A secreted bacterial protein from a human pathogen that mediates serum resistance and adherence was overexpressed, purified, refolded and crystallized. Preliminary X-ray diffraction data are presented.
Keywords: autotransporters, passenger domains, refolding, BrkA
Abstract
Autotransporters (ATs) are proteins that deliver effectors (the passenger domain) to the surface of Gram-negative bacteria by the type V secretion pathway. The passenger domain of BrkA, a Bordetella pertussis autotransporter mediating serum resistance and adherence, was cloned in a pET expression system and overexpressed in Escherichia coli. The gene product was correctly refolded, purified to homogeneity and crystallized. The crystals diffracted to 2.8 Å resolution. The space group was assumed to be P41212, with unit-cell parameters a = b = 108.19, c = 115.35 Å.
1. Introduction
Autotransporters (ATs) are a family of proteins that deliver functionally diverse effectors to the surface of Gram-negative bacteria via arguably the most common and perhaps the simplest secretion mechanism: the type V secretion pathway (Henderson et al., 2004 ▶). The precursor of an autotransporter has a modular structure composed of an N-terminal signal peptide, a passenger domain and a C-terminal translocation unit. After being produced in the cytoplasm, the precursor protein is targeted to the inner membrane by the signal peptide and secreted to the periplasm via the Sec machinery. The signal peptide is cleaved at the periplasmic side of the inner membrane. The translocation unit, which forms a β-barrel in the outer membrane, facilitates the export of the passenger domain to the cell surface. Upon delivery to the cell surface, the passenger domain is cleaved and is either released to the milieu or remains on the cell surface via noncovalent binding. The processed passenger domain bears various functions and the best characterized have been shown to include a variety of serine proteases (SPATEs), toxins, adhesins, invasins and factors that mediate serum resistance and actin polymerization (Henderson et al., 2004 ▶). Recent structural prediction and molecular-modelling studies have revealed that despite a wide diversity in size, sequence and functions, most of the passenger domains are predicted to adopt a β-helix structure (Junker et al., 2006 ▶; Kajava & Steven, 2006 ▶). Whether this fold is directly involved in AT effector function or whether the conserved β-helical structure contributes to the OM translocation and folding of ATs remains to be investigated.
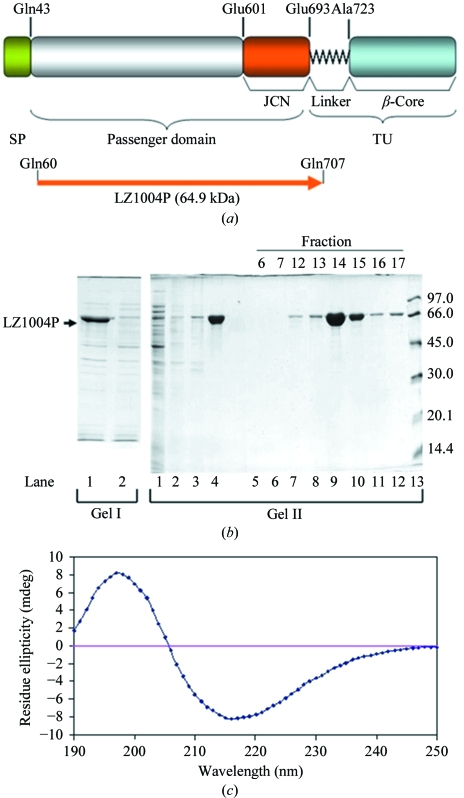
BrkA (NCBI accession No. AAA51646) is a Bordetella pertussis autotransporter that mediates serum resistance and adherence (Fernandez & Weiss, 1994 ▶; Oliver, Huang & Fernandez, 2003 ▶). BrkA is expressed as a 103 kDa precursor which is proteolytically processed to a 42-amino-acid signal peptide, a 73 kDa passenger domain and a 30 kDa translocation unit during secretion (Fig. 1 ▶ a). The primary sequence of the BrkA passenger domain shares 30% identity to that of P.69 pertactin, the passenger domain of the autotransporter pertactin, a B. pertussis adhesin, which also has a β-helix structure (Emsley et al., 1996 ▶). Determination of the structure of the BrkA passenger domain using X-ray crystallography will aid in defining possible sites involved in secretion, folding and the BrkA effector functions of complement resistance and adherence. In this study, the passenger domain of BrkA was cloned in a pET expression system and overexpressed as inclusion bodies in Escherichia coli. The gene product was correctly refolded, purified to homogeneity and crystallized. The crystals diffracted to 2.8 Å resolution. The space group was assumed to be P41212 based on preliminary molecular-replacement results.
Figure 1.
Expression and purification of the BrkA passenger domain. (a) Domain organization of BrkA (Oliver, Huang & Fernandez, 2003 ▶; Oliver, Huang, Nodel et al., 2003 ▶). SP represents the signal peptide (residues 1–42). The passenger domain contains residues 43–692 and the junction region (JCN) is located at the C-terminus of the passenger domain (residues 601–692). TU represents the translocation unit, which is composed of the linker region (residues 693–731) and the β-core (residues 732–1010). The BrkA passenger domain (LZ1004P) was expressed using a pET expression system. (b) Purification of LZ1004P. Gel I and gel II are from two reproducible purification experiments. Gel I: lane 1, whole cell lysate; lane 2, soluble form of cell lysate. Gel II: lane 1, soluble form of cell lysate. Lanes 2–4 are from the insoluble inclusion bodies after various treatments. Lane 2, supernatant of the inclusion-bodies sample after Triton X-100 wash; lane 3, precipitate removed after dialysis; lane 4, supernatant remaining after dialysis. Lanes 5–12, fractions eluted from Resource Q column. Lane 13, low-molecular-weight markers (labelled in kDa). The majority of the protein was in the supernatant after resolubilization and dialysis and was eluted between NaCl concentrations of 0.21 and 0.25 M. (c) Far-UV circular-dichroism spectrum of 0.15 mg ml−1 LZ1004P in 20 mM phosphate buffer pH 7.5.
2. Materials and methods
2.1. DNA manipulation and expression
E. coli DH5α (Invitrogen) was used for cloning and E. coli strain BL21 (DE3) (Invitrogen) was used for protein expression. Plasmid pET30b was obtained from Novagen. Plasmid pDO6935 from our laboratory collection has been described previously (Oliver, Huang & Fernandez, 2003 ▶). Kanamycin was added to the media at a concentration of 50 µg ml−1. Ampicillin was added at 100 µg ml−1.
DNA manipulations and polymerase chain reactions (PCRs) were carried out using standard techniques (Sambrook & Russell, 2001 ▶) and reagents as described previously (Oliver, Huang & Fernandez, 2003 ▶). Plasmid pDO6935 was used as a template in all PCR reactions. The primers used in this study were obtained from Alpha DNA (Montreal, Canada). DNA sequencing was performed by the Nucleic Acid and Protein Services (NAPS) Unit at the University of British Columbia.
Briefly, the gene encoding the BrkA passenger domain (Gln60–Gln707) was amplified by PCR from pDO6935 using primer pair LZ-BA4NdeI (5′-GGGGGGGCATATGCAGGAAGGAGAGTTCGACCA-3′) and G2stop (5’-CAATTTAAGCTTTCACTGGCCCGCGCGCTGC-3′). The 2.0 kb PCR product was cloned into pET30b using NdeI and HindIII sites to yield the construct pLZ1004. The gene product is LZ1004P plus an initiating methionine residue at the N-terminus. E. coli BL21(DE3) harbouring pLZ1004 was grown to an OD600 of approximately 0.6 and was induced with 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG). The cells were then continuously cultivated for 4 h.
2.2. Purification and refolding of BrkA passengers
The cells were harvested from a 100 ml culture by centrifugation and the resulting cell pellet (wet weight 140 mg) was resuspended in 30 ml sonication buffer (20 mM sodium phosphate pH 7.5, 0.1 M NaCl, 1 mM EDTA, 5 mM benzamidine). The cells were sonicated and the inclusion bodies were collected by centrifugation at 18 000g for 20 min. The pellet was then washed in the same volume of sonication buffer with 1% Triton X-100, centrifuged to remove the supernatant and solubilized using 5 ml 6 M guanidinium hydrochloride (GuHCl) dissolved in sonication buffer. The concentration of GuHCl was reduced to 2 M by dropwise addition of sonication buffer. Any insoluble debris was removed by centrifugation (15 000g for 20 min). Refolding of LZ1004P was performed at 277 K by successive dialyses against a 100× volume of pre-chilled 20 mM sodium phosphate pH 7.5, 1 mM EDTA and 5 mM benzamidine plus GuHCl reduced from 2 M by 0.5 M increments in a stepwise manner. Each dialysis step was for a minimum of 3 h. After centrifugation at 15 000g for 20 min to remove any precipitates that might have formed, LZ1004P was ultimately dialyzed into 20 mM sodium phosphate buffer pH 7.5 and loaded onto a 6 ml Resource Q anion-exchange column equilibrated with the same buffer. The protein was eluted with the same buffer containing an NaCl gradient from 0 to 0.5 M using the ÄKTA explorer 100 FPLC system (GE Healthcare). Fractions were monitored by absorption at 280 nm, collected and examined by SDS–PAGE. Fractions containing LZ1004P were collected, combined and concentrated using a Millipore centrifugal filter device (10 000 molecular-weight cutoff). SDS–PAGE was carried out as previously described (Fernandez & Weiss, 1994 ▶; Laemmli, 1970 ▶). The final yield of LZ1004P was 20 mg.
2.3. Far-UV circular-dichroism spectrometry of LZ1004P
The far-UV circular-dichroism (CD) spectra were measured at room temperature using a Jasco J-810 CD spectropolarimeter (Jasco, Easton, Maryland, USA) in a 1 mm path-length quartz cell. Individual spectra were collected by averaging ten scans collected over a spectral window of 190–250 nm. The concentration of LZ1004P was 0.15 mg ml−1 in 20 mM sodium phosphate pH 7.5.
2.4. Crystallization
Crystals of LZ1004P were obtained at room temperature by the hanging-drop vapour-diffusion method by mixing 1 µl protein solution (15 mg ml−1 in 20 mM Tris–HCl pH 8.0) and 1 µl precipitant solution (4% PEG 4000, 0.1 M sodium acetate pH 5.2). After microseeding, crystals grew to a maximal size of 130 µm within three weeks.
2.5. Data collection and processing
X-ray diffraction data were collected from these crystals on beamline 1-5 of the Stanford Synchrotron Radiation Laboratory (SSRL). The data were indexed, integrated, scaled and merged with the HKL package (Otwinowski et al., 2003 ▶). Molecular replacement was carried out with the program Phaser using the coordinates of P.69 pertactin (Read, 2001 ▶; Emsley et al., 1996 ▶). Density modification was performed with the program DM (Cowtan, 1994 ▶). The crystallographic data statistics are summarized in Table 1 ▶.
Table 1. Summary of data-collection and processing statistics.
Values in parentheses are for the highest resolution shell.
| Space group | P41212 |
| Unit-cell parameters (Å) | a = b = 108.19, c = 115.35 |
| Wavelength (Å) | 0.9794 |
| Resolution (Å) | 78.81–2.8 (2.9–2.8) |
| Measured reflections | 115431 |
| Unique reflections | 16902 |
| Redunduncy | 6.8 (5.2) |
| Completeness (%) | 97.0 (90.7) |
| Rmerge (%) | 7.4 (38.3) |
| 〈I/σ(I)〉 | 17.0 (4.0) |
3. Results and discussion
LZ1004P, corresponding to the BrkA passenger domain (Gln60–Gln707), was cloned in pET30b and overexpressed in E. coli BL21 (DE3) upon IPTG induction (Fig. 1 ▶ b; gel I lane 1). We concluded that LZ1004P was expressed as inclusion bodies, since it was not found in the soluble form of the cell lysate (Fig. 1 ▶ b; gel I lane 2). Purification of LZ1004P from the initial insoluble fraction of a subsequent induction is shown in Fig. 1 ▶(b) (gel II lanes 2–12). Washing of the pellet with 1%(v/v) Triton X-100 caused little loss of the target polypeptide (Fig. 1 ▶ b; gel II lane 2). The pellet was solubilized in 6 M GuHCl and after the GuHCl was carefully dialyzed out, 95% of the LZ1004P was found in the supernatant (Fig. 1 ▶ b; gel II lane 3 and 4). LZ1004P purified to homogeneity was eluted from a Resource Q column between 0.21 and 0.25 M NaCl (Fig. 1 ▶ b; gel II lanes 9 and 10).
A previous study of DO418P, a similar recombinant BrkA passenger with an N-terminal His tag, showed that BrkA acquires β-structure when refolded and that without correct refolding the BrkA passenger domain does not have any binding activity to host cells (Oliver, Huang, Nodel et al., 2003 ▶). Thus, to check if the purified protein refolded correctly, circular-dichroism (CD) spectroscopy was carried out for LZ1004P. The far-UV CD profile indicated that LZ1004P is enriched in β-structure, with a minimum at 218 nm (Fig. 1 ▶ c). This result is consistent with the far-UV CD spectrum of DO418P (Oliver, Huang, Nodel et al., 2003 ▶). Moreover, the correctly refolded LZ1004P was functional as it was shown to have binding activity to human HeLa cells (data not shown) using a FACS binding assay (Oliver, Huang, Nodel et al., 2003 ▶).
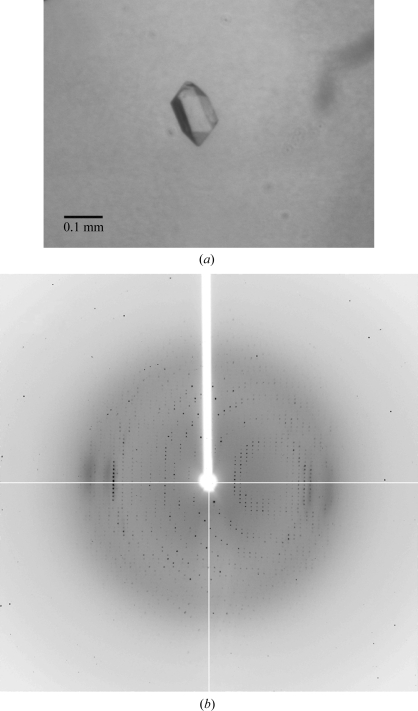
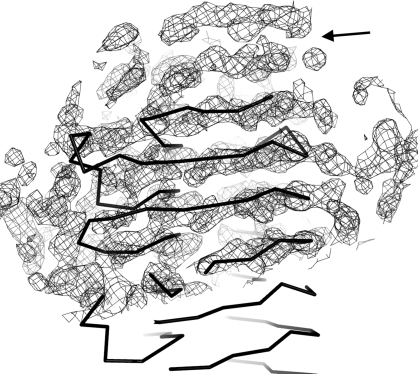
The original crystals of LZ1004P were obtained at room temperature (298 K) by the hanging-drop vapour-diffusion method using the Crystal Screen kit (Hampton Research) to determine the initial crystallization condition of 0.1 M sodium acetate pH 4.6, 8%(w/v) PEG 4000. The crystallization conditions were optimized to 0.1 M sodium acetate pH 4.2 and 4–5% PEG 4000 and the microseeding method was used to improve the crystals. The optimized crystals were grown for three weeks to dimensions of 0.13 × 0.1 × 0.1 mm (Fig. 2 ▶ a). X-ray diffraction data were collected from these crystals to a resolution of 2.8 Å (Fig. 2 ▶ b). The diffraction images were integrated and scaled assuming a tetragonal lattice and space group P41212 (unit-cell parameters a = b = 108.19, c = 115.35 Å). The Matthews coefficient was 2.59 Å3 Da−1 and the solvent content was 52.5% assuming the presence of one molecule of LZ1004P in the asymmetric unit. Molecular replacement was carried out using the structure of P.69 pertactin as a search model (Emsley et al., 1996 ▶; Read, 2001 ▶). The preliminary molecular-replacement solution was obtained in space group P41212. After density modification by the program DM the resulting electron-density map indicates that LZ1004P is predominately a β-helix structure. Interestingly, the very N-terminal region of BrkA passenger domain (Gln43–Asp267) shows no significant homology to entries in the GenBank database with a BLASTP search. The corresponding N-terminal region of LZ1004P (Met1–Asp209), which is not present in the P.69 pertactin structure, was also observed as a β-helix in the electron density extending from the C-terminal domain of the P.69 pertactin search model (Fig. 3 ▶). Currently, modelling and refinement to determine the BrkA structure are under way.
Figure 2.
X-ray diffraction of BrkA passenger crystals. (a) Crystal image of LZ1004P. Crystals of LZ1004P were obtained using the vapour-diffusion method. The crystals grew over three weeks to dimensions of 0.13 × 0.1 × 0.1 mm. (b) X-ray diffraction image of BrkA LZ1004P with an oscillation angle of 1°.
Figure 3.
Electron density at the N-terminal region of BrkA after density modification by the program DM. The map is contoured at 1.3σ and the extended density at the N-terminus is indicated by an arrow. The N-terminus of the search model is drawn as an Cα trace.
Acknowledgments
We would like to thank Dr Woo Cheol Lee for his help with the data analysis and structure modelling. This work was supported by grants from the Canadian Institute of Health Research (MOP49597 to MEPM and DOP53723 to RCF) and the Natural Sciences and Engineering Research Council of Canada (RGPIN194599 to RCF).
References
- Cowtan, K. (1994). Jnt CCP4/ESF–EACBM Newsl. Protein Crystallogr.31, 34–38.
- Emsley, P., Charles, I. G., Fairweather, N. F. & Isaacs, N. W. (1996). Nature (London), 381, 90–92. [DOI] [PubMed]
- Fernandez, R. C. & Weiss, A. A. (1994). Infect. Immun.62, 4727–4738. [DOI] [PMC free article] [PubMed]
- Henderson, I. R., Navarro-Garcia, F., Desvaux, M., Fernandez, R. C. & Ala’Aldeen, D. (2004). Microbiol. Mol. Biol. Rev.68, 692–744. [DOI] [PMC free article] [PubMed]
- Junker, M., Schuster, C. C., McDonnell, A. V., Sorg, K. A., Finn, M. C., Berger, B. & Clark, P. L. (2006). Proc. Natl Acad. Sci. USA, 103, 4918–4923. [DOI] [PMC free article] [PubMed]
- Kajava, A. V. & Steven, A. C. (2006). J. Struct. Biol.155, 306–315. [DOI] [PubMed]
- Laemmli, U. K. (1970). Nature (London), 227, 680–685. [DOI] [PubMed]
- Oliver, D. C., Huang, G. & Fernandez, R. C. (2003). J. Bacteriol.185, 489–495. [DOI] [PMC free article] [PubMed]
- Oliver, D. C., Huang, G., Nodel, E., Pleasance, S. & Fernandez, R. C. (2003). Mol. Microbiol.47, 1367–1383. [DOI] [PubMed]
- Otwinowski, Z., Borek, D., Majewski, W. & Minor, W. (2003). Acta Cryst. A59, 228–234. [DOI] [PubMed]
- Read, R. J. (2001). Acta Cryst. D57, 1373–1382. [DOI] [PubMed]
- Sambrook, J. & Russell, D. W. (2001). Molecular Cloning: A Laboratory Manual. New York: Cold Spring Harbor Laboratory Press.