A novel 15 kDa antifungal protein amaryllin has been crystallized using 30% PEG 8000 as the precipitating agent. The crystals belong to orthorhombic space group I or I212121 with cell dimensions, a = 48.6, b = 61.9 and c = 79.6–Å.
Keywords: antifungal activity, pathogenesis-related proteins, N-terminal sequence and structure analysis
Abstract
A novel antifungal protein, amaryllin, has been isolated from the underground bulbs of Amaryllis belladonna, purified to homogeneity and crystallized. The protein was extracted using ammonium sulfate fractionation. The purified protein samples indicated a molecular weight of 15 kDa on SDS–PAGE. The protein showed antifungal activity against Aspergillus flavus and Fusarium oxysporum. The N-terminal sequence of the first 15 amino-acid residues was determined using Edman degradation and did not show significant sequence identity to any known protein. The protein was crystallized using the hanging-drop vapour-diffusion method with 30% PEG 8000 as precipitating agent. The crystals diffracted to 2.7 Å resolution and belonged to the orthorhombic space group I222 or I212121, with unit-cell parameters a = 48.6, b = 61.9, c = 79.6 Å. The complete sequence and structure determination of amaryllin are in progress.
1. Introduction
Unlike humans, plants do not have an immune system, but they have evolved other powerful defence mechanisms. When exposed to pathogens such as fungi, viruses and bacteria, plants produce a variety of low-molecular-weight antimicrobial compounds and peptides, as well as small proteins. These antifungal proteins and peptides have been shown to protect economically important plants and crops from fungal attack (Ho et al., 2007 ▶). Several types of proteins and peptides from various plant sources are known for their antifungal activity and they have varying potencies. These include thionins (Bloch et al., 1998 ▶), defensins (Broekaert et al., 1995 ▶), hevein-like proteins (Segura et al., 1993 ▶) and knottin-like peptides (Segura et al., 1993 ▶). Many of these proteins are known to be protease inhibitors (Lorito et al., 1994 ▶; Chen et al., 1998 ▶; Joshin et al., 1998 ▶; Chilosi et al., 2000 ▶), amylase inhibitors (Pueyo et al., 1993 ▶), xylanase inhibitors (McLauchlan et al., 1999 ▶), glucanases (Vogelsang & Barz, 1993 ▶), chitinases (Gozia et al., 1995 ▶) and ribosome-inactivating proteins (Leah et al., 1991 ▶; Mishra et al., 2005 ▶). These proteins are collectively called plant pathogenesis-related (PR) proteins.
Here, we the report isolation, purification, N-terminal sequence determination and preliminary X-ray crystallographic studies of a novel PR protein called amaryllin from the bulbs of Amaryllis belladonna. The N-terminal sequence of amaryllin has been determined. The complete amino-acid sequence determination and structure solution of the protein are in progress.
2. Materials and methods
2.1. Purification and activity of amaryllin
Samples of the underground bulbs of A. belladonna were collected from local nurseries. The bulbs were cut into small pieces and pulverized in the presence of liquid nitrogen in a ventilated hood. The pulverized samples were stirred for 24 h at 277 K in extraction solution containing 0.2 M sodium chloride and 50 mM sodium phosphate buffer pH 7.2. 2.5 g polyvinylpyrolidine (PVP) was added to 100 ml of the above solution and homogenized. The homogenate was centrifuged at 5000g for 30 min at 277 K. The protein was precipitated with 80% saturated ammonium sulfate and was separated and solubilized in 50 mM sodium phosphate buffer pH 7.2. This solution was examined using SDS–PAGE. It showed two prominent bands corresponding to molecular weights of 66 and 15 kDa. In order to carry out further purification, the protein solution was loaded onto a DEAE-Sephadex A-50 column (50 × 2 cm) equilibrated with 50 mM sodium phosphate buffer pH 7.2. The proteins were eluted using a continuous gradient of 0.0–0.5 M NaCl in the same buffer. The two peaks were collected and pooled separately. The pooled solution containing the second peak was loaded onto a Sephadex G-50 column (150 × 1 cm) and eluted with 25 mM Tris–HCl pH 8.0 at a flow rate of 6 ml h−1. The first peak from the above elution was collected, pooled and lyophilized. This peak was further examined on SDS–PAGE, which showed a single band with a molecular weight of about 15 kDa (Fig. 1 ▶). In order to identify the protein, the sequence of the first 15 amino-acid residues from the N-terminus was determined using a PPSQ21A automatic protein sequencer (Shimadzu, Japan).
Figure 1.

SDS–PAGE showing the purity of amaryllin.
The antifungal activity of amaryllin against Aspergillus flavus and Fusarium oxysporum was analyzed in 90 × 15 mm plates with 35 ml potato dextrose agar. After the fungal colony had developed in the centre of the plate, sterile blank paper disks were kept around the colony at a distance of 5 mm away from the colony border. 10 µl of purified amaryllin solution at 20 mg ml−1 was added to the disk. The plates were incubated at 293 K for 72 h until mycelia growth had enveloped the disks in the control plate.
2.2. Crystallization
Freshly purified samples of protein were dissolved to a final concentration of 25 mg ml−1 in 20 mM sodium phosphate buffer pH 7.2. The protein was crystallized by the hanging-drop vapour-diffusion method at 293 K using 24-well Linbro crystallization plates. An initial crystallization screening was carried out using Hampton Research crystallization kits (HR2-110 and HR2-112). Crystals were obtained by equilibrating 10 µl protein drops against a reservoir solution containing 100 mM ammonium sulfate, 100 mM HEPES buffer pH 7.2 and 30% PEG 8000.
2.3. X-ray data collection and processing
A complete set of X-ray intensity data was collected at 285 K using a 345 mm diameter MAR Research DTB imaging-plate scanner mounted on a Rigaku RU-300 rotating-anode X-ray generator operating at 100 mA and 50 kV. Osmic blue confocal optics were used to focus Cu Kα radiation. Diffraction data were indexed and scaled using the programs DENZO and SCALEPACK (Otwinowski & Minor, 1997 ▶). The overall value of R merge was found to be 10.9% for the entire data set.
3. Results and discussion
Amaryllin is present in a significant amount in the bulbs of A. belladonna. The protein was purified using ammonium sulfate precipitation and anion-exchange chromatography in accordance with the acidic nature of the protein. The final SDS–PAGE analysis of the protein samples showed a single band corresponding to an approximate molecular weight of 15 kDa (Fig. 1 ▶). The protein was tested for antimicrobial activity against the fungi Aspergillus flavus and F. oxysporum. The crescent of inhibition around disks containing amaryllin suggested significant inhibition of fungal growth. The purified amaryllin sample was crystallized. The crystals grew to maximum dimensions of 0.31 × 0.15 × 0.10 mm within five weeks (Fig. 2 ▶). The crystals diffracted to 2.7 Å resolution (Fig. 3 ▶) and belonged to the orthorhombic space group I222 or I212121, with unit-cell parameters a = 48.7, b = 61.9, c = 79.6 Å. The Matthews coefficient (Matthews, 1968 ▶) was estimated to be 2.0 Å3 Da−1, suggesting the presence of four molecules in the unit cell, i.e. one per asymmetric unit. The details and statistics of data collection are summarized in Table 1 ▶. The sequence of the 15 N-terminal amino-acid residues was determined to be 1Gln-Lys-Ile-Gln-Glu-Ile-Asp-Leu-Gln-Thr-Tyr-Leu-Gln-Pro-Gln15. Surprisingly, it did not show significant sequence identity to any known protein, indicating that amaryllin is a novel protein. In the meantime, amino-acid sequence determination of the complete protein chain is in progress. Efforts to prepare heavy-atom/ion derivatives for structure determination are under way.
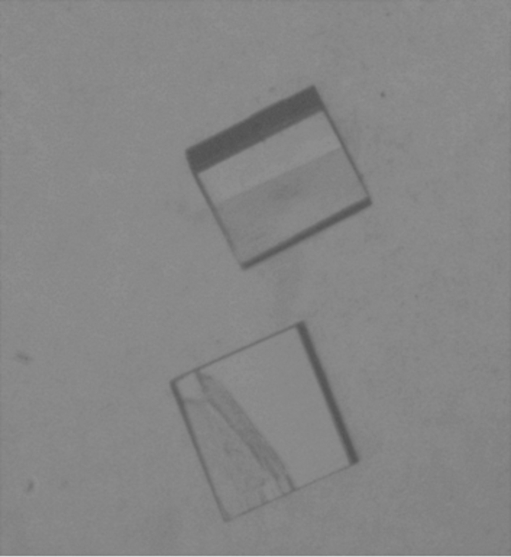
Figure 2.
Crystal of amaryllin obtained using the hanging-drop vapour-diffusion technique (approximate dimensions 0.31 × 0.15 × 0.10 mm).
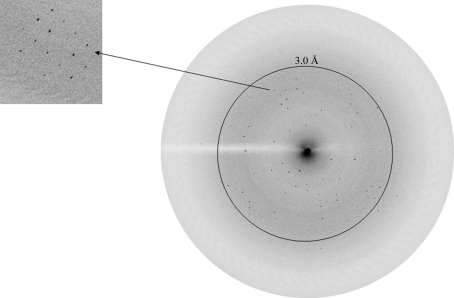
Figure 3.
Diffraction pattern from an amaryllin crystal to 2.7 Å resolution. A high-resolution section of the diffraction has been enlarged (left).
Table 1. Data-collection statistics.
Values in parentheses are for the outermost shell.
| Space group | I222 or I212121 |
| Unit-cell parameters (Å) | a = 48.6, b = 61.9, c = 79.6 |
| No. of molecules in the unit cell | 4 |
| VM (Å3 Da−1) | 2 |
| Solvent content (%) | 39 |
| Resolution range (Å) | 43.2–2.7 (2.8–2.7) |
| Total No. of measured reflections | 17521 |
| No. of unique reflections | 3285 |
| Completeness (%) | 98.7 (98.9) |
| Rmerge† (%) | 10.9 (49.7) |
| Rr.i.m.† (%) | 11.8 (59.1) |
| Rp.i.m.† (%) | 9.4 (34.2) |
| Average I/σ(I) | 5.4 (2.0) |
R
merge = 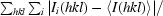
 , R
r.i.m. =
, R
r.i.m. = 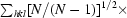
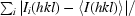
 , R
p.i.m. =
, R
p.i.m. = 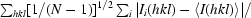
 (Weiss, 2001 ▶).
(Weiss, 2001 ▶).
Acknowledgments
The authors acknowledge the financial grant from the Department of Science and Technology, New Delhi. TPS thanks the Department of Biotechnology, New Delhi for the award of Distinguished Biotechnologist.
References
- Bloch, C. Jr, Patel, U. S., Baud, F., Zvelebil, J. M., Carr, D. M., Sadler, J. P. & Thornton, M. J. (1998). Proteins, 32, 334–349. [DOI] [PubMed]
- Broekaert, W. F., Terras, R. F., Cammue, P. B. & Osborn, R. W. (1995). Plant Physiol.108, 1353–1358. [DOI] [PMC free article] [PubMed]
- Chen, Z. Y., Brown, R. L., Lax, A. R., Guo, B. Z., Cleveland, T. E. & Russin, J. S. (1998). Phytopathology, 88, 276–281. [DOI] [PubMed]
- Chilosi, G., Caruso, C., Caporale, C., Leonardi, L., Bertini, L. & Buzi, A. (2000). J. Phytopathol.148, 477–481.
- Gozia, Q., Ciopraga, J., Bentia, T., Lungu, M., Zamfirescu, I. & Tuder, R. (1995). FEBS Lett.370, 245–249.
- Ho, V. S., Wong, J. H. & Ng, T. B. (2007). Peptides, 28, 760–766. [DOI] [PubMed]
- Joshin, B. N., Sainani, M. N., Bastawade, K. B., Gupta, V. S. & Ranjekar, P. K. (1998). Biochem. Biophys. Res. Commun.246, 382–387. [DOI] [PubMed]
- Leah, R., Tommerup, H., Svendsen, I. & Mundy, J. (1991). J. Biol. Chem.266, 1564–1573. [PubMed]
- Lorito, M., Broadway, R. M., Hayes, C. K., Woo, S. L., Noviello, C. & Williams, D. L. (1994). Mol. Plant Microb. Interact.7, 525–527.
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed]
- McLauchlan, W. R., Garcia-Conesa, M. T., Williamson, G., Roza, M., Ravestein, P. & Maat, J. (1999). Biochem. J.338, 441–446. [PMC free article] [PubMed]
- Mishra, V., Bilgrami, S., Sharma, R. S., Kaur, P., Yadav, S., Krauspenhaar, R., Betzel, C., Voelter, W., Babu, C. R. & Singh, T. P. (2005). J. Biol. Chem.280, 20712–20721. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Pueyo, J. J., Hunt, D. C. & Chrispeels, M. J. (1993). Plant Physiol.101, 1341–1348. [DOI] [PMC free article] [PubMed]
- Segura, A., Moreno, M. & Garcia-Olmedo, F. (1993). FEBS Lett.332, 243–246. [DOI] [PubMed]
- Vogelsang, R. & Barz, W. (1993). Planta, 189, 60–69. [DOI] [PubMed]
- Weiss, M. S. (2001). J. Appl. Cryst.34, 130–135.