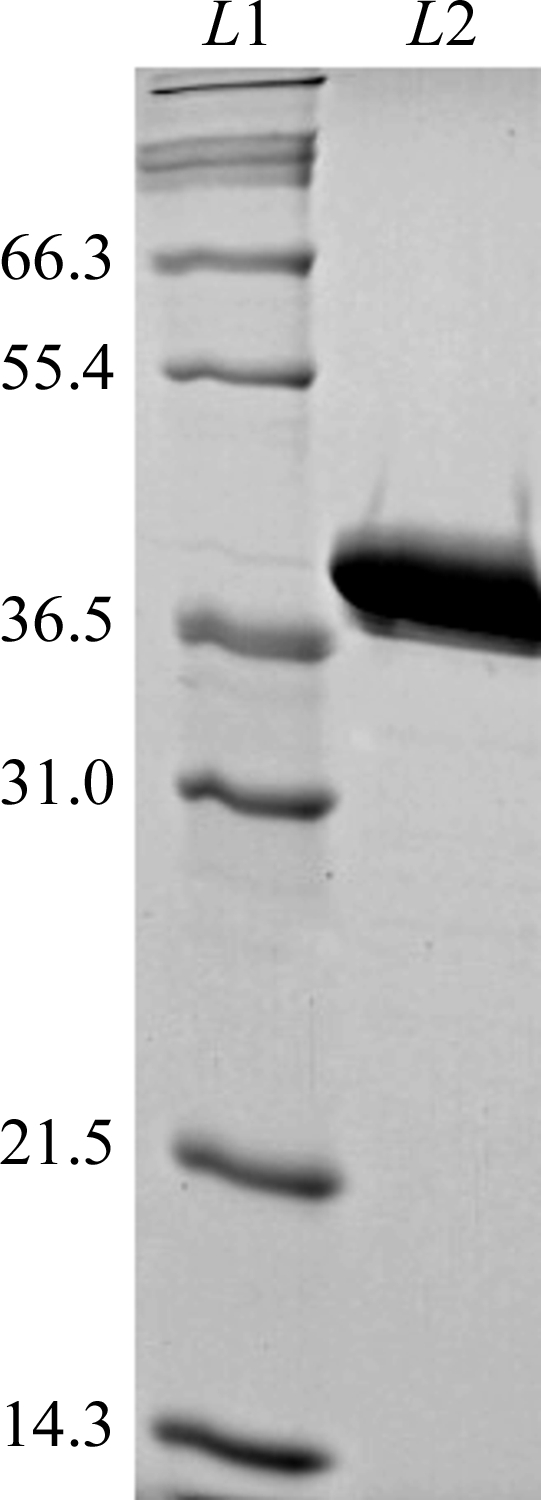
A 12% SDS–PAGE of Oih-1 under reducing conditions. Lane L1 shows molecular-weight standards with the estimated weight in kDa given at the side. Lane L2 shows 10 µg purified mature Oih-1 protein. Oih-1 appears to run aberrantly on SDS–PAGE, showing a higher apparent molecular weight.
2.2. Crystallization
Initial sitting-drop crystallization trials were performed with the commercially available sparse-matrix screens Crystal Screen I (CS I) and Crystal Screen II (CS II) from Hampton Research. These trials were carried out in Intelli-Plates (Art Robbins Instruments) using a reservoir volume of 75 µl and drops comprising 1 µl protein solution and 1 µl reservoir solution. The plates were left in incubators set at three different temperatures (277, 288 and 295 K). Additional crystallization experiments using selected conditions which gave rise to crystals from the initial screens were also set up in Intelli-Plates using 75 µl reservoir solution and drop ratios of either 1:1 or 2:2 protein:reservoir.
2.3. Data collection and preliminary X-ray analysis
The Oih-1 crystals were harvested from the crystallization drop, soaked briefly in a cryoprotectant comprising the crystallization solution with either 30%(v/v) ethylene glycol (form 1) or 25%(v/v) glycerol (form 2) and flash-cooled immediately in liquid nitrogen. The crystals were stored in a sample cassette designed for use with the Stanford Automated Mounting (SAM) system (Cohen et al., 2002 ▶). The crystals were transferred to beamline BL7-1 at the Stanford Synchrotron Radiation Laboratory (SSRL), screened for diffraction quality and found to diffract to beyond 2.0 Å resolution. X-ray diffraction data were collected from crystal form 1 on beamline BL11-1 and from crystal form 2 on BL7-1 using a single crystal in both cases maintained at 100 K with an Oxford Cryosystem. A total of 125 images were collected from the form 1 crystal using a Rayonix MX-325 CCD detector with an oscillation range of 1° per image, an exposure time of 10 s and a crystal-to-detector distance of 225 mm. For form 2, a total of 360 images were collected using an ADSC Q315r detector with an oscillation range of 0.25° per image, an exposure time of 6 s and a crystal-to-detector distance of 240 mm. The data from both crystals were processed using the XDS/XSCALE program package (Kabsch, 1993 ▶). Table 1 ▶ gives a summary of the data-collection statistics for both crystal forms.
Table 1. Data-collection statistics.
Values in parentheses are for the highest resolution shell.
| Form 1 | Form 2 | |
|---|---|---|
| Space group | P3121 | P3121 |
| X-ray source | SSRL BL11-1 | SSRL BL7-1 |
| X-ray wavelength (Å) | 0.9795 | 0.9795 |
| Unit-cell parameters | ||
| a = b (Å) | 122.7 | 56.4 |
| c (Å) | 59.9 | 165.6 |
| α = β (°) | 90.0 | 90.0 |
| γ (°) | 120.0 | 120.0 |
| Resolution (Å) | 1.75 (1.80–1.75) | 1.65 (1.70–1.75) |
| Reflections (observed/unique) | 380551/101533 | 190521/37531 |
| Rmerge† | 0.046 (0.595) | 0.037 (0.377) |
| 〈I/σ(I)〉 | 25.2 (3.5) | 25.1 (3.4) |
| Completeness (%) | 99.7 (100.0) | 99.5 (99.1) |
| Redundancy | 7.3 (6.7) | 4.4 (4.1) |
R
merge = 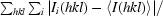
 , where I
i(hkl) is the ith observation of reflection hkl and 〈I(hkl)〉 is the weighted average intensity for all observations i of reflection hkl.
, where I
i(hkl) is the ith observation of reflection hkl and 〈I(hkl)〉 is the weighted average intensity for all observations i of reflection hkl.
3. Results and discussion
3.1. Crystallization and data collection
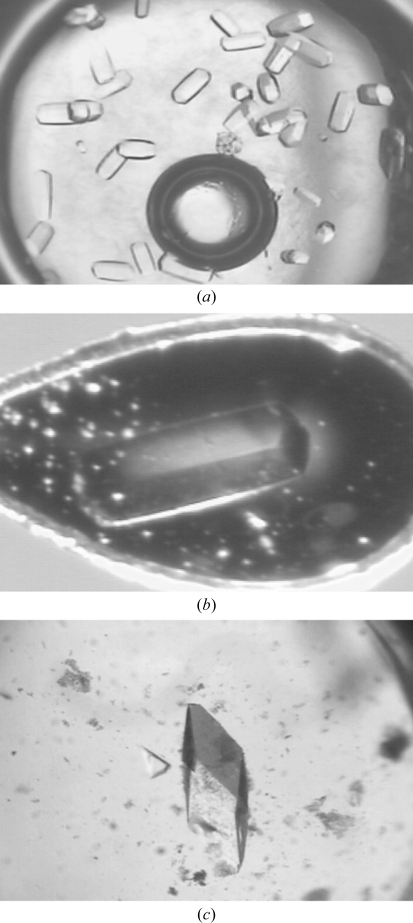
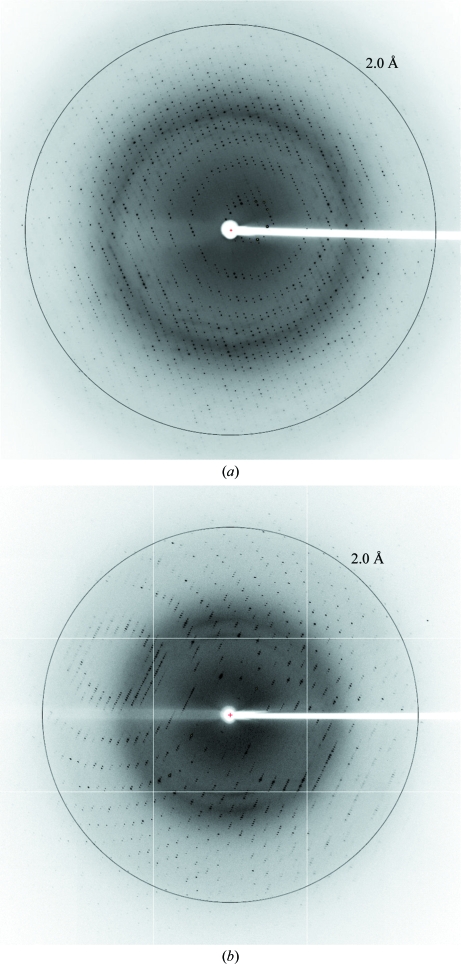
Inspection of the original sparse-matrix screens showed that Oih-1 crystallized under a range of conditions scattered across both CS I and CS II, with CS I condition No. 47 (form 1; 0.1 M sodium acetate pH 4.6, 2.0 M ammonium sulfate, 277 K) giving rise to large hexagonal rod-shaped crystals in 2–3 d (Figs. 2 ▶ a and 2 ▶ b) and CS I condition No. 14 [form 2; 0.2 M calcium chloride, 0.1 M HEPES pH 7.5, 28%(w/v) PEG 400, 288 K] giving large diamond-shaped crystals after 5–7 d (Fig. 2 ▶ c). Both crystal forms gave high-quality diffraction to a greater than 2.0 Å resolution, so it was decided that no additional fine-screening of the conditions was required. Preliminary X-ray diffraction analysis on crystal form 1 showed that they belonged to a trigonal space group with unit-cell parameters a = 122.7, c = 59.9 Å. A complete data set to approximately 1.75 Å resolution was subsequently collected from a single flash-cooled crystal (Fig. 3 ▶ a). Analysis of crystal form 2 showed that they also belonged to a trigonal space group, with unit-cell parameters a = 56.4, c = 165.6 Å. These crystals diffracted to 1.65 Å resolution and a complete data set was also collected (Fig. 3 ▶ b).
Figure 2.
(a) Hexagonal crystals of form 1 Oih-1 obtained from the original Crystal Screen I condition No. 47. (b) The form 1 crystal used for data collection with approximate dimensions 0.50 × 0.1 × 0.1 mm. (c) Diamond-shaped crystals of form 2 Oih-1 from Crystal Screen I condition No. 14 with approximate dimensions 0.6 × 0.2 × 0.2 mm.
Figure 3.
(a) Diffraction image of form 1 Oih-1. (b) Diffraction image of form 2 Oih-1.
3.2. Data processing
The form 1 crystal data were processed as P321 using the programs XDS and XSCALE (Kabsch, 1993 ▶). Inspection of the reflections along the threefold axis indicated the presence of a 31 or 32 screw axis. The Matthews coefficient (V M; Matthews, 1968 ▶) for the P321 space group, using an estimated molecular mass of 30.4 kDa, was 2.1 Å3 Da−1 (solvent content 43%) assuming the presence of two molecules in the asymmetric unit. However, initial rotation-function searches using MOLREP (Vagin & Teplyakov, 1997 ▶) indicated the presence of one solution. Recalculation of the Matthews coefficient assuming one molecule in the P321 asymmetric unit gave a value for V M of 4.3 Å3 Da−1, corresponding to a solvent content of 71%. Although this value is high, it is not without precedent, particularly in higher symmetry space groups (Chruszcz et al., 2008 ▶). For the form 2 crystals, the data were integrated and scaled in P321 using XDS and XSCALE (Kabsch, 1993 ▶).
3.3. Molecular replacement
Using the program CHAINSAW from the CCP4 suite (Collaborative Computational Project, Number 4, 1994 ▶), guided by the sequence alignment of Oih-1 and the Bacillus licheniformis BS3 β-lactamase (PDB code 1i2s), the latter model was truncated such that the conserved residues were retained and nonconserved residues were truncated to alanine. Using the 1.65 Å resolution data from the form 2 crystals, molecular-replacement (MR) calculations were performed using the program MOLREP (Vagin & Teplyakov, 1997 ▶). The V M assuming the presence of one molecule in the asymmetric unit was 2.5 Å3 Da−1 (51% solvent). The rotation-function search found a single rotation solution consistent with the calculated V M and subsequent translation searches in P321, P3121 and P3221 using this peak and the next 18 from the rotation-function list produced the best solution in the P3121 space group (R factor = 46%, correlation coefficient = 0.48), with the scores for the other two possible space groups being significantly poorer. A similar preliminary molecular-replacement calculation using the 1.75 Å resolution form 1 data indicated that the correct space group for these crystals was also P3121. Both data sets were subsequently rescaled in this space group. Refinement of the Oih-1 structure is currently under way.
Acknowledgments
This work was supported by grant No. AI057393 from the National Institutes of Health (SBV) and by grant No. 5 P41 RR001209 from the National Center for Research Resources (SSRL). Portions of this research were carried out at the Stanford Synchrotron Radiation Laboratory, a national user facility operated by Stanford University on behalf of the US Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy (BES, BER) and by the National Institutes of Health (NCRR, BTP, NIGMS). The contents of this paper are solely the responsibility of the authors and do not necessarily represent the official view of NCRR or NIH.
References
- Ambler, R. P., Coulson, A. F. W., Frere, J. M., Ghuysen, J. M., Joris, B., Forsman, M., Levesque, R. C., Tiraby, G. & Waley, S. G. (1991). Biochem. J.276, 269–270. [DOI] [PMC free article] [PubMed]
- Chruszcz, M., Potrzebowski, W., Zimmerman, M. D., Grabowski, M., Zheng, H., Lasota, P. & Minor, W. (2008). Protein Sci.17, 623–632. [DOI] [PMC free article] [PubMed]
- Cohen, A. E., Ellis, P. J., Miller, M. D., Deacon, A. M. & Phizackerley, R. P. (2002). J. Appl. Cryst.35, 720–726. [DOI] [PMC free article] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Fisher, J. F., Meroueh, S. O. & Mobashery, S. (2005). Chem. Rev.105, 395–424. [DOI] [PubMed]
- Jacoby, G. & Bush, K. (2007). Amino Acid Sequences for TEM, SHV and OXA Extended-Spectrum and Inhibitor-Resistant β-Lactamases. http://www.lahey.org/studies/webt.asp.
- Kabsch, W. (1993). J. Appl. Cryst.26, 795–800.
- Livermore, D. M. & Woodford, N. (2006). Trends Microbiol.14, 413–420. [DOI] [PubMed]
- Lu, J., Nogi, Y. & Takami, H. (2001). FEMS Microbiol. Lett.205, 291–297. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed]
- Minasov, G., Wang, X. & Shoichet, B. K. (2002). J. Am. Chem. Soc.124, 5333–5340. [DOI] [PubMed]
- O’Callaghan, C. H., Morris, A., Kirby, S. M. & Shingler, A. H. (1972). Antimicrob. Agents Chemother.1, 283–288. [DOI] [PMC free article] [PubMed]
- Poole, K. (2004). Cell. Mol. Life Sci.61, 2200–2223. [DOI] [PMC free article] [PubMed]
- Smith, C. A., Caccamo, M., Kantardjieff, K. A. & Vakulenko, S. (2007). Acta Cryst. D63, 982–992. [DOI] [PubMed]
- Strynadka, N. C. J., Adachi, H., Jensen, S. E., Johns, K., Sielecki, A., Betzel, C., Sutoh, K. & James, M. N. G. (1992). Nature (London), 359, 700–705. [DOI] [PubMed]
- Takami, H., Takaki, Y. & Uchiyama, I. (2002). Nucleic Acids Res.30, 3927–3935. [DOI] [PMC free article] [PubMed]
- Vagin, A. & Teplyakov, A. (1997). J. Appl. Cryst.30, 1022–1025.