Hibiscus chlorotic ringspot virus is a positive-sense monopartite single-stranded RNA virus that belongs to the Carmovirus genus of the Tombusviridae family. Authentic virus harvested from infected host kenaf leaves was purified and virus crystals were grown in multiple conditions. One of the crystals diffracted to 3.2 Å resolution and allowed the collection of a partial data set.
Keywords: hibiscus chlorotic ringspot virus, carmoviruses
Abstract
Hibiscus chlorotic ringspot virus (HCRSV) is a positive-sense monopartite single-stranded RNA virus that belongs to the Carmovirus genus of the Tombusviridae family, which includes carnation mottle virus (CarMV). The HCRSV virion has a 30 nm diameter icosahedral capsid with T = 3 quasi-symmetry containing 180 copies of a 38 kDa coat protein (CP) and encapsidates a full-length 3.9 kb genomic RNA. Authentic virus was harvested from infected host kenaf leaves and was purified by saturated ammonium sulfate precipitation, sucrose density-gradient centrifugation and anion-exchange chromatography. Virus crystals were grown in multiple conditions; one of the crystals diffracted to 3.2 Å resolution and allowed the collection of a partial data set. The crystal belonged to space group R32, with unit-cell parameters a = b = 336.4, c = 798.5 Å. Packing considerations and rotation-function analysis determined that there were three particles per unit cell, all of which have the same orientation and fixed positions, and resulted in tenfold noncrystallography symmetry for real-space averaging. The crystals used for the structure determination of southern bean mosaic virus (SBMV) have nearly identical characteristics. Together, these findings will greatly aid the high-resolution structure determination of HCRSV.
1. Introduction
Hibiscus chlorotic ringspot virus (HCRSV; ICTV 74.0.2.0.008) is a monopartite single-stranded RNA virus that belongs to the Carmovirus genus of the Tombusviridae family. HCRSV has been identified in hibiscus species worldwide, including the United States of America and southeast Asia (Waterworth et al., 1976 ▶; Wong & Chng, 1992 ▶). HCRSV infects kenaf (Hibiscus cannabinus L.), an annual crop of major interest to the wood-pulp industry (Johnson, 2001 ▶). It also attacks virtually all ornamental Hibiscus cultivars and has caused severe disease symptoms and resulted in a massive removal of these plants from many recreational areas in Singapore. The HCRSV virion is 30 nm in diameter and has T = 3 quasi-symmetry with 180 copies of a 38 kDa coat protein (CP). The CP has three domains: an RNA-binding (R) domain, a shell-forming (S) domain and a protruding (P) domain. Sequence alignment using online Clustal showed that the CP of HCRSV shares about 30% sequence homology with that of carnation mottle virus (CarMV) (http://www.ebi.ac.uk/tools/clustalw2; Thompson et al., 1994 ▶), with the highest amino-acid sequence divergence in the P domain. However, their host ranges differ greatly. CarMV is widespread in commercial stocks of carnations (Morgunova et al., 1994 ▶).
The structures of several carmoviruses have previously been reported, including those of turnip crinkle virus (TCV; 3.2 Å resolution; Hogle et al., 1986 ▶), CarMV (3.2 Å resolution; Morgunova et al., 1994 ▶), cowpea mottle virus (CPMoV; 7.0 Å resolution; Ke et al., 2004 ▶) and melon necrotic spot virus (MNSV; 2.8 Å resolution; Wada et al., 2008 ▶). From the genus Tombusvirus, the structure of tomato bushy stunt virus (TBSV) has been determined at 2.9 Å resolution (Harrison et al., 1978 ▶; Harrison, 1980 ▶; Olson et al., 1983 ▶). There are several common structural features among these viruses. The coat-protein subunits of all five viruses have similarly folded structures. Both the S and P domains contain a classical eight-stranded antiparallel β-sandwich fold. CPMoV, TCV and CarMV have a deletion in the β3 strand in the S domain relative to TBSV. CPMoV has an elongated C-terminus like TBSV, but it does not interact with the icosahedrally related P domain as observed in TBSV. MNSV shows a higher degree of similarity in structure to TBSV. CarMV lacks the β-annulus around the threefold icosahedral axes.
A 12 Å resolution structure of the HCRSV virion has been obtained using cryo-EM reconstruction (Doan et al., 2003 ▶). The structure has an arrangement of 90 dimers of P domains characteristic of the Tombusviridae arranged in rings of six and five above the threefold and fivefold icosahedral axes, respectively. An additional 20 Å thick internal layer of density was observed that was separated from the outer shell by about 10 Å but connected via thin strands of density. This layer corresponds to the R-domain position combined with density from the viral RNA predicted previously from neutron scattering (Harrison, 1980 ▶). Around the same time, virus crystals were grown that diffracted to 4.5 Å resolution (Lee et al., 2003 ▶). The crystals belonged to the cubic space group P23, with unit-cell parameter a = 392 Å. However, the data quality was poor beyond 5 Å and the crystals could not be improved to allow high-resolution structure determination.
In this paper, we describe the purification, crystallization and X-ray analysis of HCRSV purified from kenaf. The HCRSV crystals diffracted to 3.2 Å resolution and a high-quality data set was collected that will provide a high-resolution structure of HCRSV.
2. Purification
HCRSV was purified using saturated ammonium sulfate precipitation and sucrose density-gradient centrifugation. Frozen inoculated kenaf leaves were homogenized using a Waring Blender in three volumes (w/v) of extraction buffer (0.2 M sodium acetate pH 5.4, 50 mM NaCl, 20 mM CaCl2 and 5 mM EDTA) containing 0.1% β-mercaptoethanol. All subsequent procedures were carried out at 277 K. The slurry was centrifuged at 9000 rev min−1 for 15 min in a JA14 rotor (Beckman Coulter Inc., USA) at 277 K. The supernatant was filtered through Miracloth and kept on ice. The pellet was re-extracted with extraction buffer to maximize the virus yield. The supernatant was pooled and an equal volume of saturated ammonium sulfate solution was added before incubation for 2 h on ice. Centrifugation was performed at 9000 rev min−1 for 20 min in a JA14 rotor (Beckman Coulter Inc., USA) at 277 K and the resulting pellet was resuspended overnight at 277 K in resuspension buffer (0.05 M sodium acetate pH 5.4, 50 mM NaCl, 20 mM CaCl2, 5 mM EDTA) supplemented with 0.1% β-mercaptoethanol and 1% Triton X-100. The suspension was centrifuged at 9000 rev min−1 for 15 min. The supernatant was collected and layered onto a 10% sucrose cushion with resuspension buffer before ultracentrifugation at 30 000 rev min−1 for 3 h using an SW41 rotor (Beckman Coulter Inc., USA). A small volume of resuspension buffer was added to resuspend the pellet overnight at 277 K. This was followed by centrifugation at 14 000 rev min−1 for 3 min to remove insoluble debris. The supernatant was collected and layered onto a 10–40% sucrose gradient in resuspension buffer before ultracentrifugation at 27 000 rev min−1 for 3 h in an SW41 rotor. The visible virus band was collected from about 25–30% sucrose fractions. After threefold dilution with resuspension buffer, the purified virus was centrifuged at 30 000 rev min−1 for 3 h using an SW41 rotor. The virus pellet was resuspended in a small amount of virus buffer (10 mM sodium acetate pH 5.4, 50 mM NaCl, 5 mM CaCl2) and stored at 277 K. The virus concentration was calculated using an extinction coefficient of 5.0 at 260 nm (Morris & Carrington, 1988 ▶).
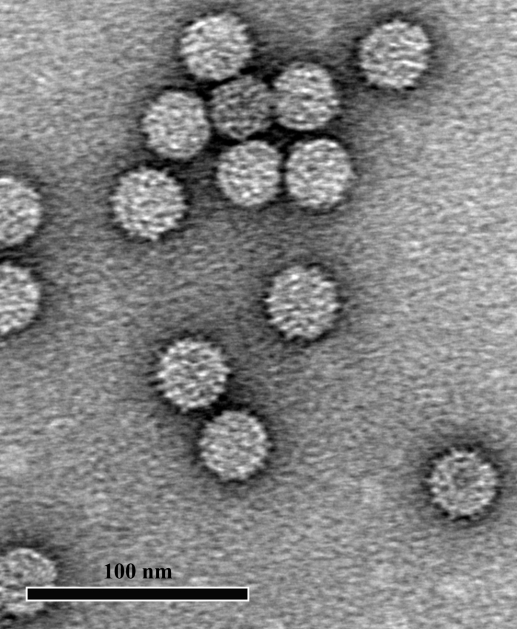
Virus yields of ∼2 mg per 100 g of infected kenaf leaves were obtained. The yield was quite low compared with that described by Lee and coworkers, who obtained yields of 48–70 mg highly purified virus from 100 g of infected leaves (Lee et al., 2003 ▶). However, their virus sample was contaminated with another ‘smooth’ virus owing to the soil that they used (Doan et al., 2003 ▶). The presence of a contaminating virus may explain the observed yield difference. The A 260/280nm value of the purified HCRSV was determined to be 1.5, compared with the value of 1.76 reported by Lee et al. (2003 ▶). Examination of the purified HCRSV virions by transmission electron microscopy (TEM) revealed isometric particles that were ∼30 nm in diameter (Fig. 1 ▶).
Figure 1.
Purified HCRSV particles negatively stained with 1% uranyl acetate and viewed at 50 000× magnification using a Jeol JEM-1030 TEM.
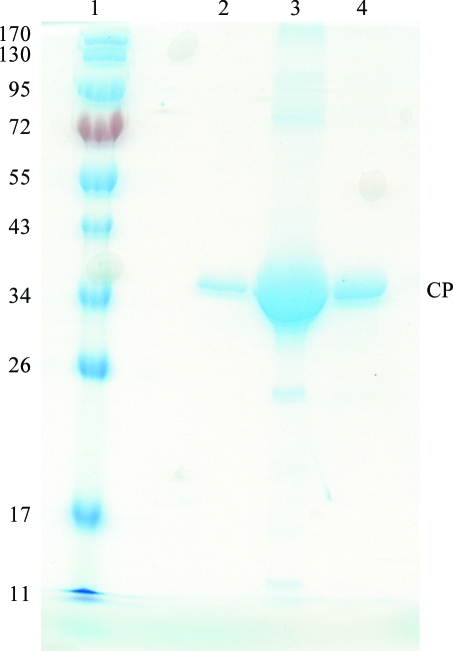
To verify the purity and homogeneity, purified HCRSV was run on a 12% SDS–PAGE gel. Only one prominent band (about 38 kDa) was observed (Fig. 2 ▶). Dynamic light scattering (DynaPro 99 Molecular Sizing Instrument) was used to examine the size homogeneity of the purified samples. The concentration of HCRSV was 100–200 µg ml−1 at temperatures of 277, 283, 293 and 303 K. The light-scattering data showed that the purified HCRSV particles were homogeneous and monodisperse. The average particle radius and molecular weight were ∼17.85 nm and ∼2857 kDa, respectively.
Figure 2.
SDS–PAGE of purified HCRSV. Lane 1, molecular-weight markers; lanes 2–4, purified HCRSV.
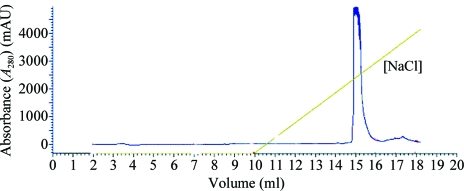
To further purify HCRSV, samples were run on a Mono Q5-50 column and eluted with an NaCl gradient. The elution curve showed that HCRSV contained only a single ionic species (Fig. 3 ▶). The purified HCRSV before further purification using Mono Q5-50 was considered to be sufficiently pure and was used for crystallization.
Figure 3.
Further purification of HCRSV using a MonoQ 5-50 column. HCRSV was eluted at ∼0.6 M NaCl concentration. The single peak shows that the purified HCRSV was homogeneous.
3. Crystallization
Lee et al. (2003 ▶) obtained large faceted bipyramidal crystals of HCRSV with 2.9% PEG 8000, 2% 2,3-butanediol and 0.1 M MES pH 6 at 288 K using a virus concentration of 60–90 mg ml−1. The crystals only formed at pH 6 and had a tendency to disintegrate within a week. When the virus concentration was decreased below 45 mg ml−1, dendrite-like crystals were observed. Repeating these conditions with the current purified HCRSV did not yield crystals. By increasing the PEG 8000 concentration to 9% at 20 mg ml−1 virus concentration, irregular-shaped crystals or crystal showers formed over a wider pH range (5–8). The crystals were of poor quality and also disintegrated. Increasing the virus concentration to 90 mg ml−1 also produced dendrite-like crystals and the addition of 0.1 M cadmium chloride improved the crystal morphology and shortened the growth time. None of these improvements using the increased virus purity and homogeneity increased the crystal quality and diffraction sufficiently for a high-resolution structure determination.
Attempts were then made to determine new crystal-growth conditions. Crystallization conditions were screened by the hanging-drop vapour-diffusion method (McPherson, 1982 ▶). The drops were set up with 1 µl purified virus and 1 µl crystallization well buffer at 293 K. Pyramidal crystals appeared (Fig. 4 ▶) using 0.4 M lithium sulfate monohydrate and 10%(w/v) polyethylene glycol (PEG) 8000, but only diffracted to 8 Å resolution. Promising rhombohedral crystals were grown using 0.1 M sodium acetate pH 4.0, 0.2 M ammonium sulfate, 2.5–6% PEG 2000 MME at 293 K with 2.5–5 mg ml−1 HCRSV in storage buffer. Decreasing the concentrations of PEG MME 2000 and HCRSV produced larger crystals that diffracted to 3.2 Å resolution and remained stable in the hanging drops.
Figure 4.
(a) HCRSV crystallized using 0.4 M Li2SO4, 10% PEG 8000, 5 mg ml−1 HCRSV; the crystals diffracted to 8 Å resolution. (b) HCRSV crystallized using 0.1 M sodium acetate pH 4.0, 0.2 M ammonium sulfate, 2.5–6% PEG 2000 MME, 2.5–5 mg ml−1 HCRSV; the crystals diffracted to 3.2 Å resolution and were suitable for data collection. These rhombohedral crystals grew to 0.4–0.5 mm within 2–3 weeks.
4. Data collection
Crystals were soaked in increasing concentrations of the cryoprotectants butanediol, glycerol and sucrose (in 5% increments up to 40%), with the best cryoprotectant found to be 2,3-butanediol at a final concentration of 25%. The crystals were then immediately frozen in liquid nitrogen for storage prior to data collection.
Data collection took place on beamline X29 at the National Synchrotron Light Source at Brookhaven National Laboratory (100 K, λ = 1.1 Å) and were processed using HKL-2000 (http://www.hkl-xray.com). The best HCRSV crystal diffracted to 3.2 Å resolution and was used to collect a total of 90 frames with an oscillation angle of 0.25° and a crystal-to-detector distance of 350 mm. The data were indexed, integrated and reduced using the HKL suite (Otwinowski & Minor, 1997 ▶). The crystal belonged to space group R32, with unit-cell parameters a = b = 336.4, c = 798.5 Å (in the hexagonal setting), and scaled well with an R merge of 6.7% (Tables 1 ▶ and 2 ▶). The data completeness was 37%, which has previously been sufficient for structure determination of an icosahedral virus with a high level of noncrystallographic symmetry (Chandrasekar & Johnson, 1998 ▶).
Table 1. Data-processing statistics for HCRSV.
Values in parentheses are for the highest resolution bin. Data are for all observations with I ≥ 0. Batch mosaicity values were between 0.35° and 0.43°.
| Space group | R32 |
| Unit-cell parameters (Å) | a = b = 336.4, c = 798.5 |
| Resolution range (Å) | 35–3.2 (3.31–3.20) |
| Total observations | 185483 (3924) |
| Unique reflections | 105460 (3506) |
| Completeness (%) | 37.2 (12.5) |
| Rmerge (%)† | 6.7 (16.4) |
| Average I/σ(I) | 6.7 (2.0) |
| Redundancy‡ | 1.8 (1.1) |
| Wilson B value (Å2) | 55.7 |
R
merge = 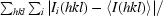
 × 100, where 〈I(hkl)〉 is the mean of the Ii(hkl) observations of reflection hkl.
× 100, where 〈I(hkl)〉 is the mean of the Ii(hkl) observations of reflection hkl.
Redundancy = 〈No. of observations/No. of unique reflections〉.
Table 2. HCRSV reduced data by resolution bin.
| Resolution range (Å) | Rmerge | Redundancy | I/σ(I) | Unique reflections | Completeness (%) |
|---|---|---|---|---|---|
| 35.0–6.88 | 0.032 | 2.2 | 24.8 | 13844 | 47.7 |
| 6.88–5.47 | 0.076 | 1.9 | 8.0 | 13791 | 48.3 |
| 5.47–4.78 | 0.071 | 2.0 | 8.8 | 13316 | 46.9 |
| 4.78–4.34 | 0.077 | 1.9 | 7.4 | 12931 | 45.6 |
| 4.34–4.03 | 0.098 | 1.8 | 5.1 | 12211 | 43.2 |
| 4.03–3.79 | 0.123 | 1.6 | 3.7 | 11097 | 39.3 |
| 3.79–3.60 | 0.124 | 1.5 | 3.4 | 10048 | 35.6 |
| 3.60–3.45 | 0.128 | 1.3 | 3.1 | 8281 | 29.3 |
| 3.45–3.31 | 0.148 | 1.2 | 2.5 | 6435 | 22.8 |
| 3.31–3.20 | 0.164 | 1.1 | 2.0 | 3506 | 12.5 |
| 35.0–3.20 | 0.067 | 1.8 | 6.7 | 105460 | 37.2 |
5. Preliminary X-ray data analysis
Two other icosahedral virus structures have had similar unit-cell parameters and space groups. The T = 3 SBMV crystallized in space group R32 with unit-cell parameters a = 337, c = 756 Å (hexagonal setting) and three particles per unit cell (Akimoto et al., 1975 ▶; Johnson et al., 1976 ▶; Abad-Zapatero et al., 1980 ▶). For the three particles to pack with R32 symmetry in this cell, one will be positioned at the origin with one threefold axis coincident with the c axis and a pair of its twofold axes, separated by 120°, coincident with the a and b axes of the crystal. This gives all three particles identical orientations relative to the unit-cell axes and divides the particle into six equivalent sections, defining tenfold noncrystallographic symmetry in the crystal asymmetric unit. The pseudo-T = 3 Seneca Valley Virus-001 crystallized in space group R3 with unit-cell parameters a = 311.5, c = 1526.4 Å (hexagonal setting) and six particles per unit cell (Venkataraman et al., 2008a ▶,b ▶). This cell is essentially the same as those of SBMV and HCRSV, but doubled in length along the c axis. Packing six particles in R3 also dictates a particle at the origin with one of its threefolds coincident with the c axis, but does not restrict the orientations of the twofold axes and the orientations of the six particles can differ (as two sets of three particles owing to R-centring). Thus, each particle is divided into thirds (20 protomers) and in order to have six particles with the R3 operators, 20 protomers from two different particles combine to make the crystal asymmetric unit (40 protomers in total). A locked rotation function confirmed that the two sets of particles differed by a mere 3.2° in orientation about the c axis, confirming the R3 assignment.
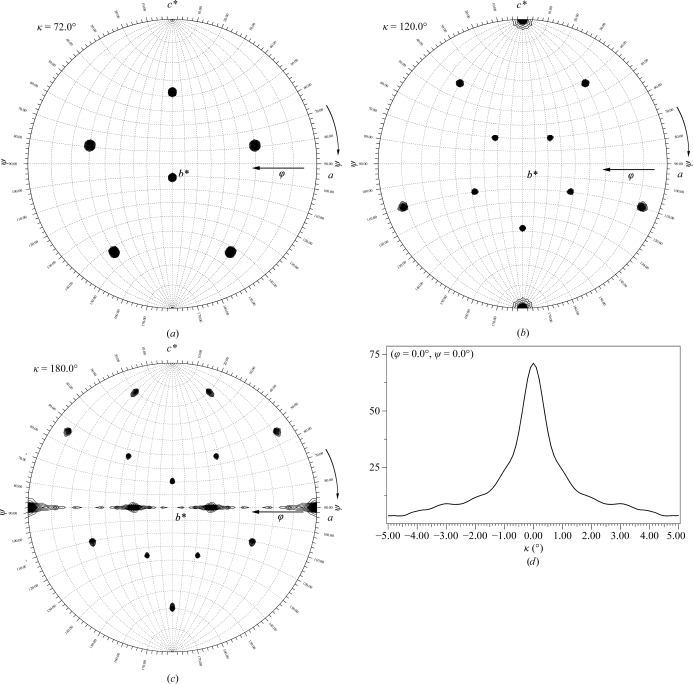
For HCRSV, the scaling R values were similar in either space group R3 or R32 and the Matthews coefficient was 3.24 Å3 Da−1 for three particles in the unit cell, suggesting a solvent content of approximately 62%. These observations are consistent with the packing found in the SBMV study. Self-rotation functions computed using the program GLRF confirmed the presence of fivefold, threefold and twofold axes arranged with icosahedral point-group symmetry (Figs. 5 ▶ a–5 ▶ c). Using data between 10 and 5 Å resolution only a single particle orientation was observed, which was identical to that in the SBMV study. To verify the single particle orientation and test the quality of the data set, a locked rotation function was calculated about the c axis by pre-aligning one threefold axis of the particle with the c axis and using only the data between 3.6 and 3.2 Å resolution and a search interval of only 0.1° (Fig. 5 ▶ d). Only a single well defined peak was observed at (ϕ = 0, ψ = 0, κ = 0). These combined results identify the position and orientation of the HCRSV particles as being the same as those found in the SBMV study and show that the high-resolution data are well measured.
Figure 5.
(a), (b), (c) Contour plots of the self-rotation function searches for fivefold, threefold and twofold rotation axes with the HCRSV X-ray data. The calculations used a total of 7690 large terms between 10 and 5 Å resolution with a radius of integration of 150 Å. All peaks for the fivefolds, threefold and twofolds predicted to be in this hemisphere are present and have signal-to-noise ratios of 6–14σ. (d) Result of the locked rotation function about the crystal c axis. The calculation used 1371 large terms between 3.6 and 3.2 Å resolution with a radius of integration of 150 Å. Only a single peak is present with a height of 3.3σ, indicating that only a single particle orientation exists in the HCRSV unit cell.
Coordinates from previously determined carmovirus structures have been positioned and oriented in the HCRSV unit cell in order to calculate initiate phases for real-space averaging. The locations of the noncrystallographic symmetry operators in rotation functions calculated with intensities from the model coordinates are consistent with the peaks from the self-rotation functions using diffraction data. Real-space averaging over the tenfold noncrystallographic symmetry will be used to refine and gradually extend the phases to 3.2 Å resolution.
Acknowledgments
We thank Dr Vijay S. Reddy and Dr J. Sivaraman, Cherlyn Ng and S. Sunita for valuable advice and help and A. Saxena and the staff of beamlines X12C and X29 at the National Synchrotron Light Source of the Brookhaven National Laboratory for assistance with data collection. This work was supported by NUS (R-154-000-295-112), the Temasek Life Sciences Laboratory and National Institutes of Health grant R01 GM054076 (JEJ).
References
- Abad-Zapatero, C., Abdel-Meguid, S. S., Johnson, J. E., Leslie, A. G. W., Rayment, I., Rossmann, M. G., Suck, D. & Tsukihara, T. (1980). Nature (London), 286, 33–39. [DOI] [PubMed]
- Akimoto, T., Wagner, M., Johnson, J. & Rossmann, M. (1975). J. Ultrastruct. Res.53, 306–318. [DOI] [PubMed]
- Chandrasekar, V. & Johnson, J. E. (1998). Structure, 6, 157–171. [DOI] [PubMed]
- Doan, D. N. P., Lee, K. C., Laurinmaki, P., Butcher, S., Wong, S.-M. & Dokland, T. (2003). J. Struct. Biol.144, 253–261. [DOI] [PubMed]
- Harrison, S. (1980). Biophys. J.32, 139–153. [DOI] [PMC free article] [PubMed]
- Harrison, S. C., Olson, A. J., Schutt, C. E., Winkler, F. K. & Bricogne, G. (1978). Nature (London), 276, 368–373. [DOI] [PubMed]
- Hogle, J. M., Maeda, A. & Harrison, S. C. (1986). J. Mol. Biol.191, 625–638. [DOI] [PubMed]
- Johnson, J. (2001). Alternative Agriculture: What Is Kenaf? Rural Enterprise and Alternative Development Initiative Report 1. Carbondale, USA: Southern Illinois University Carbondale.
- Johnson, J. E., Akimoto, T., Suck, D., Rayment, I. & Rossmann, M. (1976). Virology, 75, 394–400. [DOI] [PubMed]
- Ke, J., Schmidt, T., Chase, E., Bozarth, R. F. & Smith, T. J. (2004). Virology, 321, 349- 358. [DOI] [PubMed]
- Lee, K.-C., Lim, D., Wong, S.-M. & Dokland, T. (2003). Acta Cryst. D59, 1481–1483. [DOI] [PubMed]
- McPherson, A. (1982). Preparation and Analysis of Protein Crystals. New York: John Wiley & Sons.
- Morgunova, E. Y., Dauter, Z., Fry, E., Stuart, D. I., Stel’mashchuk, V. Y., Mikhailov, A. M., Wilson, K. S. & Vainshtein, B. K. (1994). FEBS Lett.338, 267–271. [DOI] [PubMed]
- Morris, T. J. & Carrington, J. C. (1988). The Plant Viruses, edited by R. Koenig, Vol. 3, pp. 73–112. New York: Plenum.
- Olson, A., Bricogne, G. & Harrison, S. (1983). J. Mol. Biol.171, 61–93. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Thompson, J., Higgins, D. & Gibson, T. (1994). Nucleic Acids Res.22, 4673–4680. [DOI] [PMC free article] [PubMed]
- Venkataraman, S., Reddy, S. P., Loo, J., Idamakanti, N., Hallenbeck, P. L. & Reddy, V. S. (2008a). Structure, 16, 1555–1561. [DOI] [PMC free article] [PubMed]
- Venkataraman, S., Reddy, S. P., Loo, J., Idamakanti, N., Hallenbeck, P. L. & Reddy, V. S. (2008b). Acta Cryst. F64, 293–296. [DOI] [PMC free article] [PubMed]
- Wada, Y., Tanaka, H., Yamashita, E., Kubo, C., Ichiki-Uehara, T., Nakazono-Nagaoka, E., Omura, T. & Tsukihara, T. (2008). Acta Cryst. F64, 8–13. [DOI] [PMC free article] [PubMed]
- Waterworth, H. E., Lawson, R. H. & Monroe, R. L. (1976). Phytopathology, 64, 570–575.
- Wong, S.-M. & Chng, C. G. (1992). Phytopathology, 82, 722.